Abstract
Genetic variation is critical in microbial immune evasion and drug resistance, but variation has rarely been studied in complex heterogeneous communities such as the human microbiome. To begin to study natural variation, we analyzed DNA viruses present in the lower gastrointestinal tract of 12 human volunteers by determining 48 billion bases of viral DNA sequence. Viral genomes mostly showed low variation, but 51 loci of ∼100 bp showed extremely high variation, so that up to 96% of the viral genomes encoded unique amino acid sequences. Some hotspots of hypervariation were in genes homologous to the bacteriophage BPP-1 viral tail-fiber gene, which is known to be hypermutagenized by a unique reverse-transcriptase (RT)-based mechanism. Unexpectedly, other hypervariable loci in our data were in previously undescribed gene types, including genes encoding predicted Ig-superfamily proteins. Most of the hypervariable loci were linked to genes encoding RTs of a single clade, which we find is the most abundant clade among gut viruses but only a minor component of bacterial RT populations. Hypervariation was targeted to 5′-AAY-3′ asparagine codons, which allows maximal chemical diversification of the encoded amino acids while avoiding formation of stop codons. These findings document widespread targeted hypervariation in the human gut virome, identify previously undescribed types of genes targeted for hypervariation, clarify association with RT gene clades, and motivate studies of hypervariation in the full human microbiome.
Keywords: deep sequencing, diversity-generating retroelement, mutagenesis, major tropism determinant
Key aspects of host–parasite interactions are mediated by targeted changes in DNA. The vertebrate adaptive immune system is based on covalent DNA rearrangements that diversify genes encoding Ig-domain antigen-binding proteins. In response, viral and cellular pathogens encode genetic systems that vary antigens bound by host antigen receptors (1, 2).
In this study we begin to characterize patterns of sequence variation in heterogeneous natural communities, using the human microbiome as a model. We chose to study viral samples because they represent a medically important microbiome component, but contain a smaller aggregate genome size than the full microbiome, allowing sequencing to a depth that permits empirical assessment of variation.
A newly discovered mechanism of targeted hypermutation, particularly pertinent here, involves the Bordetella bacteriophage BPP-1, which has been shown to vary the sequence of the gene encoding its phage tail fiber to bind divergent cell-surface receptors (3–6). The phage-encoded major tropism determinant (MTD) gene, which encodes the tip of the tail fiber, is subjected to targeted hypermutation by a reverse transcriptase (RT)-dependent mechanism (7, 8). The 3′ part of the tail-fiber gene is duplicated in the phage genome, and the duplicated template repeat (TR) is transcribed and reverse-transcribed in an error-prone fashion. The mutated copy is then incorporated into the MTD gene variable repeat (VR), leading to very high mutation rates. Diversity-generating systems involving related RTs and genes encoding C-type lectin folds have been inferred from prokaryotic genome sequences (3, 4), but only the BPP-1 system has been characterized functionally.
Here we have used the Solexa/Illumina HiSeq method to interrogate 48 billion bases of DNA sequence from populations of gut DNA viruses, which allowed us to identify regions of targeted hypervariation in the primary sequence data. We found that RT-associated hypervariation systems were present in 11 of 12 subjects examined, and act on a much wider range of gene types than was known previously. Analysis of the sequence information further specifies the chemical logic of the mutational targeting and suggests that the most common role of RTs in the gut virome is targeted hypervariation.
Results
Sequence and Assembly of 48 Gb of Gut Viral DNA.
To study diversity in natural populations of the human virome, we collected stool samples from 12 healthy individuals (three per subject) over a 2-mo period, then purified viral particles by sequential filtration, banding in CsCl density gradients, and treatment with nuclease, as previously described (9). DNA was isolated from viral particles, amplified, and then sequenced using the Solexa/Illumina HiSeq paired-end sequencing platform. A total of 495,053,311 reads were generated, averaging 97.2 bp in length. As an empirical error control, 153 million reads were determined for DNA from phage ΦX174, showing an accuracy of 99.94%. A total of 48 Gb of data were collected, the largest survey of viral sequences yet reported.
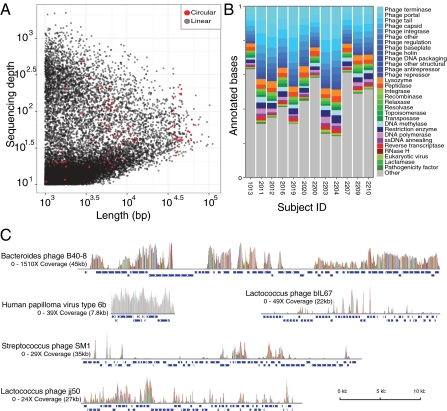
The raw sequences were assembled into contigs using the deBruijn graph-based assembler SOAPdenovo (10). The depth of sequencing for the gut viral contigs averaged 49× and ranged up to 3,000× (Fig. 1A). There were 78 contigs longer than 1 kb that assembled as complete circles, indicating probable completion of the viral genome sequence. Circular assemblies could arise either by completing the sequence of a circular genome or by sequencing concatemers, which are intermediates in the replication of many DNA viruses. The mean number of contigs per subject longer than 1 kb was 1,390, ranging from 573 to 3,390.
Fig. 1.
Assembly, functional assessment, and identification of viral sequences. (A) Summary of contigs assembled from viral sequences. Each contig is shown as a point, the length is shown on the x axis and the depth of reads mapped to each contig on the y axis. Circular contigs are shown in red. (B) Assignment of gene functions from viral contigs using the Pfam database of protein families; assignment of Pfam domains to viral functions is described in ref. 9. The proportion of sequences that were assigned for each function is indicated on the y axis. On average, 21% of each sample was assigned to a Pfam protein family. (C) The five viral RefSeq genomes with the most similarity to sequences generated in this study. Vertical lines indicate the depth of sequencing at that position, and colored lines indicate mismatches with the reference sequence. The range of coverage is noted to the left of each plot. Blue boxes below each genome indicate annotated genes.
Protein functions were inferred by comparing the conceptual translation of predicted ORFs to a curated database of protein families. A broad range of viral functions were identified in the encoded proteins (Fig. 1B), as observed previously (9, 11). On average, 72% of the ORFs did not resemble any recognizable protein family, emphasizing the immense diversity of novel genes in gut viral populations.
To assess the relationship to known viral genome sequences, contigs were compared with the National Center for Biotechnology Information (NCBI) RefSeq collection of viral genomes. The five database sequences with the most extensive similarity are shown in Fig. 1C. Most of the recognizable viral sequences showed matches to prokaryotic viruses (DNA bacteriophages). Regions of similarity were typically short patches (median alignment length 202 bp at e-value ≤ 10−5), supporting the idea that bacteriophage functions are commonly organized in genetic cassettes (12, 13). Only one well-characterized virus known to replicate on human cells was detected: human papillomavirus type 6b (Fig. 1C), which was only found within a single subject and sequenced to a depth of 23-fold. The next best hit to a eukaryotic virus was unconvincing, indicating that this viral fraction in healthy subjects is overwhelmingly composed of bacterial viruses.
Hypervariable Loci in Gut DNA Viruses.
To investigate sequence variation within each contig, we aligned the raw reads back to contigs and quantified variation at each base. Initial analysis showed that multiple small regions showed extremely high variation against a background of low variation. Comparison of filtering criteria led us to focus on regions of at least 90 bp that were sequenced to a depth of at least 5×, contained a proportion of unique sequences of at least 40%, and contained a proportion of polymorphic bases of at least 5%, yielding 36 regions of the highest variability.
Analysis of these regions revealed that 12 resembled the diversity-generating retroelement of phage BPP-1 described above (6, 8). This system is comprised of ∼100-bp repeat regions, the donor TR, the targeted VR, and an RT, which is required to mutagenize the VR at positions where the TR contains an adenine (6). Because of the central role of RT in this process, we identified all of the genes encoding RT-like sequences within the full collection of contigs, revealing 185 genes. Duplicated sequences can potentially break contig assemblies, so we manually inspected all of the RT-containing contigs to identify broken assemblies near the RT sequences suggestive of TR/VR pairs. We repaired 33 contigs through a combination of directed resequencing and manual realignment of shotgun Solexa/Illumina reads. This process increased the number of variable regions to 51, those variable regions falling within a TR/VR pair to 36, and those with both an RT and a TR/VR pair to 29 (Fig. S1 and Table S1). In every case where a variable region was near an RT, it also contained a TR/VR pair. Such elements were found in 11 of 12 subjects studied. Based on the resemblance to BPP-1, we refer to these systems as “diversity-generating retroelements” below. Of these retroelements, 18 of 29 were found in contigs that could be tentatively assigned to a specific bacteriophage family (Table S1).
Short hairpins near the VR are essential for activity in the BPP-1 system (14). Similar hairpin sequences were found in only 13 of the above 29 elements (Table S1). Evidently some of the novel diversity-generating retroelements may use different initiation mechanisms that do not involve hairpin structures. Of those 13 hairpins, 6 were found within ORFs, raising the question of how this structure may constrain amino acid evolution in the host gene.
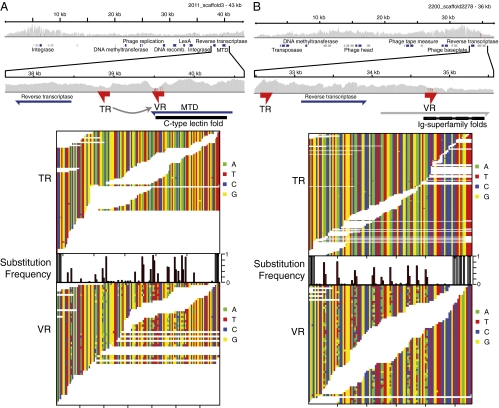
All of the 29 variable regions adjoining both an RT and a TR/VR pair were within an intact ORF longer than 500 bp, allowing the targeted genes to be analyzed. We used BLASTp alignments and the homology-based structural prediction pipeline Phyre2 to analyze each ORF (Table S1) (15). Fourteen ORFs had the hypervariable region near the 3′ end of the coding region and showed a predicted C-type lectin fold (Phyre2 confidence score 90–100%), resembling the well-studied MTD of BPP-1 (percentage of identity 15–33%) (Fig. 2A; the arrow indicates the direction of information transfer inferred from the BPP-1 model).
Fig. 2.
RT-associated hypervariable regions from the human gut virome. (A) Hypervariation in a gene predicted to encode a protein with an MTD-like C-type lectin fold. (B) Hypervariation in a gene predicted to encode an Ig-superfamily fold. In A and B, the Upper section shows the contig of origin, with gray vertical lines showing sequencing depth and boxes showing annotated proteins. The indicated area is expanded below to show the TR, the corresponding VR, RT, and the ORF that contains the targeted VR. The inferred direction of information transfer between the TR and VR is shown with an arrow. The Lower section of each plot shows an alignment of the sequences spanning the TR and VR for each element (white space indicates gaps between reads). Above the VR sequence is a barplot indicating the proportion of bases in the VR that differ from the consensus base in the TR. DNA bases are indicated by colors as indicated on the sides of the panels.
Unique Gene Types at Hypervariable Loci.
Surprisingly, six hypervariable ORFs encoded proteins that aligned to cadherins, invasins, and fibronectins, which contain Ig-superfamily β-sandwich domains (Table S1). Proteins aligned with modest percent identity (11–15%), but structure predictions suggested multiple β-sandwich domains with high confidence (Phyre2 confidence scores of 97–99%). Several of these proteins were also homologous to each other (80–90% identity), despite being isolated from different individuals. Ig-superfamily proteins are common in bacteriophage (16–18), but genes encoding Ig-superfamily proteins were not previously known to be subject to hypervariation by an RT-associated mechanism. The VRs for Ig-superfamily proteins were in the middle of the target ORF, so that the TR and VR were separated by an average of 1,938 bp. In comparison, the MTD-like TR and VR are separated by an average of only 356 bp. The greater distance between repeats in Ig-superfamily genes may have hindered their detection in previous studies based on DNA sequence comparisons (3, 6).
Three hypervariable ORFs also encoded predicted leucine-rich repeat proteins N terminal to the hypervariable region (Phyre2 confidence score of 95–97%). These proteins were all embedded in large ORFs, ranging from 1,716 to 2,181 predicted amino acids. One of these also had a C-type lectin fold in the hypervariable region. The remaining eight ORFs containing hypervariable regions did not have convincing similarity to known proteins, and may represent still further types of proteins subject to targeted hypermutagenesis.
Hypervariation at Adenine Residues.
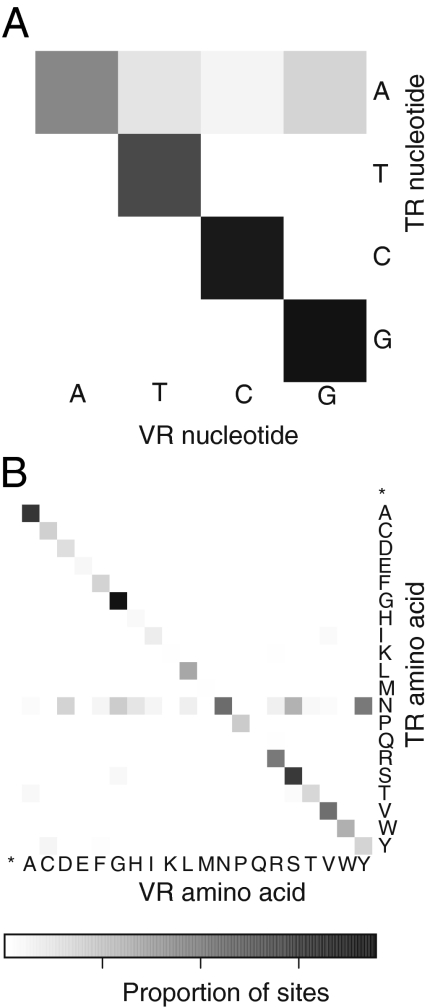
Adenine residues were targeted in all of the hypervariable regions identified here (Fig. 3A). The 5′-AAY-3′ sequences were particularly strongly affected (e.g., Fig. 2, Lower). This substitution pattern in 5′-AAY-3′, which encodes asparagine, allows access to many different chemistries in the encoded amino acid side chains while suppressing creation of stop codons, as was originally pointed out for the MTD system (3). The size of the dataset reported here allowed us to carry out statistical analysis of the placement of the 5′-AAY-3′ relative to the three possible reading frames, which showed that the 5′-AAY-3′ sequences were overwhelmingly in the asparagine-encoding frame (Fig. 3B) (P < 10−163). Thus, variable region sequences have evolved to take advantage of asparagine-codon diversification while suppressing other types of changes.
Fig. 3.
Characteristics of RT-associated hypervariation in the gut virome. (A) Heatmap showing the relationship of positions in the TR (y axis) to the resulting nucleotides in the VR (x axis). Of 15,447 mutated bases, 14,930 (97%) are located at adenine-positions relative to the TR. (B) Amino acid substitution heatmap showing the relationship of codons in the TR (y axis) to the resulting codons in the VR (x axis). Of 11,462 mutated codons, 9,212 (80%) are located at asparagine (N) codons in the TR.
RT Gene Populations in Gut DNA Viruses and Bacteria.
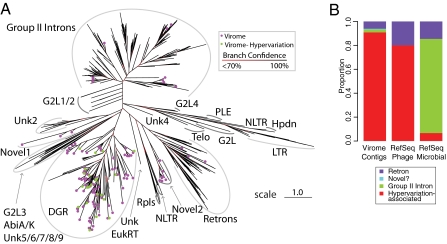
We next took advantage of the above data to annotate functions of gut virome RT genes. All of the RT sequences previously associated with hypervariable regions (3, 6, 19) cluster in a monophyletic clade containing the BPP-1 RT (Fig. 4A, cluster marked “DGR” for diversity-generating retroelements). In our dataset, we found that most of the new RTs clustered in this group (n = 99), including all of the RTs found to be associated with hypervariable regions (Fig. 4A, green symbols). There was no obvious correlation between RT phylogeny and targeted gene type. Far fewer gut virome RTs clustered with group II intron RTs (n = 8), and retron RTs (n = 6). We observed two previously undescribed groups of RT sequences. Five sequences fall into “novel 1” (Fig. 4A), which is most similar to the Unk2 family (19). Seven sequences fall into “novel 2” (Fig. 4A), which is a sister clade to the retron RTs. The average pair-wise distance of the pooled RTs associated with diversity generating systems described here is 1.14, which greatly increases the diversity of this group (previously 0.90) and rivals the diversity of the large retroviral/LTR retrotransposon RT clade (1.20).
Fig. 4.
RT sequences found in DNA viruses of the human gut. (A) Phylogentic tree of RT sequences. Each sequence was aligned to a position-specific scoring matrix to construct a multiple sequence alignment. The tree was constructed using the maximum-likelihood method. Green circles indicate RT sequences on viral contigs from this dataset that contain hypervariable regions and TR/VR pairs. Purple circles indicate other RT sequences from this dataset; the remaining leaves indicate reference sequences from the NCBI. RT clades were adapted from refs. 6 and 19, and are indicated by gray lines. The bootstrap support of internal nodes is indicated by the color of internal branches as described in the key. Clades are marked according to refs. 6 and 19: Abi, abortive-phage-infection; DGR, diversity generating retroelements; G2L, group II intron-like families; Hpdn, hepadnaviruses; LTR, LTR retrotransposons and retroviruses; NLTR, non-LTR retrotransposons; PLE, Penelope-like elements; Rpls, retroplasmid; Telo, telomerase; Unk, unknown families (19). The scale bar indicates the log-corrected distance metric used by FastTree, adapted from BLOSUM45. Distances range from 0, indicating a perfect match, to 3, indicating no overlap. (Scale bar, 1.0). (B) Relative proportions of RTs in viruses studied here, the RefSeq phage genome database, and the RefSeq bacterial genome database.
We compared the distribution of RT clades in gut DNA viruses described here to that of their bacterial hosts. The bacterial genomes were dominated by the RT clades associated with group II introns and retrons, and thus differed from the DNA viruses, where RTs associated with diversity-generating retroelements dominated (Fig. 4B).
Discussion
We report that DNA viruses of the human gut are rich in hypervariable regions, and that these are associated with template repeat/variable repeat pairs and characteristic RTs. The frequency of substitutions was so high that up to 96% of alleles in hypervariable regions encoded unique protein sequences. Most of the RT genes in the virome dataset were in the clade linked to diversity-generating retroelements. Thus, targeted hypervariation appears to be the major role of RTs in DNA viruses of the human gut.
Surprisingly, several of the genes subject to hypervariation were predicted to encode Ig-superfamily proteins. Thus, both gut viruses and vertebrate antigen receptors have evolved to use Ig domains as scaffolds for displaying highly diversified polypeptides. Evolution may have converged on these β-sheet–rich domains because they are relatively rigid and so can maintain their folds despite primary sequence diversification, as has also been suggested for the C-type lectin fold (3). The placement of diversified regions on Ig domains appears to differ between vertebrates and phage. Although more complete structural characterization is needed, modeling suggests that the phage Ig-superfamily domains may be diversified along one surface and into the adjoining linker between domains, but the vertebrate antigen receptors are diversified in loops between β-sheets within an Ig domain. The mechanism of diversification in phage clearly differs from that in the vertebrate immune system—the phage genes are diversified by error-prone reverse transcription (8), but the Gnathostomata immune system is diversified by V(D)J recombination, which involves DNA double strand breaks (20), and targeted deamination by activation-induced cytidine deaminase (21).
The functions of the previously undescribed viral hypervariable genes found here are not fully clarified. Hypervariable genes may encode viral structural proteins targeted by human IgA, which is secreted into the gut in large amounts, so that diversification of viral structural proteins may allow immune evasion. However, a role in ligand binding may be more likely—a weakness of the immune evasion model is that only specific short regions are targeted for hypermutagenesis, so the remainder of the protein could still be antigenic.
Some of the hypervariable Ig proteins may be homologs of T4 highly immunogenic outer capsid (hoc) protein, which encodes an Ig protein related in sequence to those studied here. Hoc decorates T4 heads by binding to sixfold symmetric vertices in hexameric capsomeres, thereby providing a polyvalent binding moiety on the outside of phage heads. Hoc is proposed to mediate binding of T4 to surfaces such as the Escherichia coli host cell (22), and has also been used for phage display to create new binding specificities for biotechnology applications (23). If the Ig-superfamily proteins studied here are also accessory head proteins, they may mediate binding of viral particles to candidate host cells or environmental materials, allowing selection to enrich for those binding specificities that optimize reproductive success. The most useful binding specificities may differ widely during replication in the human gut or after shedding in feces, but hypervariation allows optimization in each new environment. Further research will be needed to clarify the full biological roles of these viral diversity generating systems.
Methods
Gut Viral DNA Sequencing and Assembly.
Details can be found in the SI Methods. Briefly, stool samples were collected from healthy subjects as described previously (9, 24). Viral DNA was purified by filtration and density ultracentrifugation (9) to a purity of ∼99.9% by 16S rDNA quantitative PCR analysis, and three pooled samples per subject were sequenced on an Illumina HiSEq. 2000 using 100-bp paired-end chemistry. Sequences were trimmed to Q35 (using FASTX v0.0.13) and assembled within each subject using SOAPdenovo (v1.05) and a k-mer size of 63. Reads were mapped back to those contigs within each subject using Burrows-Wheeler Aligner (v0.5.9-r16). Functions encoded by these contigs were predicted using RPSBLAST (v2.2.20) and the NCBI Conserved Domain Database.
Analysis of Hypervariable Loci.
Variable regions were found using R scripts that analyzed the variability of reads mapped back to these novel contigs. ORFs containing hypervariable loci were translated using custom scripts and submitted to Phyre2 (15), using a confidence threshold of 95%. RT sequences associated with hypervariable loci were aligned to the curated RT position-specific scoring matrix PF00078 using hmmalign (HMMER v3.0), and the resulting approximately maximum-likelihood tree was generated by FastTree. R scripts, BAM alignment files of the 29 contigs found in Table S1, and RT sequence alignments are available upon request.
Supplementary Material
Acknowledgments
We thank members of the G.D.W., J.D.L., and F.D.B. laboratories for help and suggestions; Scott Sherril-Mix for the gift of a useful script; and the Penn Genome Frontiers Institute. This work was supported by Human Microbiome Roadmap Demonstration Project UH2DK083981 (G.D.W., F.D.B., and J.D.L. are co-Principal Investigators); National Institutes of Health (NIH) Grant T32AI060516 (to S.M.); a grant with the Pennsylvania Department of Health; NIH Grant AI39368 (to G.D.W.); the Molecular Biology Core of The Center for Molecular Studies in Digestive and Liver Diseases (P30 DK050306); the Joint Penn-Children's Hospital of Philadelphia Center for Digestive, Liver, and Pancreatic Medicine; NIH instrument Grant S10RR024525 and NIH Clinical Translational Science Award Grant UL1RR024134 from the National Center for Research Resources; and the Crohn's and Colitis Foundation of America.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.H.M. is a guest editor invited by the Editorial Board.
Data deposition: Contigs containing variable regions listed in Table S1 have been deposited in the GenBank database.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119061109/-/DCSupplemental.
References
- 1.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: ASM,; 2002. [Google Scholar]
- 2.Bushman FD. Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 3.McMahon SA, et al. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12:886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- 4.Miller JL, et al. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS Biol. 2008;6:e131. doi: 10.1371/journal.pbio.0060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai W, et al. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc Natl Acad Sci USA. 2010;107:4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doulatov S, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 7.Liu M, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, et al. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol Cell. 2008;31:813–823. doi: 10.1016/j.molcel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minot S, et al. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75:423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H, et al. Target site recognition by a diversity-generating retroelement. PLoS Genet. 2011;7:e1002414. doi: 10.1371/journal.pgen.1002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 16.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 17.Fraser JS, Maxwell KL, Davidson AR. Immunoglobulin-like domains on bacteriophage: Weapons of modest damage? Curr Opin Microbiol. 2007;10:382–387. doi: 10.1016/j.mib.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Pell LG, et al. The solution structure of the C-terminal Ig-like domain of the bacteriophage λ tail tube protein. J Mol Biol. 2010;403:468–479. doi: 10.1016/j.jmb.2010.08.044. [DOI] [PubMed] [Google Scholar]
- 19.Simon DM, Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36:7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 21.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 22.Fokine A, et al. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85:8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oślizło A, et al. Purification of phage display-modified bacteriophage T4 by affinity chromatography. BMC Biotechnol. 2011;11:59. doi: 10.1186/1472-6750-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.