Abstract
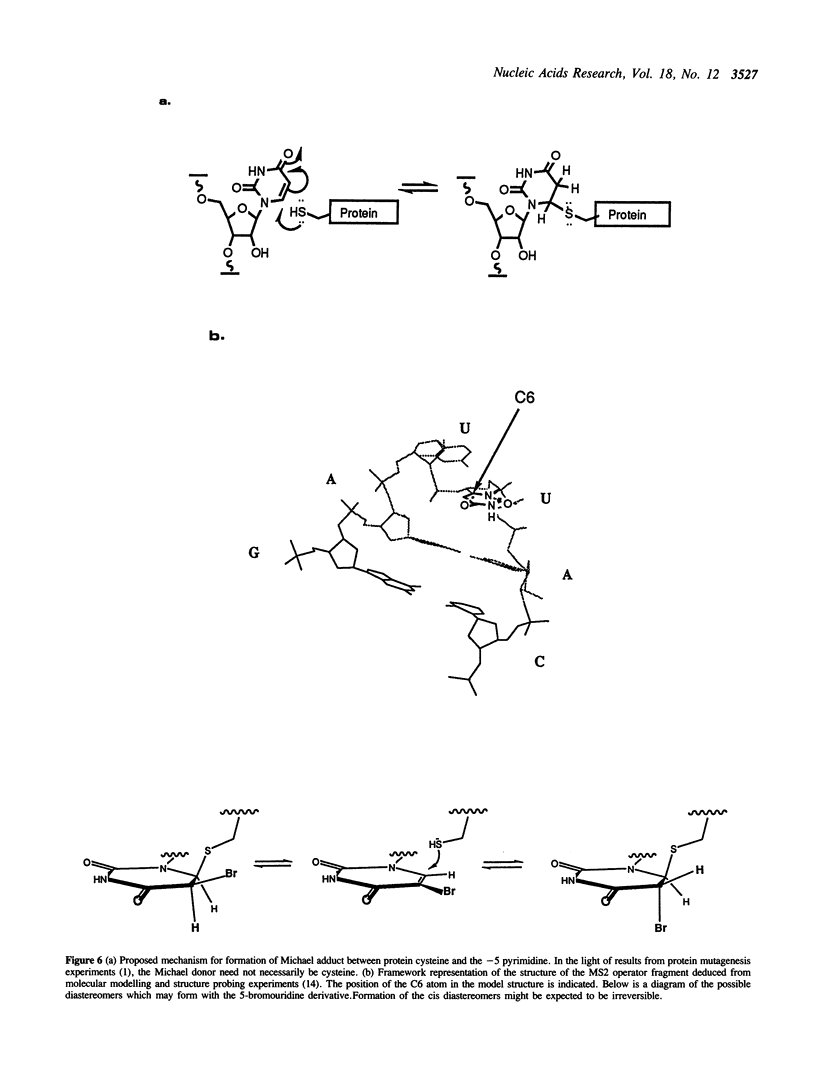
Synthetic oligoribonucleotides have been used to probe the interaction of MS2 coat protein with the translational operator of the MS2 replicase gene. We have investigated the possible formation of a transient covalent bond between the single-stranded uridine residue, at position -5, and a cysteine side-chain on the coat protein, by the incorporation of a chemically modified residue (5-BrU) at this position. This chemically synthesised operator variant has a binding constant of between 10 and 50 times greater than that of the wild type and is therefore comparable with the tight binding variant having a cytidine substituted at the -5 position. Dissociation kinetics show that the complex with the 5-BrU operator is more stable than the -5C variant; a result which is consistent with the formation of a Michael adduct at the -5 position. In addition, a number of other chemical variants of the operator have been analysed. These include operators incorporating deoxyadenine residues at each of the important single-stranded adenine sites. Recently the Michael adduct proposal has been challenged on the basis of mutagenesis of the coat protein cysteine residues. These results are discussed in the light of our data in support of Michael adduct formation.
Full text
PDF
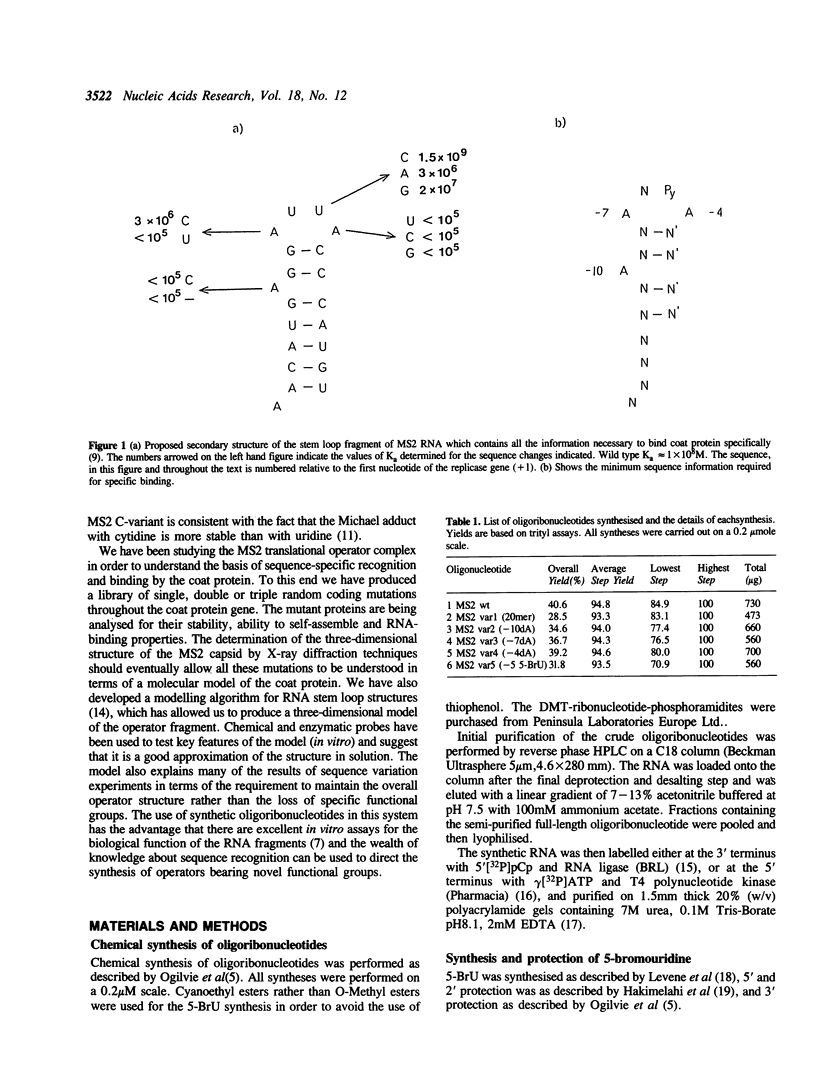
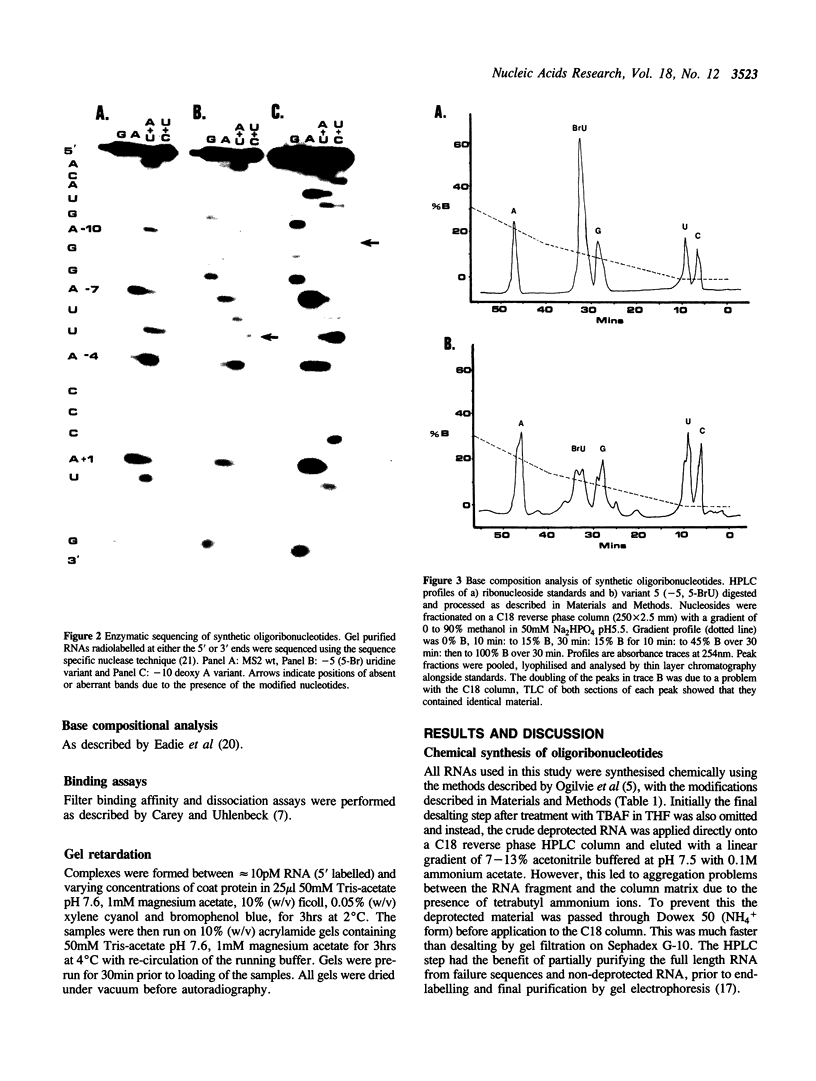

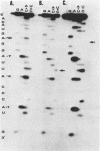
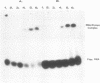
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Carey J., Lowary P. T., Uhlenbeck O. C. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry. 1983 Sep 27;22(20):4723–4730. doi: 10.1021/bi00289a017. [DOI] [PubMed] [Google Scholar]
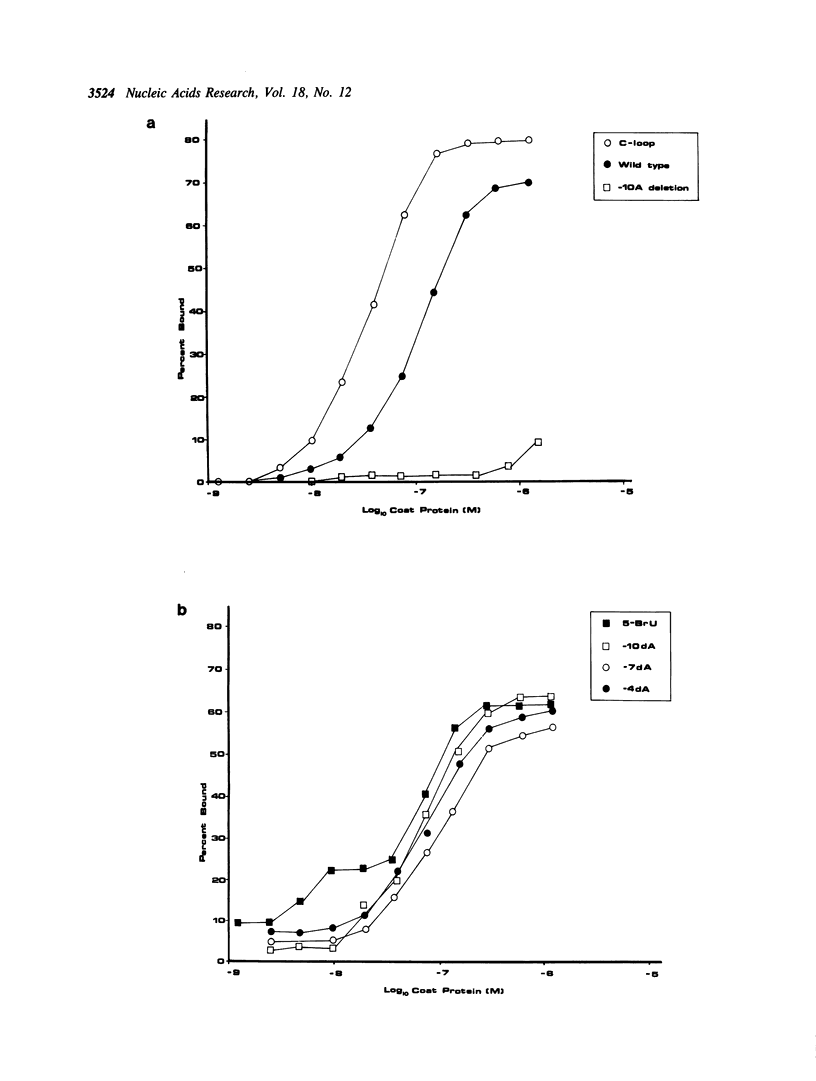
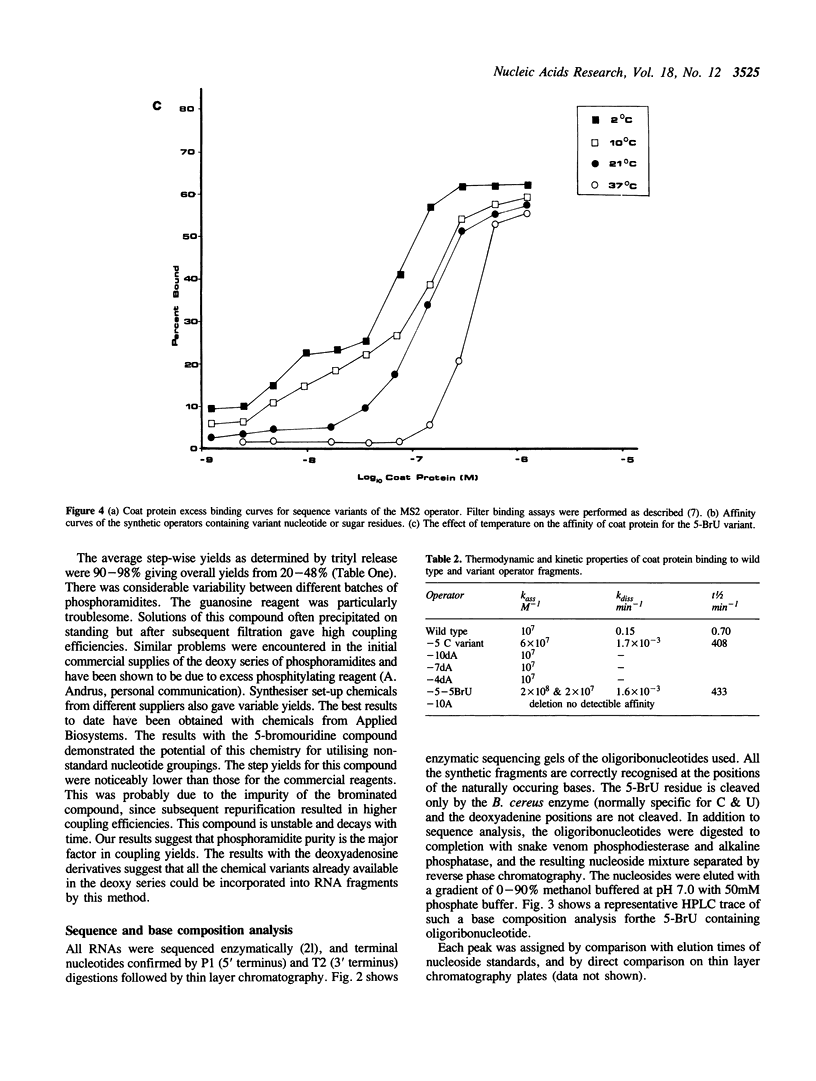
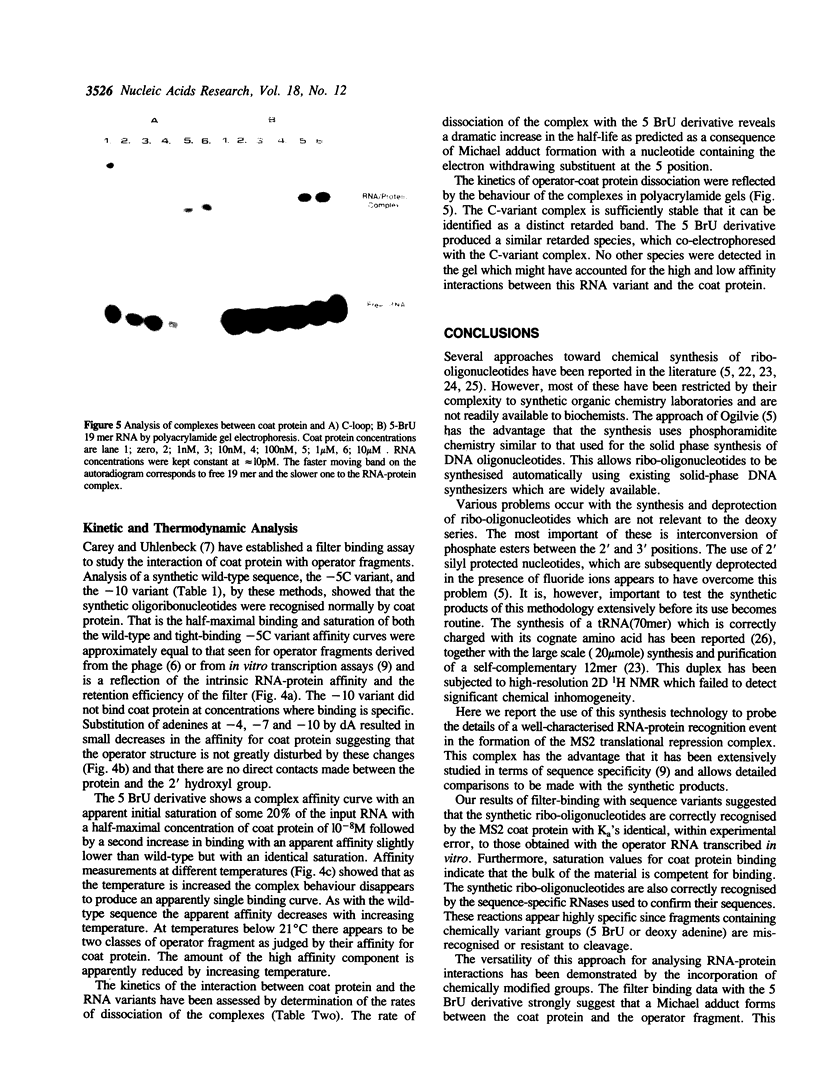
- Carey J., Uhlenbeck O. C. Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry. 1983 May 24;22(11):2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- Chaix C., Duplaa A. M., Molko D., Téoule R. Solid phase synthesis of the 5'-half of the initiator t-RNA from B. subtilis. Nucleic Acids Res. 1989 Sep 25;17(18):7381–7393. doi: 10.1093/nar/17.18.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973 Mar 25;248(6):2235–2244. [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. Solid-phase synthesis and high-resolution NMR studies of two synthetic double-helical RNA dodecamers: r(CGCGAAUUCGCG) and r(CGCGUAUACGCG). Biochemistry. 1989 Mar 21;28(6):2422–2435. doi: 10.1021/bi00432a013. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie J. S., McBride L. J., Efcavitch J. W., Hoff L. B., Cathcart R. High-performance liquid chromatographic analysis of oligodeoxyribonucleotide base composition. Anal Biochem. 1987 Sep;165(2):442–447. doi: 10.1016/0003-2697(87)90294-6. [DOI] [PubMed] [Google Scholar]
- Haneef I., Talbot S. J., Stockley P. G. Modeling loop structures in proteins and nucleic acids: an RNA stem-loop. J Mol Graph. 1989 Dec;7(4):186–195. doi: 10.1016/0263-7855(89)80001-3. [DOI] [PubMed] [Google Scholar]
- Iwai S., Ohtsuka E. 5'-Levulinyl and 2'-tetrahydrofuranyl protection for the synthesis of oligoribonucleotides by the phosphoramidite approach. Nucleic Acids Res. 1988 Oct 25;16(20):9443–9456. doi: 10.1093/nar/16.20.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek R., Caruthers M. H., Longfellow C. E., Swinton D., Turner D. H., Freier S. M. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986 Dec 2;25(24):7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary P. T., Uhlenbeck O. C. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res. 1987 Dec 23;15(24):10483–10493. doi: 10.1093/nar/15.24.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie K. K., Usman N., Nicoghosian K., Cedergren R. J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S. Translational repression by bacteriophage MS2 coat protein does not require cysteine residues. Nucleic Acids Res. 1989 Aug 11;17(15):6017–6027. doi: 10.1093/nar/17.15.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., Uhlenbeck O. C. Nucleoside and nucleotide inactivation of R17 coat protein: evidence for a transient covalent RNA-protein bond. Biochemistry. 1985 Jul 16;24(15):4239–4244. doi: 10.1021/bi00336a064. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Koontz S. W., Schimmel P. A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature. 1982 Jul 8;298(5870):136–140. doi: 10.1038/298136a0. [DOI] [PubMed] [Google Scholar]