Abstract
Understanding the chromophore maturation process in fluorescent proteins is important for the design of proteins with improved properties. Here, we present the results of electronic structure calculations identifying the nature of a blue intermediate, a key species in the process of the red chromophore formation in DsRed, TagRFP, fluorescent timers, and PAmCherry. The chromophore of the blue intermediate has a structure in which the π-system of the imidazole ring is extended by the acylimine bond, which can be represented by the model N-[(5-hydroxy-1H-imidazole-2yl)methylidene]acetamide (HIMA) compound. Ab initio and QM/MM calculations of the isolated model and protein-bound (mTagBFP) chromophores identify the anionic form of HIMA as the only structure that has absorption which is consistent with the experiment and is stable in the protein binding pocket. The anion and zwitterion are the only protonation forms of HIMA whose absorption (421 and 414 nm, or 2.95 and 3.00 eV) matches the experimental spectrum of the blue form in DsRed (the absorption maximum is 408 nm or 3.04 eV) and mTagBFP (400 nm or 3.10 eV). The QM/MM optimization of the protein-bound anionic form results in a structure which is close to the X-ray one, whereas the zwitter-ionic chromophore is unstable in the protein binding pocket and undergoes prompt proton transfer. The computed excitation energy of the protein-bound anionic form of mTagBFP-like chromophore (3.04 eV) agrees with the experimental absorption spectrum of the protein. The DsRed-like chromophore formation in red fluorescent proteins is revisited on the basis of ab initio results and verified by directed mutagenesis revealing a key role of the amino acid residue 70, which is the second after the chromophore tripeptide, in the formation process.
1 Introduction
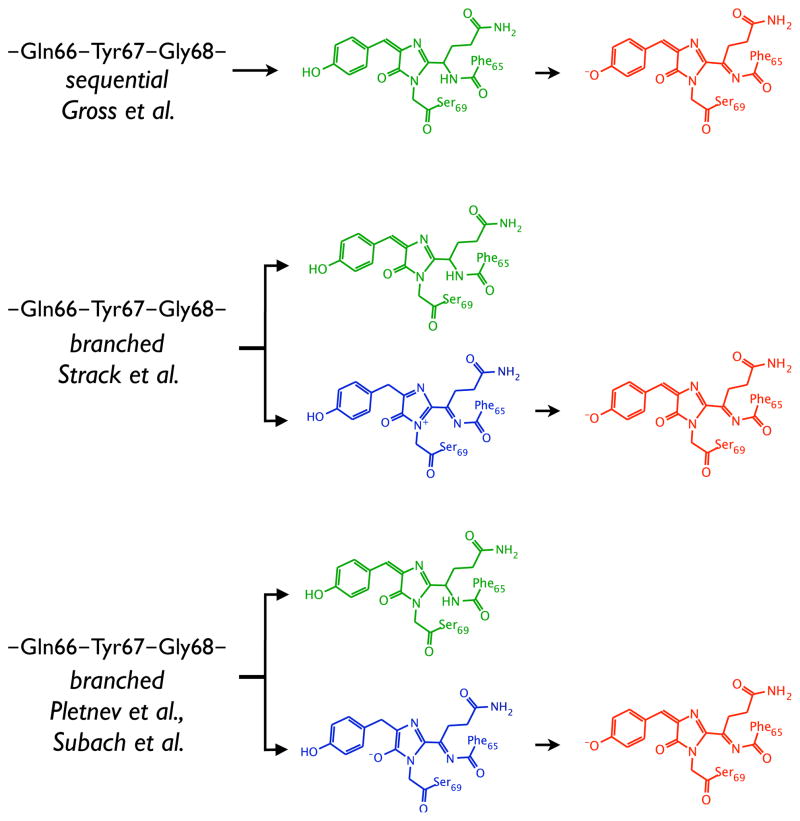
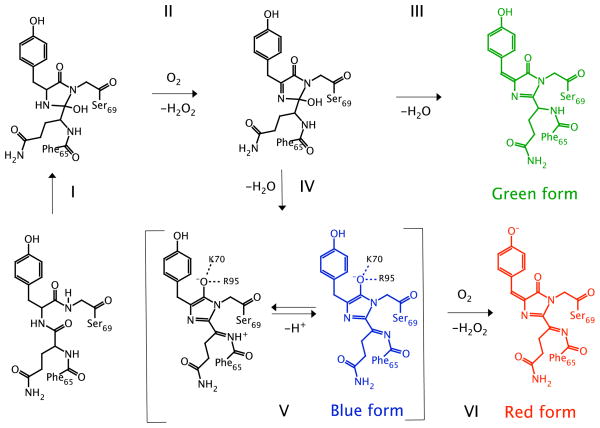
Among fluorescent proteins (FPs) used as genetically encoded fluorescent tags, the red-emitting FPs (RFPs) are of particular importance as suitable markers for mammalian deep tissue imaging.1 Photoactivatable RFPs are also valuable in super-resolution microscopy, e.g., fluorescence photoactivation localization microscopy.2 Most of the RFPs share a DsRed-like chromophore in which the π-system of the GFP chromophore is extended by an additional N-acylimine moiety, however, the mechanism of the chromophore maturation is not yet well understood. Initially, it was assumed that the red chromophore (in DsRed) is produced by oxidizing an immature green chromophore,3 however, the later studies presented strong evidence in favor of a branched maturation pathway with the two distinct end points leading to a mixture of the green and red forms in mature DsRed,4 as illustrated in Fig. 1. Mechanistic studies of DsRed maturation have identified a blue emitting intermediate (absorbance maximum at 408 nm or 3.04 eV).4 A blue-emitting form has also been observed in fluorescent timers (FTs),5 as a precursor of the mature red protein. Moreover, a dark form of PAmCherry, which emits in red upon photoactivation, absorbs in violet spectral range.6
Figure 1.
Different proposed mechanisms of the red chromophore formation: a sequential formation of the green and red chromophores proposed by Gross et al.3 (top), a branched pathway suggested by Strack et al.10 (middle), and a branched mechanism by Verkhusha and co-workers5, 9 (bottom). The blue intermediate structures from the two suggested branched mechanisms differ by their oxidation state.
To determine the X-ray structure of the intermediate, Subach et al.7 were able to convert a number of RFPs (TagRFP, mCherry, mKeima, HcRed1, M355NA) to the blue emitting forms using structure-based directed evolution and random mutagenesis. In particular, they converted TagRFP to the blue-emitting protein, mTagBFP (absorbance maximum at 400 nm or 3.10 eV) and determined its X-ray structure (2.2 Å resolution). Recently, an enhanced brighter version of the mTagBFP protein, mTagBFP2, has become available.8
Based on the similarity of the respective spectral properties, Subach et al.7 proposed that the blue form is the trapped blue intermediate in the red chromophore maturation process in DsRed. Indeed, it is reasonable to expect that the mechanism of the DsRed-like chromophore formation is similar in various RFPs and, consequently, that the blue chromophores in DsRed, dark form of PAmCherry, FTs, and mTagBFP are structurally similar (if not identical).
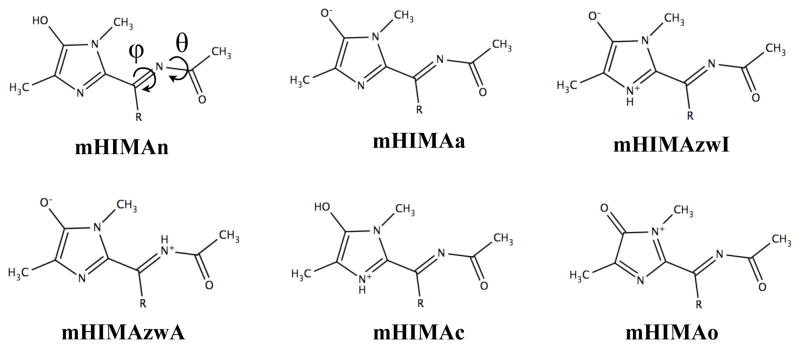
The X-ray structures mTagBFP,9 PAmCherry,6 and the blue precursor of FT,5 as well as mass-spectrometry analysis of mTagBFP and PAmCherry, support the structure of the blue-emitting chromophore in which the imidazolinone ring is conjugated with the N-acylimine bond and chromophore’s Tyr ring is separated from the π-system by the sp3-hybridized Cβ atom. However, the chromophore protonation and oxidation states are ambiguous, and a number of different structures have been proposed,5,6,9,10 as illustrated in Fig. 2 that shows model systems representing proposed structures. These six model chromophores that are considered in our study to elucidate the nature of the blue chromophore are different protonation and oxidation states of N-[(5-hydroxy-1H-imidazole-2yl)methylidene]acetamide (HIMA).
Figure 2.
Model HIMA chromophores, R=H (HIMA); R=CH3 (mHIMA), in different protonation states: neutral (mHIMAn), anionic (mHIMAa), cationic (mHIMAc), zwitterionic with protonated imidazolinone ring and acylimine (mHIMAzwI) and (mHIMAzwA), respectively, and 2-electron oxidized form (mHIMAo). The ϕ and θ dihedral angles are defined as C[Imidazole]–C–N–C[=O] and C–N–C–O, respectively.
Strack et al.10 suggested the cationic chromophore with 10 electrons in the π-system in which each formal double bond contributes 2 π-electrons (mHIMAo). In contrast, Verkhusha and coworkers considered an anionic chromophore with 12-electron π-system comprised by four formal double bonds and two lone pairs of the nitrogen and oxygen atoms (mHIMAa) as a blue emitting form in the process of FT maturation.5 A neutral chromophore (mHIMAn) was suggested for mTagBFP.9 While all these structures are chemically sound, ab initio calculations can help to discriminate between different chromophores on the basis of their computed excitation energies and optimized structures within the protein binding pocket. The oxidation state of the chromophore has important mechanistic implications, as it determines the relative order of oxidation and dehydration steps in the red chromophore maturation. The protonation state is also important, as it influences interactions between the chromophore and nearby residues and ultimately affects the pH stability.
Here we present computational and experimental evidence that the blue intermediate has anionic structure which can be formed in a single oxidation step. The assignment is based on the calculations of isolated model chromophores (Fig. 2) and the QM/MM results for mTagBFP. We choose mTagBFP as a model system because it was originally derived from RFP with an aim to trap the blue form (thus, it is structurally similar to the parental TagRFP11), and because the crystal structures of both are available.9 By identifying the protonation state of the chromophore, we were able to find the key residues for chromophore maturations and to verify their role experimentally by mutagenesis.
2 Experimental and computational methods
2.1 Computational details
The ground-state geometries of isolated model chromophores were optimized with RI-MP2/cc-pVTZ. The core electrons were frozen in the RI-MP2 calculations. The optimized structures were further used in calculations of the vertical excitation energies and energy profiles along chromophore’s twisting coordinate, see Supplementary Materials (SM). Vertical excitation energies were computed using two different approaches. First, we employed SOS-CIS(D) (scaled opposite spin variant of configuration interaction singles (CIS) with perturbative account of double excitations, Ref. 12). The initial benchmark study12 has reported mean average errors of ~0.4 eV for 0–0 transitions for a large set of organic molecules. The discrepancies between the theory and experiment were, in part, due to using Hartree-Fock and CIS optimized geometries for the ground and excited states, respectively.12 Therefore, we anticipate smaller errors in vertical excitation energies using more accurate RI-MP2-optimized ground-state geometries. The SOS-CIS(D) method has been shown to provide reliable excitation energies for the GFP chromophores13–15 and flavin.16 However, owing to its perturbative nature, SOS-CIS(D) can fail in the case of near-degenerate states or, more generally, when CIS wave function is not a good approximation for an excited state. To verify the SOS-CIS(D) results, we employed the equation-of-motion coupled-cluster for excitation energies (EOM-EE-CCSD) approach, which provides a balanced treatment of ground and excited states of different nature including (near)-degenerate cases.17–20
Excitation energies were computed using the aug-cc-pVDZ basis set. Additional data on the basis set effects is available in SM. Due to resonance (i.e., metastable with respect to electron detachment) character of the excited states of these anions,13,21 a non-augmented cc-pVDZ basis was used for mHIMAa and HIMAa.
The 6-31+G(d) basis was used in EOM-EE-CCSD calculations. These calculations are expected to yield reliable wave functions that can be compared with SOS-CIS(D), however, the absolute values of excitation energies are expected to be less accurate owing to a small basis set and the tendency of EOM-EE-CCSD to systematically overestimate excitation energies of valence states.21,22
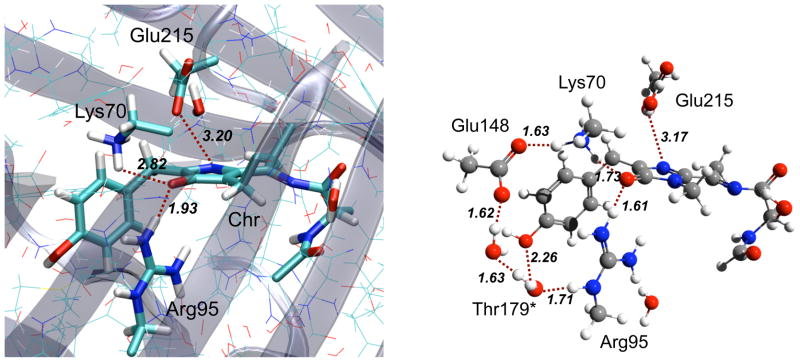
The effect of the protein environment (mTagBFP) on the excitation energies of the chromophores was estimated using the QM/MM (quantum mechanics/molecular mechanics) approach. The model system comprised the entire mTagBFP molecule with atomic coordinates of heavy atoms from the X-ray structure (PDB ID: 3M24,9 unit A). The quantum part (QM) (see Fig. 3, left panel) included the chromophore, the sidechains of Lys70, Arg95, Glu215 (DsRed numeration scheme), and the two water molecules forming hydrogen bonds to the chromophore (total 75 atoms). Similar QM part has been used in the calculations of the GFP excitation energies yielding satisfactory results.23 Positions of hydrogen atoms were optimized using the TINKER package24 and CHARMM22 molecular mechanics force field. All heavy atoms were frozen during optimization. The force-field parameters for the chromophore were adopted from the GFP chromophore25 and augmented by the parameters of the similar molecular fragments from amino-acids. Note that chromophore’s force-field only affects the hydrogen atom positions.
Figure 3.
Left panel: the X-ray structure and the quantum part selection for the mTagBFP model system used for excitation energy calculations. Right panel: optimized QM/MM structure of the mTagBFP active site (only the QM part shown) with the chromophore in the anionic form (QM:PBE0/cc-pVDZ/MM:CHARMM27). DsRed numeration is used. mTagBFP’s Lys67, Arg92, Glu215, and Thr179 correspond to the Lys70, Arg95, Glu215, and Ser179 residues of DsRed.
Vertical excitation energies were computed using SOS-CIS(D)/aug-cc-pVDZ and electrostatic embedding using CHARMM22 force field (QM:SOS-CIS(D)/aug-cc-pVDZ/MM:CHARMM22). Two sets of calculation were performed: (i) cluster model, in which interaction of the QM part with the protein environment were neglected; (ii) full QM/MM calculation, in which electrostatic effects of the protein environment on the QM part were taken into account using electrostatic embedding. In all QM/MM calculations the link atom scheme was employed to cap the QM region. Electrostatic interaction between the link atoms and MM atoms (point charges) connected through one and two bonds with the capped QM atom were turned off.
To assess the stability of different protonation forms of the protein-bound chromophore, we performed QM/MM geometry optimizations for mTagBFP with the chromophore in the anionic, zwitterionic, and 2-electron oxidized forms. The model systems for QM/MM optimizations were prepared as follows. The mTagBFP’s X-ray structure (PDB ID: 3M24,9 unit A) was used as the starting geometry. The protein was solvated by 6819 water molecules including those present in the crystallographic structure. Seven negative (Cl−) and eleven positive (Na+) counterions were added to neutralize charged surface residues. All solvent molecules (added upon solvation and crystal water molecules) and counterions were equilibrated using constant temperature (NVT ensemble) molecular dynamics (MD) simulations (10 ps trajectory with 1 fs time step) at 298 K using the Berendsen thermostat.26 All long-range electrostatic interactions were computed by using the particle mesh Ewald method. Except for the side-chains of the surface charged residues, all protein atoms were fixed in the MD simulation. The system was then annealed for 2 ps from 298 to 0 K. The obtained structure was used to extract the protein geometry as well as protein’s solvent shell (water molecules within 3 Å from the protein atoms and counterions). This geometry was further used as a starting point for the QM/MM calculations. Mechanical embedding QM/MM optimization was performed using the PBE0 hybrid functional, the cc-pVDZ basis set, and the CHARMM2227 force field. To ensure that the mechanical embedding QM/MM scheme captures all essential interactions, we employed an extended quantum part (relative to the one described above), which included all charged residues in the vicinity of the chromophore as well as the residues forming hydrogen bonds to the chromophore and to the charged residues. This quantum part (QM) (see Fig. 3, right panel) was comprised by the chromophore (anion, zwitterion or doubly-oxidized form), the sidechains of Lys70, Arg95, Glu145, Glu215, OH group of Thr179, and the three water molecules located in the active site (total 91 atoms). The partial charges of the chromophore atoms that were used to compute the QM/MM electrostatic interaction energy were computed at the X-ray geometry using Natural Bond Orbital analysis28 of the PBE0/cc-pVDZ density (extracted from the full QM calculation). Protein residues, water molecules, and counterions that were further than 10 Å away from the QM part were frozen in the QM/MM geometry optimizations.
All calculations of the excitation energies were performed with the Q-CHEM electronic structure program.29 The TINKER24 package was used for MD simulations. The QM/MM ground-state geometry optimizations were performed using the Firefly package,30 which is partially based on the GAMESS (US)31 code.
2.2 Experimental characterization of the mTagBFP and TagRFP mutants
The mTagBFP and TagRFP mutants were produced by site-directed mutagenesis using the mTagBFP and TagRFP genes in the pBAD/HisB vector as templates. The recombinant proteins with the N-terminal His6-tag were expressed in the LMG194 bacterial strain (Invitrogen) by overnight culture in RM minimal medium at 37°C in the presence of 0.005% arabinose. The culture was then centrifuged at 5,000 rpm at 4°C for 15 min, the cell pellet resuspended in phosphate buffer saline (PBS), 300 mM NaCl, pH 7.4 and lysed by sonication on ice. The recombinant protein was purified with Ni-NTA resin (Qiagen) followed by dialysis overnight against PBS.
Absorption spectra were recorded on a U-3010 spectrophotometer (Hitachi). The excitation and emission spectra were measured using a FluoroMax-3 spectrofluorometer (Jobin Yvon). For measurements the protein samples in PBS were used. Absorption of alkali denatured FPs were measured right after the mixing of the protein sample with NaOH (to the final concentration of 0.7 M) in the optical cuvette. Brightness of the proteins was calculated as a product of the quantum yield and the extinction coefficient. The quantum yield was measured using mTagBFP (quantum yield is 0.637) or TagRFP(quantum yield is 0.4811) as the reference standards, respectively. The apparent extinction coefficients were determined for the mTagBFP mutants as the ratio of the absorption at 400 nm to the absorption at 280 nm followed by normalization to the mTagBFP extinction coefficient, and for the TagRFP mutants — as the ratio of the absorption at 555 nm to the absorption at 280 nm followed by normalization to the TagRFP extinction coefficient. Equilibrium pH titrations were performed using a series of buffers (100 mM NaOAc, 300 mM NaCl for pH 2.5–4.5, and 100 mM NaH2PO4, 300 mM NaCl for pH 4.5–11.0).
3 Results and Discussion
3.1 Vertical excitation energies
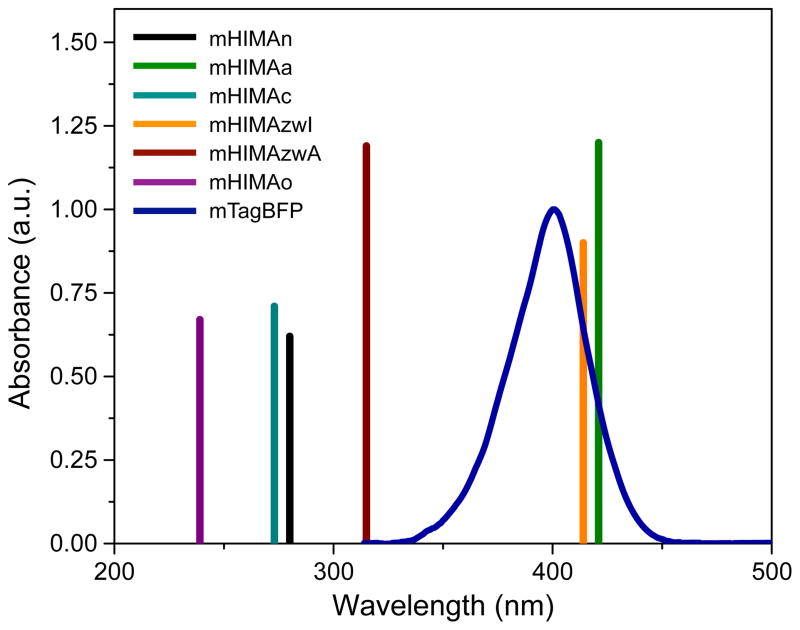
Vertical excitation energies for the lowest bright (oscillator strength, fL, greater than 0.1) excited state computed using SOS-CIS(D) are collected in Table 1 and visualized in Fig. 4. The method has been shown to provide reliable vertical excitation energies for similar chromophores.13 Moreover, the SOS-CIS(D) results were also validated against EOM-CCSD (Table S3). We estimate the uncertainties of the computational methods to be around 0.3 eV. Note that the experimental absorption band half-width is about 45 nm (0.34 eV), which gives an estimate of the anticipated broadening due to thermal motions.
Table 1.
Vertical excitation energies and oscillator strength (fL) for the lowest bright ππ* excited states in different model chromophores and the dihedral angles of their optimized structuresa.
| Molecule | Δ E, nm (eV) | fL | φ, ° | θ, ° |
|---|---|---|---|---|
| mHIMAn | 280 (4.43) | 0.62 | 176 | 67 |
| mHIMAa | 421 (2.95) | 1.20 | 170 | 20 |
| mHIMAc | 273 (4.54) | 0.71 | 174 | 60 |
| mHIMAzwI | 414 (3.00) | 0.90 | 171 | 42 |
| mHIMAzwA | 315 (3.94) | 1.19 | 155 | 6 |
| mHIMAo | 239 (5.19) | 0.65 | 174 | 75 |
|
| ||||
| QM(mHIMAa) | 375 (3.31) | 0.78 | 168 | 106 |
| QM(mHIMAa)/MM | 408 (3.04) | 0.77 | 168 | 106 |
The QM(mHIMAa) results are for the cluster model with the QM geometry corresponding to the X-ray mTagBFP structure with the chromophore in the anionic state. The QM/MM values refer to the mTagBFP excitation energies computed using an electrostatic embedding QM/MM model representing the chromophore cluster model inside the protein matrix.
Figure 4.
The SOS-CIS(D)/aug-cc-pVTZ excitation energies for HIMA in different protonation and oxidation states. The bar heights are proportional to oscillator strengths. mTagBFP absorption spectrum is from Ref. 9. Absorption maximum for the blue forms of other red FPs (FTs, DsRed, dark PAmCherry) is in the range of 403–410 nm.4–6
Despite the same number of atoms participating in the π-conjugated system the lowest π – π* excitation energies vary from 2.93 to 5.13 eV (423–242 nm) for different protonation and oxidation states (Table 1). Only anionic and zwitterionic forms (mHIMAa and mHIMAzwI) agree with the experimental absorption maxima of the DsRed blue-form (3.04 eV or 408 nm) or mTagBFP (3.10 eV or 400 nm).
Although the computed excitation energies may change at higher-level of theory or when the interactions with the protein matrix are taken into account, the gap of 2 eV is sufficiently large to rule out mHIMAo structure. Moreover, this structure has several dark states below the bright state, which can facilitate radiationless relaxation suppressing fluorescence. Finally, the QM/MM calculations show that this structure is unstable in the protein binding pocket. Note that this structure differs from other forms by oxidation state, which leads to important mechanistic implications discussed below.
The variations in the excitation energies for different protonation states are caused not only by different electronic structure, but also by variations in equilibrium geometries. The steric repulsion between the methyl group and the acylimine moiety leads to non-planarity of the chromophore, i.e., θ dihedral angle (Figs. 2 and S1) varies from 6 to 75° for different protonation states (Table 1, for more discussion see SI).
The excitation energies for the isolated model anionic and zwitterionic chromophores agree well with the experimental absorption maximum of the blue form in DsRed and mTagBFP. The protein can affect the optical properties of the blue chromophore by inducing structural deformations and by the electrostatic field of the nearby residues. Indeed, the C=O group of the acylimine in the mTagBFP-like chromophore is out of the chromophore plane, with the acylimine θ angle varying from −152 to 106° for four different protein chains in crystallographic asymmetric unit.9 As out-of-plane distortions involve conjugated π-system, one may expect notable effect on the chromophore absorption. However, our calculations of potential energy profiles along the θ angle show that the excitation energy of the anionic form is not affected by the non-planarity. Thus, the structural deformation caused by the protein environment does not significantly affect its optical properties, although it may be responsible for a minor discrepancy between the gas-phase excitation energy of mHIMAa and mHIMAzwI and the absorption maximum of the blue form (see Figs. S6–S8).
To further quantify the effect of the protein on excitation energies, we performed QM/MM calculations of the protein-bound (mTagBFP) anionic (mHIMAa) chromophores. Two models were used: a cluster model including the chromophore and several nearby residues and the full QM/MM model that takes into account the effect of the electrostatic field of the entire solvated protein (Table 1). The interactions with the nearby residues (cluster model) lead to the blue shift of 0.36 eV. However, the electrostatic field of the rest of the protein/solvent/counter-ions red-shifts the excitation energy to 3.04 eV (408 nm), which is close to the experimental absorption maximum of mTagBFP (3.10 eV or 400 nm). A relatively small shift due to the interaction with the protein is consistent with negligible protein-induced shifts of the GFP-like chromophore absorption, which has been observed both experimentally and predicted by calculations.23,32–34 Thus, it is highly unlikely that the protein can perturb the excited states of other protonation and oxidation states of the mHIMA chromophore by 0.9 eV to account for the differences between the experimental absorption maximum and the excitation energies.
Apart from the excitation energy criterion, the analysis of the chemical stability of the protein-bound chromophore in different protonation and oxidation states provides additional evidence against doubly-oxidized form of the chromophore (mHIMAo). Moreover, these calculations reveal that the zwitterionic structure (mHIMAzwI) is unstable in the protein binding pocket thus allowing us to rule out this structure as well.
To analyze the stability of the chromophore in different protonation states inside the binding pocket of mTagBFP, we performed series of the QM/MM geometry optimizations with the chromophore in the anionic (mHIMAa), zwitterionic (mHIMAzwI), and doubly-oxidized (mHI-MAo) forms. The QM/MM optimized structure of the protein-bound anionic chromophore is very close to the X-ray structure of mTagBFP (see Fig. 3). In contrast, no stationary point was found for the zwitterionic and doubly-oxidized chromophores. In the former case, we observe prompt proton transfer from the imidazole ring to deprotonated Glu215 (see Fig. S11 for the alignment of the amino acid sequences). Highly energetic structure of the doubly-oxidized chromophore was also found to be unstable: the chromophore cation undergoes a nucleophilic attack by the nearby water molecule (Fig. S11). This provides additional evidence against the cationic structure.
In sum, from the two protonation states of the chromophore with optical properties matching the experiment (anion and zwitterion), only the anionic form is stable inside the protein matrix.
The negative charge on the enol moiety of the mHIMA chromophore is consistent with the X-ray structure of mTagBFP9 and the QM/MM optimized geometries. The enol oxygen of the chromophore forms two hydrogen bonds with positively charged Arg95 and Lys70 with respective distances between the heavy atoms of 2.82–2.98 and 3.03–3.31 Å for different protein chains in the mTagBFP crystallographic unit (see Figs. 3 and S9). The active-site configuration with the two positively charged residues (Lys70 and Arg95) suggests the stabilization of these two positive centers by the negative charge of the chromophore group, in addition to the Glu145 residue (Fig. S9). Moreover, a number of RFPs including DsRed, eqFP611, eqFP578 and their derivatives such as FTs, TagRFP, mCherry, mKate and mNeptune contain Lys or Arg homologous of mTagBFP’s Lys70, whereas GFP, EGFP, and copGFP, in which the N-acylimine bond is absent, have neutral Gln at this position (Fig. S10).
The structure of the mTagBFP active site also explains extremely low effective pKa of the chromophore (2.7±0.2).7 The protein barrel and, to a larger extent, formation of salt bridges can significantly decrease chromophore’s pKa. For example, pKa of the GFP chromophore (HBDI) in solution is 8.2 (anion/neutral),35 whereas pKa of the protein-bound anionic chromophore (in EGFP) is 5.8,36 that is, 2.4-fold less than for the bare chromophore interacting with water. When chromophore’s negative charge is stabilized by a salt bridge, pKa decreases further, as illustrated by DsRed and copGFP (pKa of 4.7 and 4.3)37,38 in which the chromophore is stabilized by the salt bridges with Lys153 and Arg156, respectively. Therefore, the two positively charged residues in the proximity of the mTagBFP chromophore are likely to be responsible for low pKa. Note that the absorption of mTagBFP in the acidic media is strongly blue-shifted (335 nm or 3.70 eV),7 which is consistent with the computed higher excitation energies of the protonated forms.
3.2 Properties of the purified mTagBFP and TagRFP mutants
To determine whether Lys70 in mTagBFP7 and Arg70 in TagRFP39 are required for the formation of the blue and red chromophores, we substituted Lys70 with Ile, Val and Gln, and Arg70 with Ala, Gln, His and Lys (Tables 2 and 3). The substitution of Lys70 in mTagBFP with Ile, Val and Gln results in blue-emitting FPs with the absorption maxima at 400–416 nm and the emission maxima at 461–463 nm that are similar to those of mTagBFP (Table 2). The dramatic 9–83 fold decrease of the extinction coefficients for the Lys70 variants indicates substantially reduced efficiency of the mTagBFP-like chromophore formation. In addition, 3.5–12.6 folds decrease of the quantum yields indicates the importance of Lys70 for the rigidity and planarity of the mTagBFP chromophore. Altogether, introduction of mutations into position 70 results in 56–1000 fold decrease in the mTagBFP brightness.
Table 2.
Properties of the mTagBFP mutants.
| mTagBFP mutants | Absorption maximum, nm (eV) | Fluorescence max., nm (eV); excitation at 399 nm | Apparent extinction coefficient at 400 nm, mM−1 cm−1a 41400 | Quantum yield | Brightness relative to mTagBFP, %b | pKa |
|---|---|---|---|---|---|---|
| mTagBFP | 400 (3.10) | 455 (2.73) | 0.63 | 100 | 2.7 ± 0.2 | |
| K70I | 400 (3.10) | 462 (2.69) | 4700 | 0.10 | 1.8 | 9.0 ± 0.2 |
| K70V | 416 (2.98) | 463 (2.68) | 1400 | 0.18 | 1 | 9.4 ± 0.2 |
| K70Q | 410 (3.03) | 461 (2.69) | 500 | 0.05 | 0.1 | 10.3 ± 0.2 |
| R95K | 400 (3.10) | 455 (2.73) | 800 | 0.62 | 1.9 | 2.7 ± 0.2 |
Apparent extinction coefficients were determined as the ratio of the absorption at 400 nm to the absorption at 280 nm.
Brightness was determined as a product of the extinction coefficient and the quantum yield.
Table 3.
Properties of the TagRFP mutants.
| TagRFP mutants | Absorption maximum, nm (eV)b | Fluorescence max., nm (eV); excitation at 550 nm | Apparent extinction coefficient at 555 nm, mM−1 cm−1c | Quantum yield | Brightness relative to mTagBFP, %d | pKa |
|---|---|---|---|---|---|---|
| TagRFP | 555 (2.24) | 583 (2.13) | 100000 | 0.48 | 100 | 4.3 ± 0.2 |
| R70A | 412 (3.01), 556 (2.23) | 590 (2.10) | 5800 | 0.32 | 3.8 | 7.5 ± 0.2 |
| R70Q | 386 (3.22), 558 (2.22) | 586 (2.12) | 600 | 0.22 | 0.3 | 6.5 ± 0.2 |
| R70H | 396 (3.13), 561 (2.21)a | 590 (2.10) | 430 | 0.25 | 0.2 | 7.4 ± 0.2 |
| R70K | 387 (3.21), 512 (2.42), 555 (2.24) | 580 (2.14) | 63400 | 0.60 | 79 | 3.7 ± 0.2 |
Determined from the excitation spectrum with emission wavelength at 600 nm.
When denatured in 0.7 M NaOH, TagRFP and all its mutants have the absorption maximum at 449 nm characteristic for GFP-like green and DsRed-like red chromophores.
Apparent extinction coefficients were determined as a ratio of the absorbance at 555 nm to the absorbance at 280 nm.
Brightness was determined as a product of the extinction coefficient and the quantum yield.
Analysis of pKa of Lys70 mutants supports our hypothesis that low pKa is due to the stabilization of the negative charge of the mTagBFP-like chromophore by the two positively charged amino acids. Indeed, removing the positive charge of Lys70 in mTagBFP results in the shift of the pKa values from 2.7 to 9.0–10.3 (mTagBFP). It is interesting that the substitution of Arg95, which together with Lys70 participates in the stabilization of the mTagBFP chromophore, with another positively charged amino acid, Lys, results in the FP with identical absorption and emission wavelengths, quantum yield and pKa as the parental mTagBFP (Table 2). However, 52-fold less mTagBFP-like chromophore is formed in the case of this mutant that was expected because of absolute conservancy of Arg95 among FPs and its necessity for cyclization of the imidazolinone ring in the GFP chromophore.40 Thus, we conclude that the positive charge of Lys 70 is extremely important for the stabilization of the mTagBFP-chromophore. Lys70 influences the efficiency of the mTagBFP chromophore formation, its rigidity/planarity, and pH stability.
Introduction of Ala, Gln or His instead of Arg70 in TagRFP preserves the formation of the red chromophore with the absorption/excitation maxima at 556–561 nm and the emission maxima at 586–590 nm (Table 3). The 17–230 fold decrease of the extinction coefficients in the mutants (relative to TagRFP) indicates decreased efficiency of the chromophore formation in the mutants. The substitution of Arg70 in TagRFP has a smaller effect on the rigidity and planarity of the chromophore than the substitution of Lys70 in mTagBFP, as the quantum yields of the mutants decrease only by factor of 1.5–2.2 in comparison with TagRFP. Therefore, the overall brightness of the R70A, R70Q and R70H mutants of TagRFP drops by 26-500-fold relative to TagRFP. The R70K variant containing the positive charge at position 70 differs from the three other variants because its brightness is approximately the same as the brightness of parental TagRFP with just 1.6-fold less absorption but with 1.25-fold higher quantum yield. The three first mutants have much higher pKa values than TagRFP (6.5–7.5 versus 4.3), whereas the R70K mutant has pKa similar to that of TagRFP. Therefore, the positively charged amino acid residue at position 70 is necessary for the efficient formation of the DsRed-like chromophore and for low pKa value. The former may be due to impeded formation and stabilization of the intermediate blue form, or due to the catalytic role of the amino acid in position 70. The effect on the pH stability is likely related to the decreased delocalization of the negative charge inside the anionic DsRed-like chromophore since oxygen of the imidazolinone ring can acquire partial negative charge due to delocalization.
In the absorption spectra of the R70Q and R70H mutants of TagRFP the main peaks are observed at 386 and 396 nm (Table 3). Two approximately equal peaks at 412 nm and 556 nm are detected for the R70A mutants. In the case of the R70K mutant, in addition to the main peak at 555 nm characteristic of the DsRed-like chromophore, two peaks at 387 and 512 nm are observed. In alkaline conditions (0.7 M NaOH) all mutants exhibit a single peak at 449 nm characteristic of the GFP-like and DsRed-like chromophores but not of the mTagBFP-like chromophore.9 Thus, the formation of the green GFP-like chromophore in the protonated (peaks at 386–412 nm) or anionic (peak at 512 nm) forms is observed for all four mutants of Arg70.
In sum, the properties of the mTagBFP and TagRFP mutants at position 70 (Tables 2 and 3) reveals that the positively charged amino acid in position 70 is extremely important for the FP maturation into the blue and, consequently, into the red form. Otherwise, the formation and stabilization of the blue and red chromophores are impeded, and, in addition to the DsRed-like chromophore, the side formation of the GFP-like chromophore occurs for the TagRFP mutants.
3.3 The revised mechanism of the DsRed-like chromophore formation
Based on the computed excitation energies and the results of QM/MM geometry optimizations, the only form of the chromophore that is stable in the protein-binding pocket and has the absorption matching the experimental spectrum is the anionic form. The assignment of the chromophore protonation and oxidation state has important mechanistic implications, namely, that the anionic structure cannot be reconciled with the mechanism proposed by Strack et al.10 Since the intermediate proposed in Ref. 10 is a 2e-oxidized form with respect to mHIMAa, the mHIMAa-like blue intermediate implies that the order of the final oxidation/deprotonation stages should be reversed: the final oxidation occurs after the blue intermediate is formed rather than precedes its formation, as was suggested in Ref. 10. Final oxidation following the blue chromophore formation is also supported by the facts that blue-to-red conversion in FTs is only possible under aerobic conditions5 and the photoactivation of PAmCherry is dramatically inhibited in anaerobic conditions.6 The formation stage of the blue species in the DsRed.T7 protein10 was deduced from matching the rate and stoichiometry of the H2O2 production with the formation kinetics and yield of the matured DsRed.T7 species. Thus, this conclusion10 relies on the quantitative data for the yield of the matured DsRed.T7 molecules, which is difficult to determine with sufficient accuracy. The present results and the experimental data on FTs and PAmCherry5,6 strongly suggest that oxidation occurs at the stage of the blue-to-red conversion.
Fig. 5 presents the revised mechanism, which is similar to that proposed for TagRFP9 and FTs5 and also incorporates the present results. The first two stages follow the established mechanism of the GFP-like chromophore maturation:41 cyclization (I) and oxidation (II). The product formed at the second stage is a branching point for the formation of the green and red chromophores. Two different dehydration pathways are possible. The first one (III) corresponds to the OH− and H+ removal from the imidazolinone ring and the methylene bridge, respectively, leading to the formation of the dead-end product, the GFP-like chromophore. The second pathway (IV) assumes the OH− and H+ detachment from the imidazolinone ring and the peptide bond of Phe65, respectively, along with the rearrangement of the single/double bond pattern forming the enol form of the chromophore with the acylimine being conjugated to the imidazolinone ring. The negative charge of the product is stabilized by K70 and R95. We suggest that K70 is a key residue, a switch leading to the DsRed-like chromophore formation pathway. Stage (V) corresponds to the formation of the blue intermediate upon deprotonation. Final oxidation stage (VI) results in the H atom abstraction from the methylene bridge and the phenol moiety producing the fully conjugated red chromophore.
Figure 5.
A revised mechanism of the DsRed chromophore maturation. (I) cyclization; (II) oxidation; (III) formation of the green chromophore - dehydration (Tyr66 C); (IV and V) formation of the blue intermediate - dehydration of acylimine; (VI) formation of the red chromophore -oxidation (Tyr66 C).
The proposed scheme (Fig. 5) provides an outline of the mechanism of the DsRed-like chromophore maturation; however, more experimental and theoretical studies are required to clarify the proton shuttling pathways, characterize dark intermediates and side-way products and identify crucial amino acid residues responsible for the autocatalytic chromophore formation.
4 Conclusions
In summary, on the basis of ab initio and QM/MM calculations we assign the blue intermediate in the process of the red chromophore maturation to the anionic chromophore, whose absorption in the gas phase (421 nm or 2.95 eV) and inside the mTagBFP protein (408 nm or 3.04 eV) matches the experimental spectrum of the blue form in DsRed (the absorption maximum is 408 nm or 3.04 eV) and mTagBFP (400 nm or 3.10 eV). Unlike other forms of the chromophore, the anionic structure is stable in the protein binding pocket. We suggest a revised branched mechanism of the chromophore maturation in which the second oxidation stage occurs after the mTagBFP-like intermediate is formed. The determination of the chromophore structure reveals the key role of Lys70 for the DsRed-like chromophore maturation, which was verified experimentally by directed mutagenesis. Since the dark form of PAmCherry, the blue form of FTs, and mTagBFP seems to share the same mTag-BFP-like chromophore, the identification of the chemical structure of the blue intermediate advances our fundamental understanding of the chemistry of the red fluorescence acquisition in fluorescent proteins.
Supplementary Material
Acknowledgments
This work was supported by the grants CHE-0951634 from the National Science Foundation (A.I.K.) and GM073913 and CA164468 from the National Institutes of Health (to V.V.V.).
Footnotes
4 Tables and 11 figures illustrating relevant structural parameters of the model chromophores, the effect of the non-planarity on the excitation energies, relevant molecular orbitals and electronic densities, amino-acid sequence comparison for greed and red fluorescent proteins, as well as results of QM/MM geometry optimization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE, Adams SR, Gross LA, Ma W, Alder T, Tsien RY. Chem Biol. 2009;16:1169. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould TJ, Verkhusha VV, Hess ST. Nature Protoc. 2009;4:291. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. Proc Nat Acad Sci. 2000;97:11990. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Chem Biol. 2004;11:845. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Pletnev S, Subach FV, Dauter Z, Wlodawer A, Verkhusha A. J Am Chem Soc. 2010;132:2243. doi: 10.1021/ja908418r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subach FV, Malashkevich VN, Zencheck WD, Xiao H, Filonov GS, Almo SC, Verkhusha VV. Proc Nat Acad Sci. 2009;106:21097. doi: 10.1073/pnas.0909204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Grüenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Chem Biol. 2008;15:1116. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. PLoS One. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subach OS, Malashkevich VN, Zencheck WD, Morozova KS, Piatkevich KD, Almo SC, Verkhusha VV. Chem Biol. 2010;17:333. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strack RL, Strongin DE, Mets L, Glick BS, Keenan RJ. J Am Chem Soc. 2010;132:8496. doi: 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Nat Methods. 2007;4:555. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 12.Rhee YM, Head-Gordon M. J Phys Chem A. 2007;111:5314. doi: 10.1021/jp068409j. [DOI] [PubMed] [Google Scholar]
- 13.Epifanovsky E, Polyakov I, Grigorenko BL, Nemukhin AV, Krylov AI. J Chem Theory Comput. 2009;5:1895. doi: 10.1021/ct900143j. [DOI] [PubMed] [Google Scholar]
- 14.Epifanovsky E, Polyakov I, Grigorenko BL, Nemukhin AV, Krylov AI. J Chem Phys. 2010;132:115104. doi: 10.1063/1.3336425. [DOI] [PubMed] [Google Scholar]
- 15.Polyakov I, Grigorenko BL, Epifanovsky E, Grigorenko BL, Krylov AI, Nemukhin AV. J Chem Theory Comput. 2010;6:2377. doi: 10.1021/ct100227k. [DOI] [PubMed] [Google Scholar]
- 16.Khrenova M, Nemukhin AV, Grigorenko BL, Krylov AI, Domratcheva T. J Chem Theory Comput. 2010;6:2293. doi: 10.1021/ct100179p. [DOI] [PubMed] [Google Scholar]
- 17.Sinha D, Mukhopadhyay D, Mukherjee D. Chem Phys Lett. 1986;129:369. [Google Scholar]
- 18.Stanton JF, Bartlett RJ. J Chem Phys. 1993;98:7029. [Google Scholar]
- 19.Levchenko SV, Krylov AI. J Chem Phys. 2004;120:175. doi: 10.1063/1.1630018. [DOI] [PubMed] [Google Scholar]
- 20.Krylov AI. Annu Rev Phys Chem. 2008;59:433. doi: 10.1146/annurev.physchem.59.032607.093602. [DOI] [PubMed] [Google Scholar]
- 21.Zuev D, Bravaya KB, Crawford TD, Lindh R, Krylov AI. J Chem Phys. 2011;134:034310. doi: 10.1063/1.3516211. [DOI] [PubMed] [Google Scholar]
- 22.Epifanovsky E, Kowalski K, Fan PD, Valiev M, Matsika S, Krylov AI. J Phys Chem A. 2008;112:9983. doi: 10.1021/jp803758q. [DOI] [PubMed] [Google Scholar]
- 23.Bravaya K, Khrenova MG, Grigorenko BL, Nemukhin AV, Krylov AI. J Phys Chem B. 2011;8:8296. doi: 10.1021/jp2020269. [DOI] [PubMed] [Google Scholar]
- 24.Ponder JW. [Accessed May 7, 2009];TINKER – Software Tools for Molecular Design. URL http://dasher.wustl.edu/tinker/
- 25.Reuter N, Lin H, Thiel W. J Phys Chem B. 2002;106:6310. [Google Scholar]
- 26.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684. [Google Scholar]
- 27.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 28.Glendening ED, Badenhoop JK, Reed AE, Carpenter JE, Bohmann JA, Morales CM, Weinhold F. NBO 5.0. Theoretical Chemistry Institute. University of Wisconsin; Madison, WI: 2001. [Google Scholar]
- 29.Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown S, Gilbert ATB, Slipchenko LV, Levchenko SV, O’Neil DP, Distasio RA, Jr, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Lin CY, Van Voorhis T, Chien SH, Sodt A, Steele RP, Rassolov VA, Maslen P, Korambath PP, Adamson RD, Austin B, Baker J, Bird EFC, Daschel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu C-P, Kedziora GS, Khalliulin RZ, Klunziger P, Lee AM, Liang WZ, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Rhee YM, Ritchie J, Rosta E, Sherrill CD, Simmonett AC, Subotnik JE, Woodcock HL, III, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Herhe WJ, Schaefer HF, III, Kong J, Krylov AI, Gill PMW, Head-Gordon M. Phys Chem Chem Phys. 2006;8:3172. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- 30.Granovsky AA. Firefly version 7.1.G. http://www.classic.chem.msu.su/gran/firefly/index.html.
- 31.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Mastunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA. J Comput Chem. 1993;14:1347. [Google Scholar]
- 32.Andersen LH, Lappierre A, Nielsen SB, Nielsen IB, Pedersen SU, Pedersen UV, Tomita S. Eur Phys J D. 2002;20:597. [Google Scholar]
- 33.Sinicropi A, Anduniow T, Ferre N, Basosi R, Olivucci M. J Am Chem Soc. 2005;127:11534. doi: 10.1021/ja045269n. [DOI] [PubMed] [Google Scholar]
- 34.Bravaya K, Grigorenko BL, Nemukhin AV, Krylov AI. Acc Chem Res. 2011 doi: 10.1021/ar2001556. in press. [DOI] [PubMed] [Google Scholar]
- 35.Bell AF, Stoner-Ma D, Wachter RM, Tonge PJ. J Am Chem Soc. 2003;125:6919. doi: 10.1021/ja034588w. [DOI] [PubMed] [Google Scholar]
- 36.Haupts U, Maiti S, Schwille P, Webb WW. Proc Nat Acad Sci. 1998;95:13573. doi: 10.1073/pnas.95.23.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wall MA, Socolich M, Ranganathan R. Nature Struct Biol. 2000;7:1133. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 38.Wilmann PG, Battad J, Petersen J, Wilce MCJ, Dove S, Devenish RJ, Prescott M, Pssjohn J. J Mol Biol. 2006;359:890. doi: 10.1016/j.jmb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Nat Methods. 2007;4:741. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 40.Wachter RM. Acc Chem Res. 2007;40:120. doi: 10.1021/ar040086r. [DOI] [PubMed] [Google Scholar]
- 41.Wachter RM, Watkins JL, Kim H. Biochemistry. 2010;49:7417. doi: 10.1021/bi100901h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.