Abstract
The aim of this study was to determine whether mouse CYP2A5 and CYP2F2 play critical roles in the bioactivation of 3-methylindole (3MI), a tissue-selective toxicant, in the target tissues, the nasal olfactory mucosa (OM) and lung. Five metabolites of 3MI were identified in NADPH- and GSH-fortified microsomal reactions, including 3-glutathionyl-S-methylindole (GS-A1), 3-methyl-2-glutathionyl-S-indole (GS-A2), 3-hydroxy-3-methyleneindolenine (HMI), indole-3-carbinol (I-3-C), and 3-methyloxindole (MOI). The metabolite profiles and enzyme kinetics of the reactions were compared between OM and lung, and among wild-type, Cyp2a5-null, and Cyp2f2-null mice. In lung reactions, GS-A1, GS-A2, and HMI were detected as major products, and I-3-C and MOI, as minor metabolites. In OM reactions, all five metabolites were detected in ample amounts. The loss of CYP2F2 affected formation of all 3MI metabolites in the lung and formation of HMI, GS-A1, and GS-A2 in the OM. In contrast, loss of CYP2A5 did not affect formation of 3MI metabolites in the lung but caused substantial decreases in I-3-C and MOI formation in the OM. Thus, whereas CYP2F2 plays a critical role in the 3MI metabolism in the lung, both CYP2A5 and CYP2F2 play important roles in 3MI metabolism in the OM. Furthermore, the fate of the reactive metabolites produced by the two enzymes through common dehydrogenation and epoxidation pathways seemed to differ with CYP2A5 supporting direct conversion to stable metabolites and CYP2F2 supporting further formation of reactive iminium ions. These results provide the basis for understanding the respective roles of CYP2A5 and CYP2F2 in 3MI's toxicity in the respiratory tract.
Introduction
3-Methylindole (3MI), a metabolite of tryptophan and a constituent of tobacco smoke, is a selective respiratory tract toxicant in mice (Turk et al., 1986). 3MI can selectively induce necrosis in the bronchiolar epithelial (Clara) cells in the mouse lung (Turk et al., 1984) and necrosis in epithelial cells of Bowman's glands and olfactory sustentacular cells in mouse nasal olfactory mucosa (OM) (Turk et al., 1987; Miller and O'Bryan, 2003). Cytochrome P450 (P450)-mediated metabolic activation is believed to play a major role in 3MI-induced toxicity (Yost, 1989). Studies on the in vitro metabolism of 3MI using heterologously expressed human P450s, human lung microsomes, and cultured human bronchial epithelial cells suggested that 3MI is also a potential human respiratory tract toxicant (Ruangyuttikarn et al., 1991; Thornton-Manning et al., 1991, 1996; Lanza et al., 1999; Lanza and Yost 2001; Nichols et al., 2003; D'Agostino et al., 2009).
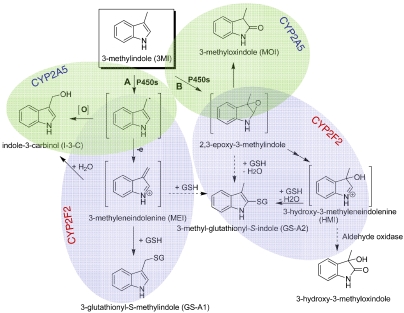
The bioactivation of 3MI can produce several potentially toxic intermediates: 3-methyleneindolenine [(MEI) derived from the 3MI radical], through the dehydrogenation pathway, and 2,3-epoxy-3MI and 3-hydroxy-3-methyleneindolenine (HMI), through the epoxidation pathway (as shown in Fig. 1). Extensive studies have shown that both pathways can lead to toxicity (Skiles and Yost, 1996; Lanza and Yost, 2001; Yost, 2001). The electrophilic intermediates produced in these pathways can deplete cellular GSH and form covalent adducts with cellular proteins (Thornton-Manning et al., 1996) and DNA (Regal et al., 2001) and induce mutagenesis in yeast (Weems et al., 2009, 2010).
Fig. 1.
Scheme for the metabolic activation of 3MI by mouse OM and lung microsomal P450s. Pathway A represents dehydrogenation and pathway B represents epoxidation [The figure has been drawn on the basis of the findings in D'Agostino et al. (2009)]. Shaded areas show metabolites preferentially formed by CYP2A5 (blue) or CYP2F2 (green).
The tissue specificity of 3MI-induced respiratory tract toxicity presumably results from the metabolic activation of 3MI by P450 enzymes selectively expressed in the target tissues, i.e., the lung and OM. Multiple human P450s with respiratory tract-selective expression have been investigated for their ability to bioactivate 3MI. In vitro studies have demonstrated that human CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2A13, CYP2E1, and CYP2F1 can all metabolize 3MI (Thornton-Manning et al., 1991, 1996; Lanza et al., 1999; Lanza and Yost 2001; D'Agostino et al., 2009). Among those P450s, CYP2F1, a lung-selective enzyme, seems to be the most efficient enzyme in catalyzing the dehydrogenation of 3MI (Lanza et al., 1999), whereas CYP2A13, also predominantly expressed in human respiratory tract (Su et al., 2000), is efficient in the metabolic activation of 3MI through both dehydrogenation and epoxidation pathways (D'Agostino et al., 2009). However, studies on the relative contributions of these enzymes to 3MI metabolism in human lung and nasal microsomes have thus far been unsuccessful, because of the lack of specific inhibitors or antibodies [see Zhang and Ding, 2008 for a recent review].
3MI metabolic activation has not been studied in detail in mice, in which the tissue-selective toxicity of 3MI in the lung and OM was first demonstrated. Although mouse CYP1A2 was found to be active in 3MI metabolism (Thornton-Manning et al., 1996), the ability of mouse CYP2A5 (orthologous to CYP2A13) and CYP2F2 (orthologous to CYP2F1) to bioactivate 3MI has not been examined. Both CYP2A5 and CYP2F2 can metabolize numerous toxicants (see Su and Ding, 2004; Zhang and Ding, 2008). CYP2A5 is abundantly expressed in OM, and at much lower levels in other tissues, including the lung (see Su and Ding, 2004), whereas CYP2F2 is expressed in the liver, lung, and OM at comparable levels (Baldwin et al., 2004; Li et al., 2011).
In the present study, we have determined whether CYP2A5 and CYP2F2 play critical roles in the bioactivation of 3MI in mouse lung and OM in vitro, by comparing the metabolite profiles and enzyme kinetics of 3MI metabolism among wild-type (WT), Cyp2a5-null (Zhou et al., 2010), and Cyp2f2-null mice (Li et al., 2011). The Cyp2a5-null and Cyp2f2-null mice have been found to be valuable for demonstrating the roles of CYP2A5 or CYP2F2 in mediating the tissue-specific toxicities in the respiratory tract induced by acetaminophen (Zhou et al., 2011), 2,6-dichlorobenzonitrile (Xie et al., 2010), and naphthalene (Li et al., 2011). Here, we show that whereas CYP2F2 plays a critical role in the 3MI metabolism in the lung, both CYP2A5 and CYP2F2 play important roles in 3MI metabolism in the OM. Furthermore, we found that the fates of the reactive metabolites produced by the two enzymes (3MI radical and 2,3-epoxy-3MI) differ with CYP2A5 supporting direct conversion to stable metabolites (which have decreased potential to cause toxicity) and CYP2F2 supporting further formation of reactive iminium ions (MEI and HMI) that can form adducts with GSH and cellular proteins (thus more likely than CYP2A5 to cause toxicity). Our findings provide the basis for understanding the respective roles of CYP2A5 and CYP2F2 in 3MI's toxicity in the respiratory tract.
Materials and Methods
Enzymes and Chemicals.
The sources of 3MI, 3-methyloxindole (MOI), indole-3-carbinol (I-3-C), 3-glutathionyl-S-methylindole (GS-A1), and NADPH were the same as those described previously (D'Agostino et al., 2009). Acetaminophen-GSH (APAP-GSH) disodium salt; and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-N-oxide (NNK-N-oxide) were purchased from Toronto Research Chemicals (North York, ON, Canada). All solvents (acetonitrile, methanol, and water) were of high-performance liquid chromatography (LC) grade (Thermo Fisher Scientific, Waltham, MA).
Microsomal Incubations for Metabolite Identification.
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the Wadsworth Center. Lung and OM microsomes were prepared as described previously (Gu et al., 1998), from adult male WT, Cyp2a5-null, or Cyp2f2-null mice, all on the C57BL/6 genetic background. Tissues were pooled from five mice for each microsomal preparation. Reaction mixtures contained 100 mM potassium phosphate buffer, pH 7.4, 50 μM 3MI, 0.5 mg/ml OM or lung microsomal protein, 0 or 5 mM GSH, and 0 or 1.0 mM NADPH, in a final volume of 0.2 ml. Reactions were performed at 37°C for 0 or 30 min and were terminated by the addition of 0.2 ml of acetonitrile. The samples were centrifuged to remove the precipitated protein. The supernatant was dried under nitrogen and then was reconstituted in 50 μl of 50% methanol/water (v/v) for liquid chromatography/mass spectrometry analysis, as described below.
Determination of Catalytic Activity.
For determination of kinetic parameters for the formation of I-3-C, HMI, and MOI by mouse OM and lung microsomal P450, reaction mixtures contained 100 mM potassium phosphate buffer, pH 7.4, 0 to 200 μM (0, 1, 2.5, 5, 10, 25, 50, 100, and 200 μM) 3MI, 0.05 mg/ml OM or 0.2 mg/ml lung microsomal protein, and 1.0 mM NADPH, in a final volume of 0.2 ml. Reactions were performed at 37°C for 5 min, and were terminated by the addition of 0.2 ml of acetonitrile containing 5 ng of the internal standard NNK-N-oxide (which was found in pilot studies to be similar to 3-MI metabolites in ionization efficiency and hydrophobicity). The samples were centrifuged, and 5-μl aliquots of the supernatant were analyzed as described below. The same protocol was used for determination of kinetic parameters for the formation of GS-A1 and 3-methyl-2-glutathionyl-S-indole (GS-A2), except that 5 mM GSH was added to the reaction mixture, and 50 ng of APAP-GSH (instead of the NNK-N-oxide) was used as the internal standard.
Liquid Chromatography/Tandem Mass Spectrometry Analytical Methods.
A liquid chromatography/tandem mass spectrometry (LC-MS/MS) system composed of an Agilent 1200 Series high-performance liquid chromatography (Agilent Technologies, Santa Clara, CA) and an ABI 4000 Q-Trap mass spectrometer (Applied Biosystems, Foster City, CA), essentially as described previously (Zhou et al., 2009), was used to identify and quantify 3MI metabolites. The metabolites were resolved by using a 5-μm Gemini C18 column (100 × 2.0 mm; Phenomenex, Torrance, CA). The mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). The samples were eluted, at a flow rate of 0.25 ml/min, with 100% solvent A for 1 min, followed by linear increases from 0 solvent B to 100% solvent B between 1 and 10 min, and then 100 solvent B for a further 2.5 min. The mass spectrometer was set for the information-dependent acquisition scan mode, using multiple ion monitoring (MIM)-dependent enhanced product ion acquisition (see Supplemental Table 1 for ion list) as described previously by Yao et al. (2008). The mass spectrometer was operated in the positive ion mode, with an electrospray ionization source. The parameters for the chamber were as follows: curtain gas, 30 psi; heated nebulizer temperature, 300°C; ion spray voltage, 4500 V; nebulizer gas, 50 psi; turbo gas, 50 psi, declustering potential, 50 V; and entrance potential, 10 V.
For quantitative analysis of 3MI metabolites, the parent/product ion pairs of m/z 130/103 [for I-3-C; the m/z 130 species results from in-source loss of water, as described in D'Agostino et al. (2009)], m/z 148/133 (for MOI and HMI), m/z 437/308 (for GS-A1 and GS-A2), m/z 224/177 (for NNK-N-oxide), and m/z 457/328 (for APAP-GSH) were monitored in the multiple reaction monitoring scan mode. The retention time for HMI, I-3-C, MOI, and their internal standard NNK-N-oxide was 7.3, 9.7, 10.1, and 7.9 min, respectively, and that for GS-A1, GS-A2, and their internal standard APAP-GSH was 8.7, 9.1, and 7.3 min, respectively. The calibration curves for I-3-C, MOI, and GS-A1 were prepared by spiking the authentic standards and internal standards to incubation mixtures containing boiled microsomes (recovery >80%).
Other Methods.
The mouse liver cytosol fraction was prepared as described previously (Zhou et al., 2010). Nonlinear regression and enzyme kinetic analyses were performed with GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA). Statistical significance of differences between two groups was examined with Student's t test.
Results and Discussion
Identification of 3MI Metabolites Formed by OM and Lung Microsomes from WT Mice.
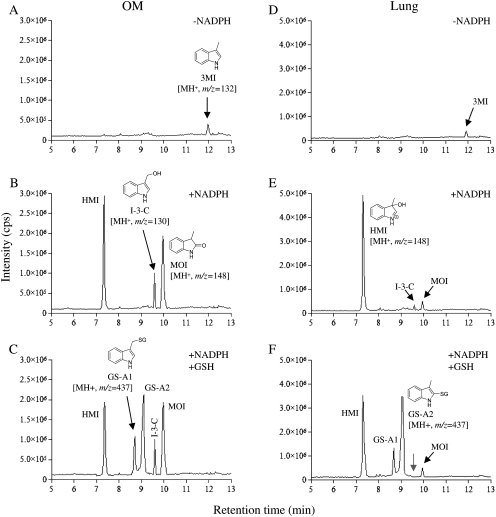
MIM for the LC-MS/MS analysis was set up for all predicted 3MI metabolites; those detected in WT mice are shown in Fig. 1. The total ion chromatograms derived from the MIM scans for mouse OM microsomal incubation samples are shown in Fig. 2, A–C. Comparisons among the three samples (complete, complete plus GSH, and complete minus NADPH) revealed three major (unconjugated) metabolite peaks (two at m/z 148, corresponding to MOI and presumably HMI, and one at m/z 130 corresponding to I-3-C) and two GSH adduct peaks (at m/z 437, corresponding to GS-A1 and presumably GS-A2). The same sets of metabolite peaks were detected in the MIM scans for mouse lung microsomal incubation samples, as shown in Fig. 2, D–F, although the relative abundance of the detected metabolites differed between the corresponding OM and lung microsomal incubation samples.
Fig. 2.
LC-MS/MS detection of metabolites of 3MI formed by mouse OM and lung microsomal P450s. Complete reaction mixtures contained 100 mM potassium phosphate buffer, pH 7.4, 50 μM 3MI, 0.5 mg/ml OM (A–C) or lung (D–F) microsomal protein, and 1.0 mM NADPH. GSH was added at 5 mM final concentration, where indicated. Reactions were performed for 0 (A and D) or 30 min (B, C, E, and F), as shown. The samples were analyzed using LC-MS/MS, as described under Materials and Methods. Typical results are shown.
The detection of the metabolite peaks was supported by subsequent MIM-dependent enhanced product ion scans, which produced MS2 spectra for the species detected (data not shown). Identification of the parent compound (3MI) and three of five of the detected metabolites (MOI, I-3-C, and GS-A1) was established on the basis of coelution under the same liquid chromatography conditions and matching of the MS2 spectra with authentic standards. Standards for HMI and GS-A2 were not available; the identification of these two metabolites was based on comparisons of their properties with those characterized previously for the same metabolites produced by heterologously expressed CYP2A13 (D'Agostino et al., 2009). The metabolite peak assigned as HMI (at m/z 148) had an MS2 fragmentation profile similar to that for MOI (data not shown). In addition, in experiments not presented, when mouse liver cytosol, representing a source of aldehyde oxidase was added to the reaction mixtures, a cytosol- and time-dependent decrease in signal intensity was observed for this (but no other) peak, and appearance of a peak corresponding to 3-hydroxyl-3-methyloxindole, the anticipated metabolite (see Fig. 1), was observed. These features are characteristic of HMI. The metabolite peak assigned as GS-A2 (at m/z 437), which was formed only in the presence of GSH, had an MS2 fragmentation pattern (data not shown) and relative retention time similar to those of GS-A2 produced by CYP2A13 (D'Agostino et al., 2009).
Although the results in Fig. 2 do not provide information on absolute levels of the various metabolites in the OM and lung, the relative abundance of the detected metabolites seems to be unique for each tissue. For example, HMI and MOI are both derived from the epoxidation pathway (Fig. 1), but the abundance ratio of MOI/HMI was much lower in the lung (Fig. 2E) than in the OM (Fig. 2B). One possible explanation for the differing abundance ratios is that different P450 enzymes are involved in the epoxidation of 3MI in the OM and lung, and the differing active site environments of these P450 enzymes lead to different partition ratios of the products derived from the reactive 2,3-epoxy-3MI. Furthermore, in the dehydrogenation pathway, the addition of GSH did not seem to change the intensity of the I-3-C peak in incubations with OM microsomes, but it caused the I-3-C peak (seen in Fig. 2E) to disappear in incubations with lung microsomes (Fig. 2F, gray arrow). This observation suggested tissue differences in the accessibility of the reactive precursor of I-3-C, produced in the dehydrogenation pathway, to the added GSH. In that regard, for both lung and OM, whereas the peak intensity for MOI was not altered by the added GSH, the intensity of the HMI peak was reduced by the inclusion of GSH in the incubations (Fig. 2, C and F), consistent with the quenching of the weakly reactive HMI by GSH. GSH is not a strong nucleophile for trapping iminium ions (Shu et al., 2008), which may explain the incomplete loss of HMI in these reactions.
Quantitative Analysis of 3MI Metabolites Formed by OM and Lung Microsomes from WT, Cyp2a5-Null, and Cyp2f2-Null Mice.
The rates of metabolite formation were determined for I-3-C, MOI, and HMI (in the absence of added GSH) and GS-A1 and GS-A2 (in the presence of saturating levels of GSH) at 3MI concentrations ranging from 0 to 200 μM, using conditions under which the rates of product formation were linear with time. For I-3-C, MOI, and GS-A1, actual rates were determined using standard curves. For HMI and GS-A2, relative activities were determined using arbitrary units, which were proportional to peak area ratios (metabolite/internal standard) in ion chromatograms from multiple reaction monitoring scans. Substrate-velocity curves for rates of formation of the five metabolites in OM and lung microsomes over the broad substrate range are shown in Supplemental Fig. 1. The calculated kinetic parameters are shown in Table 1 for OM and Table 2 for lung.
TABLE 1.
Apparent kinetic parameters for the formation of 3MI metabolites by OM microsomes from WT, Cyp2a5-null, and Cyp2f2-null mice
Rates of metabolite formation were determined as described under Materials and Methods. Values presented are the means ± S.D. of data from three separate microsomal samples, each prepared from tissues pooled from five mice. Corresponding substrate-velocity curves are shown in supplemental material. The rates of HMI, I-3-C, and MOI formation were determined in the absence of added GSH. For HMI and GS-A2, the Km and Vmax values were estimated from relative rates determined in arbitrary units.
| Metabolite | Strain | Km | Vmax | Vmax/Km |
|---|---|---|---|---|
| μM | nmol · min−1 · mg protein−1 | ml · min−1 · mg−1 · | ||
| I-3-C | WT | 7.32 ± 0.61 | 1.23 ± 0.11 | 0.17 |
| Cyp2a5-null | 7.12 ± 0.79 | 0.29 ± 0.01* | 0.04 | |
| Cyp2f2-null | 6.90 ± 0.26 | 1.14 ± 0.16 | 0.17 | |
| MOI | WT | 11.7 ± 0.5 | 1.41 ± 0.11 | 0.12 |
| Cyp2a5-null | 31.5 ± 1.4* | 0.17 ± 0.01* | 0.01 | |
| Cyp2f2-null | 11.1 ± 1.1 | 1.40 ± 0.07 | 0.13 | |
| GS-A1 | WT | 5.49 ± 0.53 | 0.79 ± 0.03 | 0.14 |
| Cyp2a5-null | 5.77 ± 0.37 | 0.73 ± 0.04 | 0.13 | |
| Cyp2f2-null | 10.3 ± 1.2* | 0.67 ± 0.09 | 0.07 | |
| Arbitrary units (analyte peak area · internal standard peak area−1 · min−1 · mg protein−1) | ||||
| HMI | WT | 6.41 ± 0.21 | 175 ± 2 | |
| Cyp2a5-null | 6.18 ± 0.41 | 171 ± 3 | ||
| Cyp2f2-null | 6.50 ± 0.33 | 49 ± 2* | ||
| GS-A2 | WT | 5.61 ± 0.91 | 194 ± 6 | |
| Cyp2a5-null | 5.56 ± 0.67 | 193 ± 6 | ||
| Cyp2f2-null | 12.8 ± 1.7* | 77 ± 5* |
P < 0.01, compared with WT mice (n = 3).
TABLE 2.
Apparent kinetic parameters for the formation of 3MI metabolites by lung microsomes from WT, Cyp2a5-null, and Cyp2f2-null mice
Rates of metabolite formation were determined as described under Materials and Methods. Values presented are the means ± S.D. of data from three separate microsomal samples, each prepared from tissues pooled from five mice. Corresponding substrate-velocity curves are shown in supplemental material. The rates of HMI, I-3-C, and MOI formation were determined in the absence of added GSH. For HMI and GS-A2, the Km and Vmax values were estimated from relative rates determined in arbitrary units.
| Metabolite | Strain | Km | Vmax | Vmax/Km |
|---|---|---|---|---|
| μM | nmol · min−1 · mg protein−1 | ml · min−1 · mg−1 | ||
| I-3-C | WT | 8.76 ± 1.30 | 0.050 ± 0.003 | 0.006 |
| Cyp2a5-null | 8.12 ± 1.40 | 0.050 ± 0.002 | 0.006 | |
| Cyp2f2-null | 8.07 ± 0.26 | 0.015 ± 0.001* | 0.002 | |
| MOI | WT | 32.7 ± 1.0 | 0.102 ± 0.003 | 0.003 |
| Cyp2a5-null | 32.8 ± 1.0 | 0.105 ± 0.002 | 0.003 | |
| Cyp2f2-null | 62.8 ± 7.2* | 0.107 ± 0.006 | 0.002 | |
| GS-A1 | WT | 11.7 ± 2.6 | 0.459 ± 0.026 | 0.039 |
| Cyp2a5-null | 11.6 ± 2.2 | 0.480 ± 0.010 | 0.042 | |
| Cyp2f2-null | 24.1 ± 4.3* | 0.112 ± 0.009* | 0.005 | |
| Arbitrary units (analyte peak area · internal standard peak area−1 · min−1 · mg protein−1) | ||||
| HMI | WT | 7.32 ± 1.3 | 107 ± 6 | |
| Cyp2a5-null | 7.10 ± 0.27 | 112 ± 7 | ||
| Cyp2f2-null | 99.5 ± 4.8* | 14 ± 1* | ||
| GS-A2 | WT | 6.55 ± 0.50 | 143 ± 5 | |
| Cyp2a5-null | 6.38 ± 0.61 | 147 ± 3 | ||
| Cyp2f2-null | 71.7 ± 6.4* | 13 ± 1* |
P < 0.01, compared with WT mice (n = 3).
In WT mice, the maximal rates (Vmax) of formation of I-3-C and MOI were much higher in OM than in lung, but the maximal rates of formation of HMI, GS-A1, and GS-A2 were only slightly higher in OM than in lung. Apparent Km values for I-3-C, HMI, and GS-A2 formation were comparable between the two tissues but were 2- to 3-fold lower for OM than for lung, for MOI and GS-A1.
The roles of CYP2A5 and CYP2F2 in the bioactivation of 3MI in OM and lung microsomes were determined by comparing kinetic parameters between WT and Cyp2a5-null or Cyp2f2-null mice. For OM microsomes, loss of CYP2A5 led to a decreased Vmax value for the formation of I-3-C (∼4-fold), with no change in the apparent Km value, which indicated a major role by CYP2A5 in the formation of I-3-C, as well as the presence of other low-Km P450s in the mouse OM microsomes for this reaction. Decreases in Vmax for the formation of MOI (∼8-fold) were also seen, with concomitant increases (∼3-fold) in Km values. Thus, CYP2A5 is the major enzyme for the formation of both MOI (through the epoxidation pathway) and I-3-C (via the dehydrogenation pathway) in mouse OM microsomes. Consistent with the results from the Cyp2a5-null mice, the loss of CYP2F2 did not lead to noteworthy changes in the apparent Km or Vmax values in OM microsomal formation of I-3-C and MOI. In contrast, the loss of CYP2F2, but not CYP2A5, led to substantial decreases in the rates of formation of HMI (also from the epoxidation pathway) (Supplemental Fig. 1C), even though the apparent Km values were not changed (Table 1). Thus, CYP2F2 is the main enzyme for HMI formation in OM microsomes.
The loss of either CYP2F2 or CYP2A5 did not decrease the Vmax values for the formation of GS-A1 in OM microsomes; however, loss of CYP2F2 caused an increase (∼2-fold) in Km value, suggesting that CYP2F2 is a low-Km enzyme for the formation of GS-A1. The metabolite-specific effect of CYP2A5 loss on the formation of I-3-C suggested that CYP2A5-catalyzed 3MI dehydrogenation favors the production of I-3-C, via the 3MI radical, over the production of GS-A1, via MEI (Fig. 1). The opposite was true for CYP2F2, which seemed to favor MEI production and GS-A1 formation.
Loss of CYP2F2, but not CYP2A5, also decreased the rates for the formation of GS-A2 in mouse OM microsomes (Supplemental Fig. 1E), although only relative activities, rather than actual rates, were determined. The results suggested that CYP2A5-catalyzed epoxidation of 3MI favors production of MOI, rather than HMI and GS-A2, whereas CYP2F2-catalyzed epoxidation of 3MI leads primarily to formation of HMI and GS-A2, but not MOI. In addition, it can be concluded that in mouse OM microsomes, both CYP2A5 and CYP2F2 are involved in 3MI metabolism through both dehydrogenation and epoxidation pathways.
For lung microsomes, loss of CYP2A5 did not lead to any notable changes in the formation of any of the metabolites analyzed (Table 2). In contrast, loss of CYP2F2 led to decreased Vmax for the formation of I-3-C (∼3-fold), with no change in the apparent Km, as well as decreased Vmax (∼4-fold) and increased Km (∼2-fold) for the formation of GS-A1. Thus, CYP2F2 is the major enzyme for the formation of both GS-A1 and I-3-C in mouse lung microsomes through the dehydrogenation pathway. The loss of CYP2F2 also led to increased Km for the formation of MOI (∼2-fold) in lung microsomes, as well as a near total loss of HMI formation and GS-A2 formation, with dramatic increases in Km (>10-fold). The results demonstrated that in mouse lung microsomes, CYP2F2, but not CYP2A5, is the major P450 for the bioactivation of 3MI in both epoxidation and dehydrogenation pathways, which implies that CYP2F2 is essential for mediating 3MI-induced lung toxicity in mice.
As was found for CYP2F2 activities in OM microsomes, CYP2F2 in lung microsomes also favors production of iminium ions and GSH adducts, rather than MOI and I-3-C. Thus, though both CYP2A5 and CYP2F2 can metabolize 3MI through either epoxidation or dehydrogenation pathway, the end products are enzyme-specific. This result may indicate differences at the active site of the two enzymes, which dictate the fate of the proximate reactive intermediates (3MI radical and 2,3-epoxy-3MI).
The finding that CYP2F2 favors the formation of the reactive iminium ions, whereas CYP2A5 favors the transition directly to more stable species, I-3-C and MOI (Fig. 1), has interesting implications. The information is valuable for subsequent toxicity studies, because it will allow dissection of the specific role of MOI in 3MI toxicity, to be revealed by comparing WT and Cyp2a5-null mice, versus contributions by the iminium ions and the associated GSH depletion and/or protein adduction, to be revealed by comparing WT and Cyp2f2-null mice. It can also be predicted that loss of CYP2A5 may not affect the toxicity of 3MI in either OM or lung, but the loss of CYP2F2 will, at least in the lung. Although I-3-C and MOI, the preferred metabolites by CYP2A5, are potentially toxic, their toxicity requires further bioactivation; whereas the iminium ions preferentially formed by CYP2F2 can directly react with GSH or protein thiol. On the other hand, the metabolism of 3MI by the abundant CYP2A5 in the OM might divert 3MI away from CYP2F2, thus serving as a protective mechanism.
Finally, CYP2A5 seems to differ from human CYP2A13 in that although CYP2A5 does not produce much GS-A1, CYP2A13 is highly efficient in that reaction (D'Agostino et al., 2009). Thus, CYP2A5 may not be a good model for CYP2A13 for predicting consequences of CYP2A-mediated 3MI metabolism in vivo. Likewise, CYP2F2 also differs from human CYP2F1, which is active in catalyzing only the dehydrogenation of 3MI (Lanza et al., 1999), in that CYP2F2 catalyzes both dehydrogenation and epoxidation. In that regard, direct studies of CYP2A13 and CYP2F1 in a CYP2A13- and/or CYP2F1-humanized mouse model would be very useful.
In summary, 3MI bioactivation in mouse OM and lung microsomes showed differential participation of P450 enzymes and unique metabolite profiles. In OM microsomes, CYP2A5 was critical for the formation of both MOI and I-3-C, whereas CYP2F2 was critical for the formation of HMI and the GSH conjugates. However, in lung microsomes, CYP2F2, but not CYP2A5, is the critical enzyme. Although both CYP2A5 and CYP2F2 can metabolize 3MI through either epoxidation or dehydrogenation, the end products are enzyme-specific; CYP2F2 favors the transition of the proximate intermediates to reactive iminium ions, which form conjugates with GSH, whereas CYP2A5 favors the transition directly to more stable species, I-3-C and MOI. Our findings provide the basis for understanding the respective roles of CYP2A5 and CYP2F2 in 3MI's respiratory toxicity in mice and, ultimately, for predicting the toxic consequences of 3MI metabolism by human CYP2A13 and CYP2F1 in exposed individuals.
Supplementary Material
Acknowledgment
We thank Weizhu Yang for technical assistance.
This work was supported by the National Institutes of Health National Institute of Environmental Health Science [Grant ES007462]; the National Institutes of Health National Cancer Institute [Grant CA092596] (to X.D.); the National Institutes of Health National Heart, Lung, and Blood Institute [Grant HL13645]; and the National Institutes of Health National Institute of General Medical Sciences [Grant GM074249] (to G.S.Y.).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- 3MI
- 3-methylindole
- OM
- olfactory mucosa
- P450
- cytochrome P450
- MEI
- 3-methyleneindolenine
- HMI
- 3-hydroxy-3-methyleneindolenine
- MOI
- 3-methyloxindole
- I-3-C
- indole-3-carbinol
- WT
- wild type
- GS-A1
- 3-glutathionyl-S-methylindole (GSH adduct 1)
- APAP-GSH
- acetaminophen-GSH
- GS-A2
- 3-methyl-2-glutathionyl-S-indole (GSH adduct 2)
- NNK-N-oxide
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-N-oxide
- LC-MS/MS
- liquid chromatography/tandem mass spectrometry
- MIM
- multiple ion monitoring.
Authorship Contributions
Participated in research design: Zhou, D'Agostino, Yost, and Ding.
Conducted experiments: Zhou and Li.
Contributed new reagents or analytic tools: Moore and Yost.
Performed data analysis: Zhou and Ding.
Wrote or contributed to the writing of the manuscript: Zhou, Yost, and Ding.
References
- Baldwin RM, Jewell WT, Fanucchi MV, Plopper CG, Buckpitt AR. (2004) Comparison of pulmonary/nasal CYP2F expression levels in rodents and rhesus macaque. J Pharmacol Exp Ther 309:127–136 [DOI] [PubMed] [Google Scholar]
- D'Agostino J, Zhuo X, Shadid M, Morgan DG, Zhang X, Humphreys WG, Shu YZ, Yost GS, Ding X. (2009) The pneumotoxin 3-methylindole is a substrate and a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab Dispos 37:2018–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Zhang QY, Genter MB, Lipinskas TW, Negishi M, Nebert DW, Ding X. (1998) Purification and characterization of heterologously expressed mouse CYP2A5 and CYP2G1: role in metabolic activation of acetaminophen and 2,6-dichlorobenzonitrile in mouse olfactory mucosal microsomes. J Pharmacol Exp Ther 285:1287–1295 [PubMed] [Google Scholar]
- Lanza DL, Code E, Crespi CL, Gonzalez FJ, Yost GS. (1999) Specific dehydrogenation of 3-methylindole and epoxidation of naphthalene by recombinant human CYP2F1 expressed in lymphoblastoid cells. Drug Metab Dispos 27:798–803 [PubMed] [Google Scholar]
- Lanza DL, Yost GS. (2001) Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome p450 enzymes. Drug Metab Dispos 29:950–953 [PubMed] [Google Scholar]
- Li L, Wei Y, Van Winkle L, Zhang QY, Zhou X, Hu J, Xie F, Kluetzman K, Ding X. (2011) Generation and characterization of a Cyp2f2-null mouse and studies on the role of CYP2F2 in naphthalene-induced toxicity in the lung and nasal olfactory mucosa. J Pharmacol Exp Ther 339:62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, O'Bryan MA. (2003) Ultrastructural changes and olfactory deficits during 3-methylindole-induced olfactory mucosal necrosis and repair in mice. Ultrastruct Pathol 27:13–21 [DOI] [PubMed] [Google Scholar]
- Nichols WK, Mehta R, Skordos K, Macé K, Pfeifer AM, Carr BA, Minko T, Burchiel SW, Yost GS. (2003) 3-methylindole-induced toxicity to human bronchial epithelial cell lines. Toxicol Sci 71:229–236 [DOI] [PubMed] [Google Scholar]
- Regal KA, Laws GM, Yuan C, Yost GS, Skiles GL. (2001) Detection and characterization of DNA adducts of 3-methylindole. Chem Res Toxicol 14:1014–1024 [DOI] [PubMed] [Google Scholar]
- Ruangyuttikarn W, Appleton ML, Yost GS. (1991) Metabolism of 3-methylindole in human tissues. Drug Metab Dispos 19:977–984 [PubMed] [Google Scholar]
- Shu YZ, Johnson BM, Yang TJ. (2008) Role of biotransformation studies in minimizing metabolism-related liabilities in drug discovery. AAPS J 10:178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiles GL, Yost GS. (1996) Mechanistic studies on the cytochrome P450-catalyzed dehydrogenation of 3-methylindole. Chem Res Toxicol 9:291–297 [DOI] [PubMed] [Google Scholar]
- Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, Ding X. (2000) Human cytochrome P450 CYP2A13: predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res 60:5074–5079 [PubMed] [Google Scholar]
- Su T, Ding X. (2004) Regulation of the cytochrome P450 2A genes. Toxicol Appl Pharmacol 199:285–294 [DOI] [PubMed] [Google Scholar]
- Thornton-Manning J, Appleton ML, Gonzalez FJ, Yost GS. (1996) Metabolism of 3-methylindole by vaccinia-expressed P450 enzymes: correlation of 3-methyleneindolenine formation and protein-binding. J Pharmacol Exp Ther 276:21–29 [PubMed] [Google Scholar]
- Thornton-Manning JR, Ruangyuttikarn W, Gonzalez FJ, Yost GS. (1991) Metabolic activation of the pneumotoxin, 3-methylindole, by vaccinia-expressed cytochrome P450s. Biochem Biophys Res Commun 181:100–107 [DOI] [PubMed] [Google Scholar]
- Turk MA, Flory W, Henk WG. (1984) Dose response in 3-methylindole-induced bronchiolar epithelial necrosis in mice. Res Commun Chem Pathol Pharmacol 46:351–362 [PubMed] [Google Scholar]
- Turk MA, Flory W, Henk WG. (1986) Chemical modulation of 3-methylindole toxicosis in mice: effect on bronchiolar and olfactory mucosal injury. Vet Pathol 23:563–570 [DOI] [PubMed] [Google Scholar]
- Turk MA, Henk WG, Flory W. (1987) 3-Methylindole-induced nasal mucosal damage in mice. Vet Pathol 24:400–403 [DOI] [PubMed] [Google Scholar]
- Weems JM, Cutler NS, Moore C, Nichols WK, Martin D, Makin E, Lamb JG, Yost GS. (2009) 3-Methylindole is mutagenic and a possible pulmonary carcinogen. Toxicol Sci 112:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weems JM, Lamb JG, D'Agostino J, Ding X, Yost GS. (2010) Potent mutagenicity of 3-methylindole requires pulmonary cytochrome P450-mediated bioactivation: a comparison to the prototype cigarette smoke mutagens B(a)P and NNK. Chem Res Toxicol 23:1682–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Zhou X, Behr M, Fang C, Horii Y, Gu J, Kannan K, Ding X. (2010) Mechanisms of olfactory toxicity of the herbicide 2,6-dichlorobenzonitrile: essential roles of CYP2A5 and target-tissue metabolic activation. Toxicol Appl Pharmacol 249:101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Ma L, Humphreys WG, Zhu M. (2008) Rapid screening and characterization of drug metabolites using a multiple ion monitoring-dependent MS/MS acquisition method on a hybrid triple quadrupole-linear ion trap mass spectrometer. J Mass Spectrom 43:1364–1375 [DOI] [PubMed] [Google Scholar]
- Yost GS. (1989) Mechanisms of 3-methylindole pneumotoxicity. Chem Res Toxicol 2:273–279 [DOI] [PubMed] [Google Scholar]
- Yost GS. (2001) Bioactivation of toxicants by cytochrome p450-mediated dehydrogenation mechanisms. Adv Exp Med Biol 500:53–62 [DOI] [PubMed] [Google Scholar]
- Zhang QY, Ding X. (2008) The CYP2F, CYP2G and CYP2J subfamilies, in Cytochromes P450: Role in the Metabolism and Toxicity of Drugs and Other Xenobiotics (Ioannides C, ed) pp 309–353, RSC Publishing, Cambridge, UK [Google Scholar]
- Zhou X, Zhang X, Weng Y, Fang C, Kaminsky L, Ding X. (2009) High abundance of testosterone and salivary androgen-binding protein in the lateral nasal gland of male mice. J Steroid Biochem Mol Biol 117:81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhuo X, Xie F, Kluetzman K, Shu YZ, Humphreys WG, Ding X. (2010) Role of CYP2A5 in the clearance of nicotine and cotinine: insights from studies on a Cyp2a5-null mouse model. J Pharmacol Exp Ther 332:578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wei Y, Xie F, Laukaitis CM, Karn RC, Kluetzman K, Gu J, Zhang QY, Roberts DW, Ding X. (2011) A novel defensive mechanism against acetaminophen toxicity in the mouse lateral nasal gland: role of CYP2A5-mediated regulation of testosterone homeostasis and salivary androgen-binding protein expression. Mol Pharmacol 79:710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.