Abstract
Ginger has received extensive attention because of its antioxidant, anti-inflammatory, and antitumor activities. However, the metabolic fate of its major components is still unclear. In the present study, the metabolism of [6]-shogaol, one of the major active components in ginger, was examined for the first time in mice and in cancer cells. Thirteen metabolites were detected and identified, seven of which were purified from fecal samples collected from [6]-shogaol-treated mice. Their structures were elucidated as 1-(4′-hydroxy-3′-methoxyphenyl)-4-decen-3-ol (M6), 5-methoxy-1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one (M7), 3′,4′-dihydroxyphenyl-decan-3-one (M8), 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (M9), 5-methylthio-1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one (M10), 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one (M11), and 5-methylthio-1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (M12) on the basis of detailed analysis of their 1H, 13C, and two-dimensional NMR data. The rest of the metabolites were identified as 5-cysteinyl-M6 (M1), 5-cysteinyl-[6]-shogaol (M2), 5-cysteinylglycinyl-M6 (M3), 5-N-acetylcysteinyl-M6 (M4), 5-N-acetylcysteinyl-[6]-shogaol (M5), and 5-glutathiol-[6]-shogaol (M13) by analysis of the MSn (n = 1–3) spectra and comparison to authentic standards. Among the metabolites, M1 through M5, M10, M12, and M13 were identified as the thiol conjugates of [6]-shogaol and its metabolite M6. M9 and M11 were identified as the major metabolites in four different cancer cell lines (HCT-116, HT-29, H-1299, and CL-13), and M13 was detected as a major metabolite in HCT-116 human colon cancer cells. We further showed that M9 and M11 are bioactive compounds that can inhibit cancer cell growth and induce apoptosis in human cancer cells. Our results suggest that 1) [6]-shogaol is extensively metabolized in these two models, 2) its metabolites are bioactive compounds, and 3) the mercapturic acid pathway is one of the major biotransformation pathways of [6]-shogaol.
Introduction
Ginger (Zingiber officinale Rosc.), a member of the Zingiberaceae family, has been cultivated for thousands of years as a spice and for medicinal purposes. Ginger has received extensive attention because of its antioxidant, anti-inflammatory, and anticancer activities (Kawai et al., 1994; Surh, 2002; Shukla and Singh, 2007; Zick et al., 2008; Wang et al., 2009). The major pharmacologically active components of ginger are gingerols and shogaols (Masada et al., 1974; Jiang et al., 2005, 2006, 2007; Yu et al., 2007). Shogaols, the dehydrated products of gingerols, are the predominant pungent constituents in dried ginger. It has been reported that shogaols are minor components in fresh ginger, and the ratio of [6]-shogaol to [6]-gingerol is approximately 1:1 in dried ginger (Govindarajan, 1982a,b; Sang et al., 2009; Wu et al., 2010).
Shogaols have gained interest because of recent discoveries revealing their higher anticancer potencies over gingerols. It is reported that [6]-, [8]-, and [10]-gingerols had little to no effect but [6]-shogaol significantly inhibited the growth of A-2780 ovarian cancer cells (Rhode et al., 2007). Kim et al. (2008) reported that [6]-shogaol exhibited much stronger growth-inhibitory effects on A-549 human lung cancer cells, SK-OV-3 human ovarian cancer cells, SKMEL-2 human skin cancer cells, and HCT-15 human colon cancer cells than [4]-, [6]-, [8]-, and [10]-gingerols. A study from our group has also demonstrated that [6]-, [8]-, and [10]-shogaols exhibited much higher antiproliferative potency than [6]-, [8]-, and [10]-gingerols against H-1299 human lung cancer cells with IC50 values of 8 μM for [6]-shogaol and 150 μM for [6]-gingerol (Sang et al., 2009). Along with our collaborators, we have reported that [6]-shogaol was more effective than [6]-gingerol in inhibiting 12-O-tetradecanoylphorbol-13-acetate-induced tumor promotion in mice (Wu et al., 2010). Furthermore, Dugasani et al. (2010) found that [6]-shogaol showed the most potent antioxidative activity with an IC50 value of approximately 8 μM, whereas [6]-, [8]-, and [10]-gingerols had IC50 values of 28, 20, and 12 μM, respectively.
The pharmacokinetics of [6]-shogaol in mice and in humans have been investigated (Zick et al., 2008, 2010; Wang et al., 2009; Asami et al., 2010; Iwabu et al., 2010). Yu et al. (2007) reported that free [6]-shogaol and glucuronidated and sulfated metabolites of [6]-shogaol were detected in the plasma with peak concentrations of 13.6 ± 6.9 ng/ml, 0.73 ± 0.54 μg/ml, and 0.047 ± 0.035 μg/ml, respectively, 1 h after oral administration of 2.0 g of ginger extracts (containing 45.04 mg of [6]-shogaol) in humans. Asami et al. (2010) used 14C-labeled [6]-shogaol and unlabeled [6]-shogaol to determine the pharmacokinetic parameters of [6]-shogaol in rats, and the results suggested that [6]-shogaol is mostly metabolized in the body and excreted as metabolites.
So far, limited data have been reported on the metabolism of [6]-shogaol. Koh and Lee (1983) noted the transformation of [6]-shogaol by Aspergillus niger to produce 1-(4′-hydroxy-3′-methoxyphenyl)-decan-10-ol-3-one and 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3,10-diol. In addition, 6-(4′-hydroxy-3′-methoxyphenyl)-4-hydroxy-hexanoic acid and homovanillic acid have been isolated from the fermentation broth of A. niger with [6]-shogaol (Takahashi et al., 1993). It has also been reported that a saturated ketone 1-(4′-hydroxy-3′-methoxyphenyl)-4-decan-3-one ([6]-paradol) was produced after in vitro incubation of [6]-shogaol in rat livers and was further reduced to an alcohol (Surh and Lee, 1992, 1994). However, the complete metabolic profile of [6]-shogaol in vivo has not been reported. Identification of [6]-shogaol metabolite structures and fully understanding their formations are essential in clarifying their bioactivities.
The objective of the present study is to elucidate the metabolic profile of [6]-shogaol in mice and in different cancer cell lines (HCT-116, HT-29, H-1299, and CL-13) and to investigate the bioactivity of the newly identified metabolites.
Materials and Methods
Materials.
[6]-Shogaol was purified from ginger extract in our laboratory (Sang et al., 2009). Sephadex LH-20, reverse-phase C18 silica gels, analytical and preparative thin-layer chromatography (TLC) plates (250- and 2000-μm thickness, 2–25-μm particle size), and CDCl3 were purchased from Sigma-Aldrich (St. Louis, MO). High-performance liquid chromatography (HPLC)-grade solvents and other reagents were obtained from VWR Scientific (South Plainfield, NJ). Liquid chromatography/mass spectrometry (LC/MS)-grade MeOH and water were obtained from Thermo Fisher Scientific (Waltham, MA). HCT-116 and HT-29 human colon cancer cells, H-1299 human lung cancer cells, and CL-13 mouse lung cancer cells were obtained from the American Type Culture Collection (Manassas, VA). McCoy's 5A medium was purchased from Mediatech (Herndon, VA). Proteinase K was obtained from Ambion (Austin, TX). Apoptag Plus Peroxydase In Situ Apoptosis Detection Kit was purchased from Millipore Corporation (Billerica, MA).
Treatment of Mice and Sample Collections.
Experiments with mice were performed according to protocols approved by the Institutional Review Board for the Animal Care and Facilities Committee at North Carolina Research Campus or North Carolina Central University. Female C57BL/6J mice and female A/J mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and were allowed to acclimate for at least 1 week before the start of the experiment. Mice were housed five per cage and maintained in air-conditioned quarters with a room temperature of 20 ± 2°C, relative humidity of 50 ± 10%, and an alternating 12-h light/dark cycle. Mice were fed Purina Rodent Chow number 5001 (Research Diets; Purina, St. Louis, MO) and water and were allowed to eat and drink ad libitum. In experiment 1, 24-h urinary and fecal samples were collected using metabolic cages for metabolic profile analysis. In brief, [6]-shogaol in corn oil or corn oil only was administered to C57BL/6J mice by oral gavage (200 mg/kg). Fecal and urinary samples were collected in metabolic cages (five mice per cage) for 24 h after administration of vehicle (control group, n = 5) or [6]-shogaol (treated group, n = 5). In experiment 2, A/J mice were administrated [6]-shogaol by oral gavage (200 mg/kg per day) for 10 days. Fecal samples were collected from mouse cages every 5 days. The combined fecal samples were used to purify the major metabolites of [6]-shogaol. These samples were stored at −80°C before analysis. In experiment 3, A/J mice were treated with either 200 mg/kg [6]-shogaol in corn oil or corn oil only by oral gavage. Blood was collected from anesthetized mice by cardiac puncture at 2 or 6 h after administration of vehicle or [6]-shogaol (five mice per time point), and plasma was isolated by centrifugation at 5000 rpm for 15 min in a refrigerated centrifuge. Plasma samples were then stored at −80°C until analysis.
Fecal, Urinary, and Plasma Sample Preparation.
For acquisition of the metabolic profile, six pieces of each fecal sample (control and treated) were chosen and put into 2-ml tubes. Each set was weighted (control, 128 mg; treated, 130 mg), and 1.2 ml of MeOH/H2O (50/50) + 0.1% acetic acid was added to each sample. Samples were sonicated for 90 min and then centrifuged at 17,000 rpm for 10 min. The supernatant (250 μl) was collected and diluted five times for analysis. Enzymatic deconjugation was performed as described previously with slight modifications (Shao et al., 2010). In brief, 250 μl of supernatant were dried under reduced pressure at 37°C, and the residue was resuspended in sodium phosphate buffer (50 mM, pH 6.8). Samples were then treated with β-glucuronidase (250 units) and sulfatase (3 units) for 24 h at 37°C and were extracted twice with ethyl acetate. The ethyl acetate fraction was dried under vacuum, and the solid was resuspended in 1.25 ml of 80% aqueous methanol with 0.1% acetic acid for further analysis. For preparation of the urinary and plasma samples, 50 μl from each group (control group and [6]-shogaol treated group) were added to 1.2 ml of MeOH to precipitate proteins. After centrifugation at 17,000 rpm for 10 min, the supernatants were transferred into vials for analysis. Enzymatic deconjugation of the urinary and plasma samples was performed as described above. In brief, 50 μl from each group (control group and [6]-shogaol-treated group) were treated with β-glucuronidase (250 units) and sulfatase (3 units) for 24 h at 37°C and were extracted twice with ethyl acetate. The ethyl acetate fraction was dried under vacuum, and the solid was resuspended in 1.25 ml (for urine) or 250 μl (for plasma) of 80% aqueous methanol with 0.1% acetic acid for further analysis.
Purification of the Major Mouse Fecal Metabolites of [6]-Shogaol.
The mouse feces (228.29 g) collected from experiment 2 were extracted with MeOH/H2O (50/50, 1000 ml each time) twice and then were extracted with MeOH five times (1000 ml each time). The extract was dried under reduced pressure at 37°C, and the residue (40.06 g) was dissolved in water (800 ml) and partitioned successively with ethyl acetate (5 × 500 ml) and 1-butanol (2 × 600 ml). The ethyl acetate-soluble portion (5.5 g) was subjected to a reverse-phase C18 column eluted with a MeOH/H2O gradient system (3:7, 4:6, 5:5, 6:4, 7:3, 8:2, 9:1; v/v; 800 ml for each gradient), giving 12 fractions. Fraction 7 was separated by preparative HPLC to give fractions 7a and 7b. Fraction 7a was successively separated on a preparative silica gel TLC plate (developed with CHCl3/MeOH, 100:1) and Sephadex LH-20 (eluted with EtOH) column chromatography (CC) to give M11 (17 mg). Fraction 7b was purified on a preparative silica gel TLC plate (developed with CHCl3/MeOH, 100:1) to yield two subfractions (7b1 and 7b2). Fraction 7b1 was first loaded on a preparative silica gel TLC plate (developed with n-hexane/EtOAc, 10:1) and then on Sephadex LH-20 (eluted with EtOH) CC to give M9 (0.5 mg) and M10 (0.8 mg). Fraction 7b2 was subjected to preparative HPLC to give M12 (0.6 mg). Fraction 8 was loaded on a preparative silica gel TLC plate (developed with n-hexane/EtOAc, 10:1) to give fractions 8a through 8c. Fraction 8a was subjected to preparative HPLC to give one major fraction, which was then successively separated on a preparative silica gel TLC plate (developed with CHCl3/MeOH, 100:1) and Sephadex LH-20 (eluted with EtOH) CC to give M6 (0.5 mg). Fraction 8b was subjected to Sephadex LH-20 (eluted with EtOH) CC and then preparative HPLC to give M7 (0.5 mg). Fraction 8c was first loaded on a preparative silica gel TLC plate (developed with n-hexane/EtOAc, 10:1) and then on preparative HPLC to give M8 (4.0 mg). 1H and 13C NMR data of M6 through M12 are listed in Tables 1 and 2.
TABLE 1.
1H and 13C NMR spectroscopic data of M6 through M9 and M11
| No. | M6a |
M7b |
M8b |
M9b |
M11b |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | |
| 1′ | 133.6 | 133.0 | 134.1 | 134.0 | 133.1 | |||||
| 2′ | 6.77 br s | 111.6 | 6.71 d (1.2) | 111.0 | 6.72 br s | 115.4 | 6.71 d (1.5) | 111.0 | 6.70 br s | 111.0 |
| 3′ | 147.4 | 146.3 | 143.6 | 146.4 | 146.4 | |||||
| 4′ | 144.0 | 143.9 | 141.9 | 143.7 | 143.9 | |||||
| 5′ | 6.70 br d (7.8) | 114.6 | 6.83 d (7.8) | 114.3 | 6.78 d (7.8) | 115.4 | 6.84 d (7.9) | 114.2 | 6.82 br d (7.9) | 114.3 |
| 6′ | 6.63 br d (7.8) | 120.3 | 6.69 dd (7.8, 1.2) | 120.8 | 6.61 d (7.8) | 120.5 | 6.69 dd (8.0, 1.5) | 120.9 | 6.67 br d (7.9) | 120.8 |
| 1 | 2.58 m | 31.1 | 2.85 t (7.8) | 29.2 | 2.79 t (7.2) | 29.2c | 2.74 m; 2.62 m | 31.7 | 2.84 t (7.5) | 29.5 |
| 2 | 1.73 | 39.3 | 2.76 m | 45.8 | 2.70 | 43.4 | 1.78 m; 1.71 m | 39.4 | 2.70 t (7.5) | 44.6 |
| 3 | 3.99 m | 71.6 | 209.0 | 211.5 | 3.64 m | 71.4 | 210.6 | |||
| 4 | 5.47 dd (15.4, 7.1) | 131.3 | 2.66 dd (16.2, 7.8); 2.42 dd (16.2, 4.8) | 47.6 | 2.38 t (7.4) | 43.2 | 1.48 m | 37.6 | 2.38 t (7.4) | 43.1 |
| 5 | 5.64 dt (15.4, 7.1) | 132.9 | 3.67 m | 77.1 | 1.56 m | 23.8 | 1.30 m | 29.4 | 1.56 m | 23.8 |
| 6 | 2.06 m | 31.9 | 1.49 m; 1.43 m | 33.8 | 1.31 m | 29.0c | 1.30 m | 29.4 | 1.26 m | 29.0 |
| 7 | 1.29 m | 28.8 | 1.31 m | 24.7 | 1.31 m | 29.1c | 1.44 m; 1.32 m | 25.6 | 1.26 m | 29.0 |
| 8 | 1.31 m | 31.2 | 1.31 m | 31.9 | 1.31 m | 31.6 | 1.28 m | 31.7 | 1.26 m | 31.6 |
| 9 | 1.34 m | 22.2 | 1.31 m | 22.6 | 1.31 m | 22.6 | 1.30 | 22.6 | 1.29 m | 22.5 |
| 10 | 0.92 t (7.2) | 13.1 | 0.90 t (7.2) | 13.9 | 0.89 t (7.2) | 14.1 | 0.91 t (7.1) | 14.0 | 0.89 t (7.1) | 14.0 |
| 3′-OMe | 3.84 s | 54.8 | 3.89 s | 55.9 | 3.90 s | 55.9 | 3.87 s | 55.9 | ||
| 5-OMe | 3.30 s | 56.9 | ||||||||
| 4′-OH | 5.47 s | 5.47 s | ||||||||
Data were measured in CD3OD at 600 (1H) and 150 MHz (13C).
Data were measured in CDCl3 at 600 (1H) and 150 MHz (13C). Chemical shifts (δ) are in ppm being relative to CD3OD and CDCl3.
Data can be exchanged with each other.
TABLE 2.
1H and 13C NMR spectroscopic data of [6]-shogaol, M10, M12, and synthetic 5-N-acetylcysteinyl-[6]-shogaol
| No. | [6]-Shogaola |
M10b |
M12b |
5-N-acetylcysteinyl-[6]-shogaola |
||||
|---|---|---|---|---|---|---|---|---|
| δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | δH multi (J in Hz) | δC | |
| 1′ | 132.6 | 132.9 | 134.0 | 134.0 | ||||
| 2′ | 6.80 d (1.8) | 111.8 | 6.71 d (1.5) | 111.0 | 6.73 d (2.2) | 111.0 | 6.81 d (1.8) | 111.7 |
| 3′ | 147.6 | 146.4 | 146.4 | 149.0 | ||||
| 4′ | 144.5 | 143.9 | 143.7 | 145.9 | ||||
| 5′ | 6.71 d (8.0) | 114.7 | 6.84 d (8.0) | 114.3 | 6.84 d (7.9) | 114.2 | 6.71 d (8.0) | 114.6 |
| 6′ | 6.63 dd (8.0, 1.8) | 120.4 | 6.69 dd (8.0, 1.5) | 120.8 | 6.71 dd (7.9, 2.2) | 120.9 | 6.65 dd (8.0, 1.8) | 120.2 |
| 1 | 2.88 m | 31.1 | 2.86 t (7.5) | 29.3 | 2.64 m; 2.75 m | 31.7 | 2.80 m | 27.5 |
| 2 | 2.66 m | 41.3 | 2.75 m | 45.5 | 1.79 m; 1.74 m | 39.8 | 2.74 dd (17.1, 6.3); 2.67 dd (17.1, 6.3) | 49.7 |
| 3 | 201.5 | 208.4 | 4.00 m | 68.9 | 211.2 | |||
| 4 | 6.12 br d (15.9) | 130.0 | 2.59 dd (16.6, 6.5); 2.69 dd (16.6, 6.5) | 48.5 | 1.71 m; 1.62 m | 40.6 | 2.74 m | 46.1 |
| 5 | 6.90 dt (15.9, 7.02) | 148.7 | 3.04 m | 41.6 | 2.75 m | 43.2 | 3.14 m | 42.6 |
| 6 | 2.22 m | 32.1 | 1.50 m | 34.4 | 1.61m | 34.8 | 1.51 m | 33.7 |
| 7 | 1.48 m | 27.6 | 1.42 m; 1.38 m | 26.5 | 1.44 m | 26.6 | 1.34 m | 26.6 |
| 8 | 1.34 m | 29.8 | 1.27 m | 31.6 | 1.31 m | 31.9 | 1.27 m | 30.9 |
| 9 | 1.34 m | 22.1 | 1.31 m | 22.6 | 1.31 m | 22.6 | 1.33 m | 22.2 |
| 10 | 0.93 t (7.1) | 13.0 | 0.90 t (7.1) | 14.0 | 0.91 t (7.1) | 14.1 | 0.92 | 14.4 |
| 3′-OMe | 3.84 s | 3.89 s | 55.9 | 3.90 s | 55.7 | 56.4 | ||
| 5-SMe | 2.04 s | 13.3 | 2.05 | 12.3 | ||||
| 4′-OH | 5.45 s | 5.47 s | ||||||
| 3-OH | 2.25 d (4.9) | |||||||
| 1′′ | 3.00 dd (4.7, 13.7); 2.92 dd (7.1, 13.7) | 30.4 | ||||||
| 2′′ | 4.58 dd (7.1, 4.7) | 54.6 | ||||||
| 3′′ | 173.1 | |||||||
| 4′′ | 173.1 | |||||||
| 5′′ | 2.01 s | 22.6 | ||||||
Data were measured in CD3OD at 600 (1H) and 150 MHz (13C).
Data were measured in CDCl3 at 600 (1H) and 150 MHz (13C). Chemical shifts (δ) are in ppm being relative to CD3OD and CDCl3.
Synthesis of 5-N-Acetylcysteinyl-[6]-Shogaol.
[6]-Shogaol (235 mg, 0.8 mmol) was dissolved in ethanol (40 ml) and added dropwise to a solution of N-acetylcysteine [1076 mg, 6.6 mmol in 100 ml of phosphate-buffered saline (PBS) at pH 7.4] at 37°C. After stirring for 24 h, the reaction mixture was extracted with ethyl acetate. The organic phase was then separated and dried, and the residue (520 mg) was redissolved in MeOH. The reconstituted solution was subjected to a reverse-phase C18 column and was eluted with a mobile phase of MeOH/H2O (70:30, v/v) at a flow rate of 2 ml/min. The samples were combined on the basis of the TLC analysis and were dried to obtain 240 mg (yield 64%) of final product. 1H and 13C NMR data of 5-N-acetylcysteinyl-[6]-shogaol are listed in Table 2.
Nuclear Magnetic Resonance.
1H (600 MHz), 13C (150 MHz), and all two-dimensional (2D) NMR spectra were acquired on a Bruker AVANCE 600 MHz NMR spectrometer (Brucker, Inc., Silberstreifen, Rheinstetten, Germany). Compounds were analyzed in CDCl3 or CD3OD.
HPLC Analysis.
An HPLC ESA electrochemical detector (ECD) (ESA, Chelmsford, MA) consisting of an ESA model 584 HPLC pump, an ESA model 542 autosampler, an ESA organizer, and an ESA ECD coupled with two ESA model 6210 four sensor cells was used for analyzing the metabolic profile of [6]-shogaol. A Gemini C18 column (150 × 4.6 mm, 5 μm; Phenomenex, Torrance, CA) was used for chromatographic analysis at a flow rate of 1.0 ml/min. The mobile phases consisted of solvent A (30 mM sodium phosphate buffer containing 1.75% acetonitrile and 0.125% tetrahydrofuran, pH 3.35) and solvent B (15 mM sodium phosphate buffer containing 58.5% acetonitrile and 12.5% tetrahydrofuran, pH 3.45). The gradient elution had the following profile: 20% solvent B from 0 to 3 min; 20 to 55% solvent B from 3 to 11 min; 55 to 60% solvent B from 11 to 12 min; 60 to 65% solvent B from 12 to 13 min; 65 to 100% solvent B from 13 to 40 min; 100% solvent B from 40 to 45 min; and then 20% solvent B from 45.1 to 50 min. The cells were then cleaned at a potential of 1000 mV for 1 min. The injection volume of the sample was 10 μl. The eluent was monitored by the Coulochem electrode array system (ESA) with potential settings at −100, 0, 100, 200, 300, 400, and 500 mV. Data for Fig. 1 was from the channel set at 300 mV of the Coulochem electrode array system.
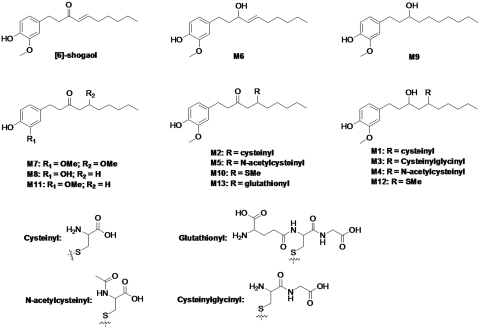
Fig. 1.
Structures of [6]-shogaol and its major metabolites.
Waters preparative HPLC system (Waters, Milford, MA) with 2545 binary gradient module, Waters 2767 sample manager, Waters 2487 autopurification flow cell, Waters fraction collector III, dual injector module, and a 2489 UV/visible detector was used to purify metabolites M6 through M8 and M12. A Gemini-NX C18 column (250 × 30.0 mm i.d., 5 μm; Phenomenex) was used with a flow rate of 20.0 ml/min, and the separation was performed with a mobile phase of MeOH/H2O. The gradient elution had the following profile: 70% solvent B from 0 to 30 min; 70 to 100% solvent B from 30 to 31 min; 100% solvent B from 31 to 36 min; 100 to 70% solvent B from 36 to 37 min; and then 70% solvent B from 37 to 42 min. The wavelength of the UV detector was set at 230 nm. Water and methanol were used as mobile phases A and B, respectively.
Liquid Chromatography/Electrospray Ionization-Mass Spectrometry Method.
LC/MS analysis was performed with a Thermo-Finnigan Spectra System, which consisted of an Accela high-speed mass spectrometry (MS) pump, an Accela refrigerated autosampler, and an LTQ Velos ion trap mass detector (Thermo Fisher Scientific) incorporated with heated electrospray ionization (H-ESI) interfaces. A Gemini C18 column (50 × 2.0 mm i.d., 3 μm; Phenomenex) was used for separation at a flow rate of 0.2 ml/min. The column was eluted from 100% solvent A (5% aqueous methanol with 0.2% acetic acid) for 3 min, followed by linear increases in solvent B (95% aqueous methanol with 0.2% acetic acid) to 40% from 3 min to 15 min, to 85% from 15 to 45 min, to 100% from 45 to 50 min, and then with 100% solvent B from 50 to 55 min. The column was then re-equilibrated with 100% solvent A for 5 min. The liquid chromatography (LC) eluent was introduced into the H-ESI interface. The positive ion polarity mode was set for the H-ESI source with the voltage on the H-ESI interface maintained at approximately 4.5 kV. Nitrogen gas was used as the sheath gas and auxiliary gas. Optimized source parameters, including ESI capillary temperature (300°C), capillary voltage (50 V), ion spray voltage (3.6 kV), sheath gas flow rate (30 units), auxiliary gas flow rate (5 units), and tube lens (120 V), were tuned using authentic [6]-shogaol. The collision-induced dissociation was conducted with an isolation width of 2 Da and normalized collision energy of 35 for MS2 and MS3. Default automated gain control target ion values were used for MS, MS2, and MS3 analyses. The mass range was measured from 50 to 1000 m/z. Data acquisition was performed with Xcalibur 2.0 version (Thermo Fisher Scientific).
Metabolism of [6]-Shogaol in Cancer Cells.
Cells (1.0 × 106) were plated in six-well culture plates and were allowed to attach for 24 h at 37°C in 5% CO2 incubator. [6]-Shogaol [in dimethyl sulfoxide (DMSO)] was added to McCoy's 5A medium (containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% glutamine) to reach a final concentration of 10 μM and was incubated with different cancer cell lines (HCT-116, HT-29, H-1299, and CL-13). At different time points (0, 30 min, 1, 2, 4, 6, 8, and 24 h), 190-μl samples of supernatant were taken and transferred to vials containing 10 μl of 0.2% ascorbic acid to stabilize [6]-shogaol and its metabolites. The metabolites were extracted from media by addition of equal volume of acetonitrile and centrifugation, in which the supernatant was harvested. The samples were then diluted 5-fold in acetonitrile and were analyzed by HPLC ECD.
Growth Inhibition of Human Cancer Cells.
Cell growth inhibition was determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (Mosmann, 1983). Human colon cancer (HCT-116) and human lung cancer (H-1299) cells (3000 cells/well) were plated in 96-well microtiter plates and were allowed to attach for 24 h at 37°C. The test compounds (in DMSO) were added to cell culture medium to desired final concentrations (0–80 μM; final DMSO concentrations for control and treatments were 0.1%). After the cells were cultured for 24 h, the medium was aspirated, and the cells were treated with 200 μl of fresh medium containing 2.41 mM MTT. After incubation for 3 h at 37°C, the medium containing MTT was aspirated, 100 μl of DMSO was added to solubilize the formazan precipitate, and the plates were shaken gently for an hour at room temperature. Absorbance values were derived from the plate reading at 550 nm on a microtiter plate reader. The reading reflected the number of viable cells and was expressed as a percentage of viable cells in the control. Both HCT-116 and H-1299 cells were cultured in McCoy's 5A medium. All of the above media were supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% glutamine, and the cells were kept in a 37°C incubator with 95% humidity and 5% CO2.
Terminal Deoxynucleotidyl Transferase Deoxyuridine Triphosphate Nick-End Labeling Assay.
HCT-116 and H-1299 cells were seeded in six-well plates at 1.0 × 105 cells/well and were incubated at 37°C in a 5% CO2 incubator. After 24 h, fresh media supplemented with DMSO (control), [6]-shogaol (10 or 20 μM), M9 (40 or 80 μM), or M11 (40 or 80 μM) were added to the wells. After 24-h incubation, cells were washed and pretreated for 15 min at room temperature with a solution of 20 μg/ml proteinase K. Cells were then washed twice with PBS pH 7.4 and were fixed for 10 min at room temperature using 10% neutral formaldehyde solution. After two washes in distilled H2O, cells were resuspended in 100 μl of distilled H2O and were applied on silanized microscope slides. Slides were incubated overnight at 37°C and were washed twice with PBS. Terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick-end labeling (TUNEL) assay was then performed according to the manufacturer's protocol. Cells were observed under 400× power using a Zeiss A1 microscope (Carl Zeiss, Inc., Thornwood, NY). Ten fields per slide were evaluated, and TUNEL-positive cells (with brown coloration in the nucleus) were expressed as a percentage of the total number of cells contained in a field.
Statistical Analysis.
For simple comparisons between two groups, two-tailed Student's t test was used. A p value of less than 0.05 was considered statistically significant in all the tests.
Results
Metabolism of [6]-Shogaol in Mice.
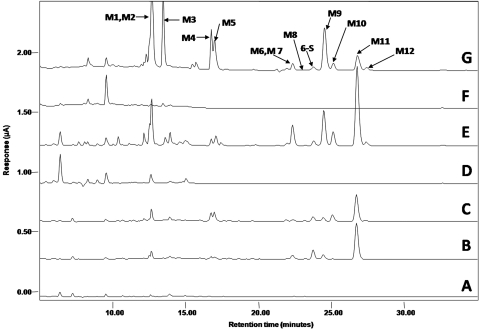
In this study, we used HPLC-ECD and LC/ESI-MS to analyze the major metabolites of [6]-shogaol in our samples. Representative HPLC chromatograms of the metabolites detected in mouse fecal, urinary, and plasmatic samples collected after the administration of 200 mg/kg of [6]-shogaol through oral gavage are shown in Fig. 2. Compared with the samples collected from control mice (Fig. 2F), 12 major metabolites (M1–M12) were observed in fecal samples collected from [6]-shogaol-treated mice (Fig. 2G). These metabolites were numbered according to their chromatographic retention times. Incubation of the fecal sample extracts with glucuronidase and sulfatase did not change the peak areas of all the metabolites (data not shown), suggesting these compounds do not exist in glucuronidated and/or sulfated forms, whereas in urinary and plasma samples, most of the metabolites were not detectable without incubation with glucuronidase and sulfatase. These results indicate the metabolites in the urine and plasma were in the glucuronidated and/or sulfated forms (data not shown). After hydrolysis, the plasma samples (Fig. 2, A–C) and urine samples (Fig. 2, D and E) showed similar metabolic profiles to those of fecal samples. This was confirmed by LC/MS analysis (data not shown).
Fig. 2.
Representative HPLC-ECD chromatograms of plasma samples from control mice (A) and mice treated with 200 mg/kg [6]-shogaol at the time points of 2 h (B) and 4 h (C); urinary samples from control mice (D) and mice treated with 200 mg/kg [6]-shogaol (E); and fecal samples from control mice (G) and mice treated with 200 mg/kg [6]-shogaol (H).
We purified seven major metabolites (M6–M12) from fecal samples collected from mice treated with 200 mg/kg [6]-shogaol using oral gavage. Their structures were elucidated on the basis of analysis of their 1H, 13C, and 2D NMR spectra. For the metabolites that we were unable to purify from mouse fecal samples (M1–M5), their structures were determined using LC/ESI tandem mass spectrometry (MS/MS) by analyzing the MSn (n = 1–3) spectra as well as by comparison with authentic standards. Among all the metabolites, M1 through M5, M10, and M12 are the thiol conjugates of [6]-shogaol and its metabolite M6. Therefore, we describe the structure elucidation of M6 through M9 and M11 first and then that of M1 through M5, M10, and M12.
Structure Elucidation of Nonthiol-Conjugated Metabolites (M6 through M9 and M11).
Metabolite M6.
M6 had the molecular formula C17H26O3 according to ESI-MS at m/z 261 [M + H − H2O]+ and its 1H and 13C NMR data. The molecular weight of M6 was 2 mass units higher than that of [6]-shogaol. In addition to the distinguishable resonance for a methoxyl group (δH 3.84, 3 H, s), the 1H NMR spectrum of M6 (Table 1) also indicated the presence of a 1,3,4-tri-substituted phenyl group [δH 6.77 (1 H, br s); 6.70 (1 H, br d, J = 7.8 Hz); and 6.63 (l H, br d, J = 7.8 Hz)], and a double bond [δH 5.47 (1 H, dd, J = 15.4, 7.1 Hz) and 5.64 (l H, dt, J = 15.4, 7.1 Hz)], and a methyl group (δH 0.92, 3 H, t, J = 7.2 Hz). Its 13C NMR spectrum (Table 1) displayed 17 carbon resonances, which were classified by heteronuclear single quantum correlation experiments as two methyls, six methylenes, six methines, and three quaternary carbons. The aforementioned NMR data implied the structure of M6 was closely related to that of [6]-shogaol. The only difference was that C-3 of M6 was assigned as an oxymethine (δH 3.99, 1 H, m; δC 71.6) instead of the expected ketone carbonyl in [6]-shogaol (δC 201.5). This was confirmed by the heteronuclear multiple-bond correlations (HMBC) (see Supplemental Fig. 1) of H-3/C-1, H-3/C-2, H-4/C-3, and H-5/C-3. Therefore, the structure of M6 was determined as shown in Fig. 1.
Metabolite M7.
M7 showed the molecular formula C18H28O4 on the basis of ESI-MS at m/z 291 [M + H − H2O]+ and its 1H and 13C NMR data. The molecular weight of M7 was 32 mass units higher than that of [6]-shogaol. Compared with [6]-shogaol, the NMR spectra of M7 gave the appearance of an oxygenated methine (δH 3.67, 1 H, m; δC 77.1), a methylene (δH 2.66, dd, J = 16.2, 7.8 Hz; 2.42, dd, J = 16.2, 4.8 Hz), and a methoxyl (δH 3.30, 3 H, s; δC 56.9) groups instead of the expected double bond at C-4 and C-5 of [6]-shogaol, which indicated that the α,β-unsaturated keto-structure of [6]-shogaol was reduced to a saturated ketone. This was further confirmed by the observation of the HMBCs (see Supplemental Fig. 1) between δH 2.66 and 2.42 (the methylene group) and C-2 (δC 45.8), C-3 (δC 209.0), and C-6 (δC 33.8), indicating that the methylene group was located at position C-4. The oxygenated methine was located at C-5 by the observation of the HMBCs between H-4 and δC 77.1. The HMBC between δH 3.30 (the methoxyl group) and C-5 (δC 77.1) suggested that the methoxyl group was directly linked with C-5. Thus, M7 was identified as shown in Fig. 1.
Metabolite M8.
M8 showed the molecular formula C16H24O3 on the basis of ESI-MS at m/z 265 [M + H]+ and its 1H and 13C NMR data. Compared with [6]-shogaol, the NMR spectra of M8 showed the disappearance of the double bond at C-4 and C-5, as well as the methoxyl group at C-3′ (Table 1), which indicated M8 was 3′, 4′-dihydroxyphenyl-decan-3-one (Fig. 1).
Metabolite M9.
M9 was obtained as a white amorphous powder. M9 was shown to have the molecular formula C17H28O3 on the basis of ESI-MS at m/z 263 [M + H− H2O]+ and its 1H and 13C NMR data. The molecular weight of M9 was 2 mass units higher than that of M6, indicating that M9 was the double-bond-reduced product of M6. This was further confirmed by the observation of the appearance of two methene groups (δH 1.48, 2 H and δC 37.6 and δH 1.30, 2 H and δC 29.4) and the disappearance of the double-bond signals in M9 (Table 1). Therefore, the structure of M9 was determined as 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (Fig. 1).
Metabolite M11.
M11 was obtained as a white amorphous powder. It showed a protonated molecular ion at m/z 279 [M + H]+, which was 2 mass units higher than that of [6]-shogaol. The 1H and 13C NMR data of M11 were very similar to those of [6]-shogaol, and the major difference was that M11 had two methene groups (δH 2.38, 2 H; δH 1.56, 2 H) (Table 1) instead of the double bond in [6]-shogaol, clearly indicating that M11 was the double-bond-reduced metabolite of [6]-shogaol. This was further confirmed by the key correlations observed in the HMBC spectrum (see Supplemental Fig. 1). Therefore, M11 was identified as 1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one, also known as [6]-paradol, which is one of the components reportedly found in ginger.
Structure Elucidation of Thiol-Conjugated Metabolites (M1–M5, M10, and M12).
Metabolite M5.
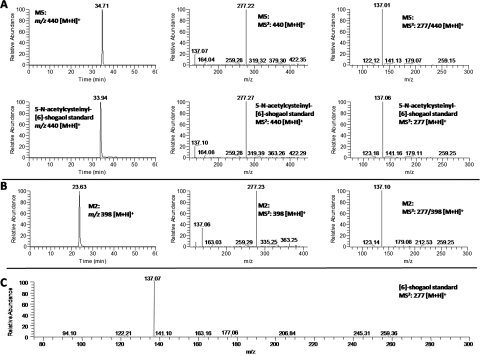
The mass spectrum of metabolite M5 exhibited [M + H]+ ions at m/z 440 in the positive mode, which was 163 mass units higher than that of [6]-shogaol, indicating that M5 was the N-acetylcysteine conjugated [6]-shogaol (molecular weight of N-acetylcysteine is m/z 163). The MS2 spectrum of M5 showed a major product ion at m/z 277 (Fig. 3A). The MS3 spectrum of this product ion had the same fragment ions as those of the authentic [6]-shogaol (Fig. 3, A and C), which indicated M5 was an N-acetylcysteine conjugate of [6]-shogaol. To further elucidate the structure of M5, it was synthesized by reacting N-acetylcysteine with 6-shogaol. The structure of the synthesized N-acetylcysteine conjugate (5-N-acetylcysteinyl-[6]-shogaol) was determined using its 1H, 13C, and 2D NMR data. The 1H and 13C NMR spectra showed very similar patterns to those of [6]-shogaol (Table 2). Compared with the 1H NMR spectrum of [6]-shogaol, the major differences were the appearance of a methine (δH 2.74, m, 2 H) and a methylene (δH 3.14, m, 1 H) group in 5-N-acetylcysteinyl-[6]-shogaol in lieu of the expected double bond of [6]-shogaol, as well as four additional proton signals for a N-acetylcysteine group (δH 3.00 dd and 2.92 dd, H-1′′; δH 4.58 dd, H-2′′; and δH 2.01 s, H-5′′). The major differences between the 13C spectra of 5-N-acetylcysteinyl-[6]-shogaol and [6]-shogaol were the presence of carbons observed at δC 46.1 (C-4) and 42.6 (C-5) instead of the double bond of [6]-shogaol, as well as the presence of five additional carbons at δC 30.4 (C-1′′), 54.6 (C-2′′), 173.1 (C-3′′ and C-4′′), and 22.6 (C-5′′) for a N-acetylcysteine group (Table 2). The HMBC spectrum of 5-N-acetylcysteinyl-[6]-shogaol had cross-peaks between H-1′′ (δH 3.00 and 2.92) and C-5 (δC 42.6), indicating that N-acetylcysteine was conjugated at C-5 of [6]-shogaol (see Supplemental Fig. 1). All of these spectral features supported the structure of 5-N-acetylcysteinyl-[6]-shogaol as shown in Fig. 1. M5 had almost the same retention time as well as the same molecular mass and fragment ion mass spectra as those of the synthetic 5-N-acetylcysteinyl-[6]-shogaol. Therefore, M5 was identified as 5-N-acetylcysteinyl-[6]-shogaol (Fig. 1).
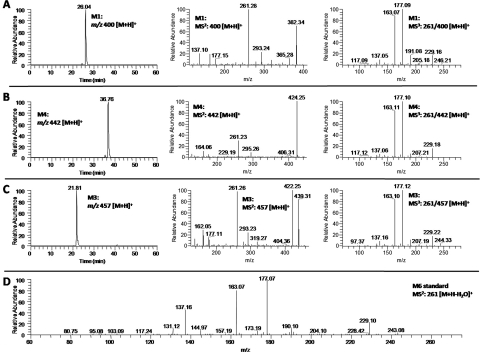
Fig. 3.
LC/MS2 and MS3 (positive) spectra of 5-N-acetyl cysteinyl-[6]-shogaol (A) and 5-cysteinyl-[6]-shogaol (B); and MS2 spectra of authentic [6]-shogaol (C).
Metabolite M2.
M2 had a molecular weight of 397 as determined by the mass ion at m/z 398 [M + H]+, which was 121 mass units higher than that of [6]-shogaol and 42 mass units lower than that of M5, indicating that M2 was the cysteine conjugated metabolite of [6]-shogaol. The major product ion of M2 showed a fragment ion at m/z 277 (Fig. 3B), and the tandem mass of this product ion was almost identical to the tandem mass of authentic [6]-shogaol (Fig. 3, B and C). All of these spectra features tentatively identified M2 as 5-cysteinyl-[6]-shogaol (Fig. 1).
Metabolites M1, M3, and M4.
M1 exhibited [M + H]+ ions at m/z 400 in the ESI-positive mode, which was 2 mass units higher than that of M2 and 121 mass units higher than that of M6, indicating that M1 was the cysteine conjugated metabolite of M6. This was confirmed by the observation of m/z 261 [M-121 − H2O + H]+ as one of the major product ions in the MS2 spectrum of M1. The tandem mass spectrum of this product ion (MS3: 261/400) was almost identical to the MS2 spectrum of authentic M6 (Fig. 4, A and D). Thus, M1 was identified as 5-cysteinyl-M6.
Fig. 4.
LC/MS2 and MS3 (positive) spectra of (A) 5-cysteinyl-M6, (B) 5-N-acetylcysteinyl-M6, and (C) 5-cysteinylglycinyl-M6, and (D) MS2 spectra of authentic M6.
M4 showed [M + H]+ ions at m/z 442 in the positive mode, which was 42 mass units higher than that of M1, 163 mass units higher than that of M6, and 2 mass units higher than that of M5, suggesting that M4 was the N-acetylcysteine conjugated metabolite of M6. Its MS2 spectrum also had product ion m/z 261 [M-121 minus] H2O + H]+, and the tandem mass spectrum of this product ion (MS3: 261/442) was almost identical to the MS2 spectrum of authentic M6 (Fig. 4, B and D). Therefore, M4 was tentatively identified as 5-N-acetylcysteinyl-M6 (Fig. 1).
M3 had a molecular weight of 456 on the basis of the observation of the [M + H]+ ions at m/z 457 in the positive mode, which was 178 mass units higher than that of M6 and 57 mass units higher than that of M1. This corresponded with the predicted molecular weight of the cysteinylglycine-conjugated metabolite of M6. Similar to that of M1 and M4, the MS3 spectrum of the product ion m/z 261 of M3 was almost identical to the MS2 spectrum of authentic M6 (Fig. 4, C and D). Thus, M3 was identified as the cysteinylglycine conjugate of M6 (Fig. 1).
Metabolite M10.
M10 had the molecular formula C18H28O3S on the basis of ESI-MS at m/z 325 [M + H]+ and its 1H and 13C NMR data, which was 48 mass units higher than that of [6]-shogaol. Compared with the NMR spectra of [6]-shogaol, the NMR spectra of M10 showed signals for a methine (δH 3.04, m, 1 H; δC 41.6), a methene (δH 2.59, dd and 2.69 dd, 2 H; δC 48.5), and a methyl (δH 2.04, 3 H, s; δC 13.3) group (Table 2) instead of the expected double bond of [6]-shogaol. The chemical shifts of the methine and methane groups were similar to those of positions 4 and 5 of M5, and the chemical shift of the methyl group was similar to that reported for the methylthiol group (Gardiner et al., 2004). All of these spectra features suggested that M10 was a methylthiol-conjugated metabolite of [6]-shogaol (Fig. 1). This was further confirmed by the observation of the cross-peak in the HMBC spectrum between δH 2.04 (the methyl group) and δC 41.6 (the methane group) (see Supplemental Fig. 1). Thus, M10 was identified as the methylthiol-conjugated [6]-shogaol (Fig. 1).
Metabolite M12.
The positive ion ESI-MS of M12 displayed a molecular ion peak at m/z 327 [M + H]+, supporting a molecular formula of C18H30O3S. The molecular weight of M12 was 2 mass units higher than that of M10, which was similar to the difference between M6 and [6]-shogaol. Compared with the NMR spectra of M10, M12 showed the signal of an oxygenated methine (δH 4.00, 1 H, m; δC 68.9) in lieu of the expected ketone group of M10, which indicated the ketone group at C-3 of M10 was reduced to a hydroxyl group of M12, which was further confirmed by the HMBCs of OH-3/C-3 and OH-3/C-4 (see Supplemental Fig. 1). Therefore, M12 was identified as the methylthiol-conjugated M6 (Fig. 1).
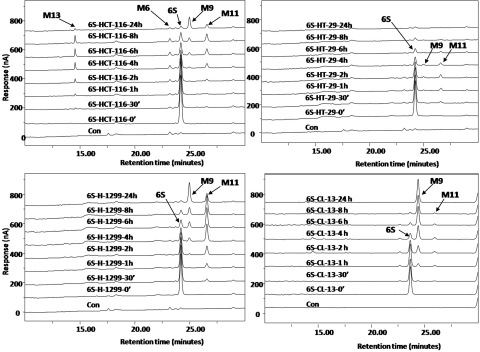
Metabolism of [6]-Shogaol in Cancer Cells.
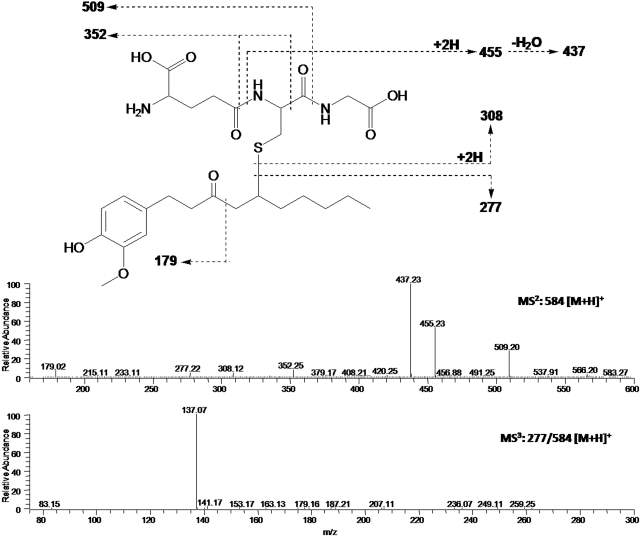
After incubation of [6]-shogaol with four different cancer cell lines (HCT-116, HT-29, H-1299, and CL-13), the culture media were collected at different time points and analyzed by HPLC-ECD. Our results indicate that [6]-shogaol was extensively metabolized in all four cancer cell lines (Fig. 5). After 24-h incubation, four major metabolites appeared in HCT-116 human colon cancer cells. Three of them were identified as M6, M9, and M11 by comparing their retention times and tandem mass fragments with those of our purified authentic standards (data not shown). The fourth metabolite (M13) was a newly revealed compound at the retention time of 14.50 min. The mass spectrum of metabolite M13 exhibited [M + H]+ ions at m/z 584 in the positive mode, which was 307 mass units higher than that of [6]-shogaol, indicating that M13 was the GSH-conjugated [6]-shogaol (molecular weight of GSH is m/z 307). Its MS2 spectrum showed product ions of m/z 277 (−307 Da, neutral loss of GSH), m/z 455 (−129 Da, neutral loss of pyroglutamic acid), m/z 437 (−147 Da, dehydrolyzation of m/z 455), and m/z 509 (−75 Da, neutral loss of glycine) (Fig. 6). The MS3 spectrum of its product ion m/z 277 was almost identical to the MS2 spectrum of authentic [6]-shogaol (Figs. 3C and 6). All of the above evidence indicates M13 is the glutathiol conjugate of [6]-shogaol (Fig. 1). Both M9 and M11 were detected as the major metabolites of [6]-shogaol in HT-29 human colon cancer cells, H-1299 human lung cancer cells, and CL-13 mouse lung cancer cells (Figs. 5). At 24 h, [6]-shogaol was almost completely converted to M9 and M11 in H-1299 cells and to M9 in CL-13 cells.
Fig. 5.
HPLC-ECD chromatograms of [6]-shogaol treated with HCT-116, HT-29, CL-13, and H-1229 cell lines.
Fig. 6.
ESI-MS2 and MS3 fragment spectra of protonated 5-glutathionyl-[6]-shogaol (M13).
M9 and M11 Inhibit the Growth of Human Cancer Cells.
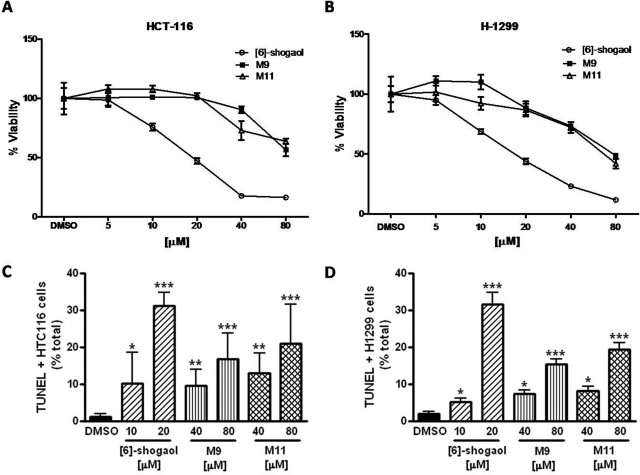
Two cancer cell lines, HCT-116 and H-1299, were treated with [6]-shogaol, M9, or M11, with concentrations ranging from 0 to 80 μM. In HCT-116 cells, [6]-shogaol exhibited the strongest inhibitory activity with an IC50 of 18.7 μM. The major metabolites M9 and M11 had decreasing potencies of 82.2 and 84.0 μM, respectively. In H-1299 cells, the IC50 values for [6]-shogaol, M9, and M11 were 16.9, 77.7, and 66.5 μM, respectively. These data demonstrate that [6]-shogaol has the greatest inhibitory activity against cancer cell lines but still shows some efficacy after metabolic biotransformation.
M9 and M11 Trigger Apoptosis in Human Cancer Cells.
Apoptosis, or programmed cell death, is a major mechanism of regulation allowing cells to undergo cell death upon activation of specific external and/or internal pathways. We investigated the role of [6]-shogaol, M9, and M11 on the induction of apoptosis in human cancer cells using the TUNEL assay, which detects breaks of DNA strands in early and late apoptotic cells. In HCT-116 and H-1299 cells, exposure to 10 μM [6]-shogaol yielded 10.3 and 5.2% of apoptotic cells, respectively, whereas 20 μM [6]-shogaol yielded 31.2 and 31.6% (Fig. 7, C and D). Exposure to 40 μM metabolite M9 for 24 h led to the observation of 9.6 and 7.4% of apoptotic cells, respectively (16.9 and 15.4% for the 80 μM dose, respectively) (Fig. 7, C and D). Exposure to 40 μM metabolite M11 led to the observation of 12.9 and 8.3% of apoptotic cells in HTC-116 and H-1299 cancer cells (21.1 and 19.4% for the 80 μM dose), respectively) (Fig. 7, C and D). All of these results are significantly different from the DMSO control. Overall, these results show that M9 and M11 are bioactive compounds and can specifically trigger apoptosis in both human colon and lung cancer cells but are not as efficient as [6]-shogaol.
Fig. 7.
Growth inhibitory effects of [6]-shogaol, M9, or M11 on HCT-116 (A) and H-1299 (B) cells; and effects of [6]-shogaol, M9, or M11 on the induction of apoptosis in HCT-116 (C) and H-1299 (D) cells. MTT assay was used to measure the growth inhibitory effect and each value in A and B represents the mean ± S.D. (n = 6). TUNEL assay was used to measure the induction of apoptosis and each value in C and D represents the mean ± S.E. (n = 10). TUNEL-positive cells have been observed at 400× power. Ten fields per slide have been counted and averaged. Significantly different from DMSO control according to the two-tailed Student's t test (*, p < 0.05, **, p < 0.001, and ***, p < 0.0001).
Discussion
The possible cancer-preventive activity of ginger is receiving a great deal of attention. Information on the metabolism of ginger components such as [6]-shogaol is important for understanding the biological effects of ginger. The mouse and cancer cells are frequently used as experimental models to study the cancer-preventive effects of ginger and its bioactive components. Our results indicate that [6]-shogaol is extensively metabolized in mice and in cancer cells. In the present study, 13 metabolites were identified, with 12 in mice and 4 in cancer cells (Fig. 1).
Reduction of xenobiotic carbonyls is a significant metabolic route to produce more hydrophilic and often less toxic compounds, which can be substrates for phase II conjugation by UDP-glucuronosyltransferases or sulfotransferases, leading ultimately to excretion of the products (Oppermann, 2007). In this investigation, reduced metabolites were formed in which M11 is the double-bond-reduced metabolite of [6]-shogaol, and M9 and M6 are ketone group-reduced metabolites of M11 and [6]-shogaol, respectively. It has been reported that α,β-unsaturated ketones can be rapidly reduced to saturated ketones by leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase (LTB4 12-HD/PGR) (Dick et al., 2001; Itoh et al., 2008). Ketones can be further reduced by carbonyl-reducing enzymes, which are grouped into two large protein superfamilies: the aldo-keto reductases and the short-chain dehydrogenases/reductases (Oppermann, 2007). It is worthwhile to further investigate whether LTB4 12-HD/PGR can reduce the double bond of [6]-shogaol to generate M11 and whether aldo-keto reductases and/or short-chain dehydrogenases/reductases can reduce [6]-shogaol and M11 to form M6 and M9, respectively.
M8, an O-demethylated metabolite of M11, was observed in our study. Cytochrome P450 isozymes are reported to be responsible for O-demethylad reactions (Honda et al., 2011). For instance, 5,7-dimethoxyflavone was reported metabolized primarily to 5-methoxy-7-hydroxyflavone by recombinant CYP1A1 (Tsuji et al., 2006). In addition, tangeretin, a major flavonoid in citrus fruits, was purportedly metabolized to two O-demethylated metabolites by CYP1A2 and CYP3A4 (Breinholt et al., 2003). Thus, the roles that individual cytochrome P450 enzymes may play in mediating the formation of M8 need to be addressed in further detail. Because of the lack of data on metabolic pathways similar to that of M7, we are unable to discuss the formation of this metabolite in the current study.
The mercapturic acid pathway has been reported as one of the major routes to metabolize endogenous and xenobiotic electrophiles, such as 4-hydroxy-2-nonenal, an α,β-unsaturated aldehyde generated from lipid peroxidation, and sulforphane (SFN), a naturally occurring isothiocyanate present in cruciferous vegetables (Kassahun et al., 1997; Hayes and McLellan, 1999; Falletti et al., 2007; Pernice et al., 2009; Ahn et al., 2010; Rudd et al., 2011). The α,β-unsaturated keto group of [6]-shogaol makes it a good substrate for thiol conjugation. In the present study, we identified eight thiol conjugates of [6]-shogaol (Fig. 1). Metabolites M10 and M12 were isolated from mouse fecal samples and were identified by their NMR data. Metabolites M1 through M5 and M13 were identified by MS/MS. The structure of metabolite M5 was further confirmed by comparison of the retention time and spectral data with those of a synthesized reference substance (5-N-acetylcysteinyl-[6]-shogaol) using LC/MS/MS. Our results clearly indicate that [6]-shogaol is mainly metabolized through the mercapturic acid pathway. Initial conjugation with GSH promoted by glutathione transferase gives rise to the corresponding conjugate, and the GSH conjugate undergoes further enzymatic modification: first modification by γ-glutamyltranspeptidase to form the cysteinylglycine conjugate; then alteration by cysteinyl-glycine dipeptidase or aminopeptidase M to form the cysteine conjugate; and finally conversion by N-acetyltransferase to form the N-acetylcysteine conjugate (Knapen et al., 1999). Then, both the cysteine and the N-acetylcysteine conjugates act as substrates of cysteine S-conjugate β-lyase, a mainly renal and hepatic enzyme that cleaves the S-C bond in the cysteinyl moiety, thus liberating a thiolated metabolite, which can be further S-methylated by thiol S-methyltransferase to form 5-methylthio-1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-one (M10) or 5-methylthio-1-(4′-hydroxy-3′-methoxyphenyl)-decan-3-ol (M12) (Ferroni et al., 1996; Kishida et al., 2001).
Electrophiles in foods have attracted great attention because of their protection against toxicity and many chronic pathological conditions (Nakamura and Miyoshi, 2010). Numerous studies have found that dietary electrophiles can activate transcription factor nuclear factor-E2-related factor 2 (Nrf2) through modifying cysteine residues in Kelch-like ECH-associated protein 1 (KEAP1) and therefore stimulating the overproduction of GSH to detoxify electrophiles as well as carcinogens (Juge et al., 2007; Higgins et al., 2009; MacLeod et al., 2009; Nakamura and Miyoshi, 2010). It has been reported that the level of GSH in mouse embryonic fibroblast cells were reduced to approximately 25% of the basal level, returned to the basal level, and rose between 1.75- and 1.9-fold higher than the basal level at 2 to 4, 8 to 12, and 18 and 24 h after treatment with 3 μM SFN, respectively (Higgins et al., 2009). The initial rapid depletion of GSH observed within the first 4 h of treatment with SFN was due to formation of a dithiocarbamate between SFN and GSH, and the overproduction of GSH 18 to 24 h after treatment with SFN was due to the activation of Nrf2-Keap1 pathway by SFN (Higgins et al., 2009). Whether [6]-shogaol can stimulate the overproduction of GSH by activating the Nrf2-Keap1 pathway is a topic for future study. In addition, studies have found that electrophiles have harmful effects at high doses (Nakamura and Miyoshi, 2010). Further studies on the optimized effective doses of ginger extract and its active components are necessary. More attention should be paid to the dose administrated as a supplement of a condensed ginger extract.
We studied the metabolism of [6]-shogaol in HCT-116 and HT-29 human colon cancer cells, H-1299 human lung cancer cells, and CL-13 mouse lung cancer cells (Fig. 5). Our results show that [6]-shogaol in cancer cells has a similar metabolic pathway as that in mice. We detected 5-glutathionyl-[6]-shogaol in treated HCT-116 cells, which gave further evidence to the existence of the mercapturic acid pathway, as deduced in the mouse study. However, secondary metabolites such as cysteinyl, N-acetylcysteinyl, and cysteinylglycinyl conjugates were not observed in the cancer cell lines likely because of the absence of the enzymes that lead to the loss of the individual amino acids from the GSH conjugate of [6]-shogaol. Over time, it seemed that the double-bond-reduced product (M11) was formed and the ketone group of M11 was further reduced to form M9. At 24 h, [6]-shogaol was almost completely converted to M9 and M11 in HCT-116 and H-1299 cells and to M9 in CL-13 cells. This result prompted us to investigate whether M9 and M11 retained the biological effects of [6]-shogaol. Our results indicate that M9 and M11 both exhibit measureable antiproliferative activity in HCT-116 and H-1299 cancer cells, albeit with less potency than [6]-shogaol (Fig. 7, A and B). In addition, we demonstrate that M9 and M11 are capable of triggering apoptosis in human colon and lung cancer cells (Fig. 7, C and D). We noted that [6]-shogaol demonstrated a superior apoptotic effect, so M9 and M11 are at least partially implicated in the stimulation of apoptosis. These findings give evidence that [6]-shogaol continues to be somewhat pharmacologically effective after being metabolized in vitro. It is possible that other metabolites described in this study participate in the inhibition of cancer cell growth and initiation of apoptosis, in either an additive or a synergistic way, to achieve the full response. There is also the possibility that metabolites can target other metabolic pathways, ultimately resulting in cell death. This merits further consideration and presents us with new tools to identify novel molecular targets of [6]-shogaol. We are synthesizing the metabolites identified in this study and will further elucidate their anticancer and anti-inflammatory activities in future studies.
In conclusion, results from this work are important for understanding the metabolism of [6]-shogaol and related analogs in humans and provide useful information that may act as a reference for the clinical pharmacology. The knowledge of the metabolism of [6]-shogaol may help in understanding the mechanism of action and therapeutic effects of [6]-shogaol as well as ginger extract.
Supplementary Material
Acknowledgment
We acknowledge Kevin Knagge of David H. Murdock Research Institute for technical assistance in collecting NMR spectra.
This work was supported by the National Institutes of Health National Cancer Institute [Grant CA138277]; and the National Institutes of Health National Cancer Institute and Office of Dietary Supplements [Grant CA138277S1].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- TLC
- thin-layer chromatography
- HPLC
- high-performance liquid chromatography
- LC/MS
- liquid chromatography/mass spectrometry
- LC/MS/MS
- liquid chromatography/tandem mass spectrometry
- MS
- mass spectrometry
- ESI
- electrospray ionization
- ESI-MS
- electrospray ionization-mass spectrometry
- CC
- column chromatography
- PBS
- phosphate-buffered saline
- H-ESI
- heated electrospray ionization
- 2D
- two dimensional
- ECD
- electrochemical detector
- DMSO
- dimethyl sulfoxide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TUNEL
- terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick-end labeling
- HMBC
- heteronuclear multiple-bond correlation
- LTB4 12-HD/PGR
- leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase
- SFN
- sulforphane
- Nrf2
- nuclear factor-E2-related factor 2
- KEAP1
- Kelch-like ECH-associated protein 1.
Authorship Contributions
Participated in research design: Sang.
Conducted experiments: H. Chen, Lv, Soroka, Warin, Parks, Hu, Zhu, and X. Chen.
Contributed new reagents or analytic tools: H. Chen, Lv, and Sang.
Performed data analysis: H. Chen and Sang.
Wrote or contributed to the writing of the manuscript: H. Chen, Soroka, Warin, and Sang.
References
- Ahn YH, Hwang Y, Liu H, Wang XJ, Zhang Y, Stephenson KK, Boronina TN, Cole RN, Dinkova-Kostova AT, Talalay P, et al. (2010) Electrophilic tuning of the chemoprotective natural product sulforaphane. Proc Natl Acad Sci USA 107:9590–9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami A, Shimada T, Mizuhara Y, Asano T, Takeda S, Aburada T, Miyamoto K, Aburada M. (2010) Pharmacokinetics of [6]-shogaol, a pungent ingredient of Zingiber officinale Roscoe (Part I). J Nat Med 64:281–287 [DOI] [PubMed] [Google Scholar]
- Breinholt VM, Rasmussen SE, Brøsen K, Friedberg TH. (2003) In vitro metabolism of genistein and tangeretin by human and murine cytochrome P450s. Pharmacol Toxicol 93:14–22 [DOI] [PubMed] [Google Scholar]
- Dick RA, Kwak MK, Sutter TR, Kensler TW. (2001) Antioxidative function and substrate specificity of NAD(P)H-dependent alkenal/one oxidoreductase. A new role for leukotriene B4 12-hydroxydehydrogenase/15-oxoprostaglandin 13-reductase. J Biol Chem 276:40803–40810 [DOI] [PubMed] [Google Scholar]
- Dugasani S, Pichika MR, Nadarajah VD, Balijepalli MK, Tandra S, Korlakunta JN. (2010) Comparative antioxidant and anti-inflammatory effects of [6]-gingerol, [8]-gingerol, [10]-gingerol and [6]-shogaol. J Ethnopharmacol 127:515–520 [DOI] [PubMed] [Google Scholar]
- Falletti O, Cadet J, Favier A, Douki T. (2007) Trapping of 4-hydroxynonenal by glutathione efficiently prevents formation of DNA adducts in human cells. Free Radic Biol Med 42:1258–1269 [DOI] [PubMed] [Google Scholar]
- Ferroni MA, Giulianotti PC, Pietrabissa A, Mosca F, Gomeni R, Pacifici GM. (1996) Captopril methylation in human liver and kidney: interindividual variability. Xenobiotica 26:877–882 [DOI] [PubMed] [Google Scholar]
- Gardiner J, Anderson KH, Downard A, Abell AD. (2004) Synthesis of cyclic beta-amino acid esters from methionine, allylglycine, and serine. J Org Chem 69:3375–3382 [DOI] [PubMed] [Google Scholar]
- Govindarajan VS. (1982a) Ginger–chemistry, technology, and quality evaluation: part 1. Crit Rev Food Sci Nutr 17:1–96 [DOI] [PubMed] [Google Scholar]
- Govindarajan VS. (1982b) Ginger-chemistry, technology, and quality evaluation: part 2. Crit Rev Food Sci Nutr 17:189–258 [DOI] [PubMed] [Google Scholar]
- Hayes JD, McLellan LI. (1999) Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res 31:273–300 [DOI] [PubMed] [Google Scholar]
- Higgins LG, Kelleher MO, Eggleston IM, Itoh K, Yamamoto M, Hayes JD. (2009) Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol Appl Pharmacol 237:267–280 [DOI] [PubMed] [Google Scholar]
- Honda M, Muroi Y, Tamaki Y, Saigusa D, Suzuki N, Tomioka Y, Matsubara Y, Oda A, Hirasawa N, Hiratsuka M. (2011) Functional characterization of CYP2B6 allelic variants in demethylation of antimalarial artemether. Drug Metab Dispos 39:1860–1865 [DOI] [PubMed] [Google Scholar]
- Itoh K, Yamamoto K, Adachi M, Kosaka T, Tanaka Y. (2008) Leukotriene B4 12-hydroxydehydrogenase/15-ketoprostaglandin Delta 13-reductase (LTB4 12-HD/PGR) responsible for the reduction of a double-bond of the alpha,beta-unsaturated ketone of an aryl propionic acid nonsteroidal anti-inflammatory agent CS-670. Xenobiotica 38:249–263 [DOI] [PubMed] [Google Scholar]
- Iwabu J, Watanabe J, Hirakura K, Ozaki Y, Hanazaki K. (2010) Profiling of the compounds absorbed in human plasma and urine after oral administration of a traditional Japanese (kampo) medicine, daikenchuto. Drug Metab Dispos 38:2040–2048 [DOI] [PubMed] [Google Scholar]
- Jiang H, Sólyom AM, Timmermann BN, Gang DR. (2005) Characterization of gingerol-related compounds in ginger rhizome (Zingiber officinale Rosc.) by high-performance liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 19:2957–2964 [DOI] [PubMed] [Google Scholar]
- Jiang H, Timmermann BN, Gang DR. (2007) Characterization and identification of diarylheptanoids in ginger (Zingiber officinale Rosc.) using high-performance liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom 21:509–518 [DOI] [PubMed] [Google Scholar]
- Jiang H, Xie Z, Koo HJ, McLaughlin SP, Timmermann BN, Gang DR. (2006) Metabolic profiling and phylogenetic analysis of medicinal Zingiber species: Tools for authentication of ginger (Zingiber officinale Rosc). Phytochemistry 67:1673–1685 [DOI] [PubMed] [Google Scholar]
- Juge N, Mithen RF, Traka M. (2007) Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell Mol Life Sci 64:1105–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahun K, Davis M, Hu P, Martin B, Baillie T. (1997) Biotransformation of the naturally occurring isothiocyanate sulforaphane in the rat: identification of phase I metabolites and glutathione conjugates. Chem Res Toxicol 10:1228–1233 [DOI] [PubMed] [Google Scholar]
- Kawai T, Kinoshita K, Koyama K, Takahashi K. (1994) Anti-emetic principles of Magnolia obovata bark and Zingiber officinale rhizome. Planta Med 60:17–20 [DOI] [PubMed] [Google Scholar]
- Kim JS, Lee SI, Park HW, Yang JH, Shin TY, Kim YC, Baek NI, Kim SH, Choi SU, Kwon BM, et al. (2008) Cytotoxic components from the dried rhizomes of Zingiber officinale Roscoe. Arch Pharm Res 31:415–418 [DOI] [PubMed] [Google Scholar]
- Kishida K, Saida N, Yamamura N, Iwai Y, Sasabe T. (2001) Cysteine conjugate of methazolamide is metabolized by beta-lyase. J Pharm Sci 90:224–233 [DOI] [PubMed] [Google Scholar]
- Knapen MF, Zusterzeel PL, Peters WH, Steegers EA. (1999) Glutathione and glutathione-related enzymes in reproduction. A review. Eur J Obstet Gynecol Reprod Biol 82:171–184 [DOI] [PubMed] [Google Scholar]
- Koh IK, Lee SS. (1983) Biodegradation mechanism of shogaols by Aspergillus niger. Yakhak Hoeji 27:29–36 [Google Scholar]
- MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, Hayes JD. (2009) Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis 30:1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masada Y, Inoue T, Hashimoto K, Fujioka M, Uchino C. (1974) [Studies on the constituents of ginger (Zingiber officinale Roscoe) by GC-MS (author's transl)]. Yakugaku Zasshi 94:735–738 [DOI] [PubMed] [Google Scholar]
- Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Miyoshi N. (2010) Electrophiles in foods: the current status of isothiocyanates and their chemical biology. Biosci Biotechnol Biochem 74:242–255 [DOI] [PubMed] [Google Scholar]
- Oppermann U. (2007) Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol 47:293–322 [DOI] [PubMed] [Google Scholar]
- Pernice R, Hauder J, Koehler P, Vitaglione P, Fogliano V, Somoza V. (2009) Effect of sulforaphane on glutathione-adduct formation and on glutathione_S_transferase-dependent detoxification of acrylamide in Caco-2 cells. Mol Nutr Food Res 53:1540–1550 [DOI] [PubMed] [Google Scholar]
- Rhode J, Fogoros S, Zick S, Wahl H, Griffith KA, Huang J, Liu JR. (2007) Ginger inhibits cell growth and modulates angiogenic factors in ovarian cancer cells. BMC Complement Altern Med 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd LP, Kabler SL, Morrow CS, Townsend AJ. (2011) Enhanced glutathione depletion, protein adduct formation, and cytotoxicity following exposure to 4-hydroxy-2-nonenal (HNE in cells expressing human multidrug resistance protein-1 (MRP1) together with human glutathione S-transferase-M1 (GSTM1). Chem Biol Interact 194:113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang S, Hong J, Wu H, Liu J, Yang CS, Pan MH, Badmaev V, Ho CT. (2009) Increased growth inhibitory effects on human cancer cells and anti-inflammatory potency of shogaols from Zingiber officinale relative to gingerols. J Agric Food Chem 57:10645–10650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Chen X, Badmaev V, Ho CT, Sang S. (2010) Structural identification of mouse urinary metabolites of pterostilbene using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 24:1770–1778 [DOI] [PubMed] [Google Scholar]
- Shukla Y, Singh M. (2007) Cancer preventive properties of ginger: a brief review. Food Chem Toxicol 45:683–690 [DOI] [PubMed] [Google Scholar]
- Surh YJ. (2002) Anti-tumor promoting potential of selected spice ingredients with antioxidative and anti-inflammatory activities: a short review. Food Chem Toxicol 40:1091–1097 [DOI] [PubMed] [Google Scholar]
- Surh YJ, Lee SS. (1992) Enzymatic reduction of shogaol: a novel biotransformation pathway for the alpha,beta-unsaturated ketone system. Biochem Int 27:179–187 [PubMed] [Google Scholar]
- Surh YJ, Lee SS. (1994) Enzymatic reduction of xenobiotic alpha,beta-unsaturated ketones: formation of allyl alcohol metabolites from shogaol and dehydroparadol. Res Commun Chem Pathol Pharmacol 84:53–61 [PubMed] [Google Scholar]
- Takahashi H, Hashimoto T, Noma Y, Asakawa Y. (1993) Biotransformation of 6-gingerol and 6-shogaol by Aspergillus niger. Phytochemistry 34:1497–1500 [Google Scholar]
- Tsuji PA, Winn RN, Walle T. (2006) Accumulation and metabolism of the anticancer flavonoid 5,7-dimethoxyflavone compared to its unmethylated analog chrysin in the Atlantic killifish. Chem Biol Interact 164:85–92 [DOI] [PubMed] [Google Scholar]
- Wang W, Li CY, Wen XD, Li P, Qi LW. (2009) Simultaneous determination of 6-gingerol, 8-gingerol, 10-gingerol and 6-shogaol in rat plasma by liquid chromatography-mass spectrometry: Application to pharmacokinetics. J Chromatogr B Analyt Technol Biomed Life Sci 877:671–679 [DOI] [PubMed] [Google Scholar]
- Wu H, Hsieh MC, Lo CY, Liu CB, Sang S, Ho CT, Pan MH. (2010) 6-Shogaol is more effective than 6-gingerol and curcumin in inhibiting 12-O-tetradecanoylphorbol 13-acetate-induced tumor promotion in mice. Mol Nutr Food Res 54:1296–1306 [DOI] [PubMed] [Google Scholar]
- Yu Y, Huang T, Yang B, Liu X, Duan G. (2007) Development of gas chromatography-mass spectrometry with microwave distillation and simultaneous solid-phase microextraction for rapid determination of volatile constituents in ginger. J Pharm Biomed Anal 43:24–31 [DOI] [PubMed] [Google Scholar]
- Zick SM, Djuric Z, Ruffin MT, Litzinger AJ, Normolle DP, Alrawi S, Feng MR, Brenner DE. (2008) Pharmacokinetics of 6-gingerol, 8-gingerol, 10-gingerol, and 6-shogaol and conjugate metabolites in healthy human subjects. Cancer Epidemiol Biomarkers Prev 17:1930–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick SM, Ruffin MT, Djuric Z, Normolle D, Brenner DE. (2010) Quantitation of 6-, 8- and 10-gingerols and 6-shogaol in human plasma by high-performance liquid chromatography with electrochemical detection. Int J Biomed Sci 6:233–240 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.