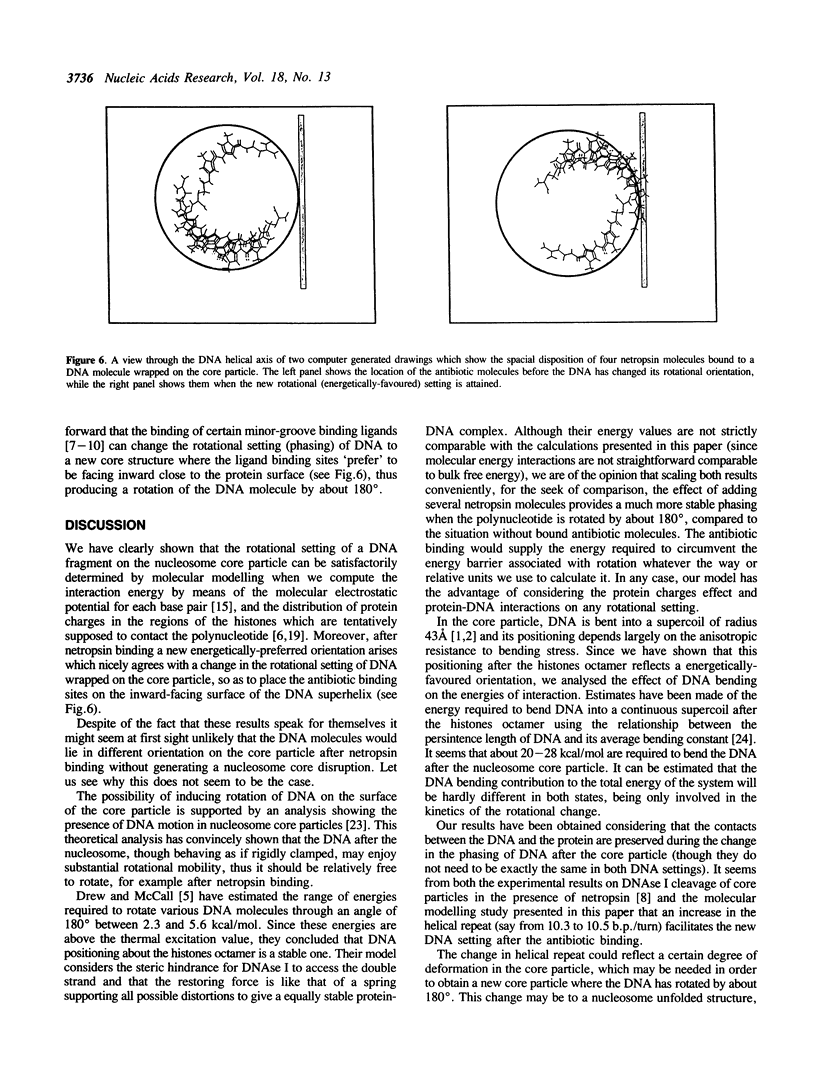
Abstract
It is well known that certain sequence-dependent modulators in structure appear to determine the rotational positioning of DNA on the nucleosome core particle. That preference is rather weak and could be modified by some ligands as netropsin, a minor-groove binding antibiotic. We have undertaken a molecular modelling approach to calculate the relative energy of interaction between a DNA molecule and the protein core particle. The histones particle is considered as a distribution of positive charges on the protein surface that interacts with the DNA molecule. The molecular electrostatic potentials for the DNA, simulated as a discontinuous cylinder, were calculated using the values for all the base pairs. Computing these parameters, we calculated the relative energy of interaction and the more stable rotational setting of DNA. The binding of four molecules of netropsin to this model showed that a new minimum of energy is obtained when the DNA turns toward the protein surface by about 180 degrees, so a new energetically favoured structure appears where netropsin binding sites are located facing toward the histones surface. The effect of netropsin could be explained in terms of an induced change in the phasing of DNA on the core particle. The induced rotation is considered to optimize non-bonded contacts between the netropsin molecules and the DNA backbone.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bavykin S. G., Usachenko S. I., Lishanskaya A. I., Shick V. V., Belyavsky A. V., Undritsov I. M., Strokov A. A., Zalenskaya I. A., Mirzabekov A. D. Primary organization of nucleosomal core particles is invariable in repressed and active nuclei from animal, plant and yeast cells. Nucleic Acids Res. 1985 May 24;13(10):3439–3459. doi: 10.1093/nar/13.10.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J., Kollman P. A molecular mechanical study of netropsin-DNA interactions. Biopolymers. 1986 Feb;25(2):249–266. doi: 10.1002/bip.360250207. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Calladine C. R. Sequence-specific positioning of core histones on an 860 base-pair DNA. Experiment and theory. J Mol Biol. 1987 May 5;195(1):143–173. doi: 10.1016/0022-2836(87)90333-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., McCall M. J. Structural analysis of a reconstituted DNA containing three histone octamers and histone H5. J Mol Biol. 1987 Oct 5;197(3):485–511. doi: 10.1016/0022-2836(87)90560-2. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Gago F., Reynolds C. A., Richards W. G. The binding of nonintercalative drugs to alternating DNA sequences. Mol Pharmacol. 1989 Feb;35(2):232–241. [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S. F., Thomas J. O. Lysine-containing DNA-binding regions on the surface of the histone octamer in the nucleosome core particle. Eur J Biochem. 1986 Oct 1;160(1):191–201. doi: 10.1111/j.1432-1033.1986.tb09957.x. [DOI] [PubMed] [Google Scholar]
- Lavery R., Pullman B. The dependence of the surface electrostatic potential of B-DNA on environmental factors. J Biomol Struct Dyn. 1985 Feb;2(5):1021–1032. doi: 10.1080/07391102.1985.10507618. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Echinomycin and distamycin induce rotation of nucleosome core DNA. Nucleic Acids Res. 1986 Sep 11;14(17):6785–6801. doi: 10.1093/nar/14.17.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Portugal J. Footprinting analysis of sequence-specific DNA-drug interactions. Chem Biol Interact. 1989;71(4):311–324. doi: 10.1016/0009-2797(89)90107-5. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Analysis of the effects of antibiotics on the structure of nucleosome core particles determined by DNAase I cleavage. Biochimie. 1987 Aug;69(8):825–840. doi: 10.1016/0300-9084(87)90209-4. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Antibiotics which can alter the rotational orientation of nucleosome core DNA. Nucleic Acids Res. 1986 Nov 25;14(22):8735–8754. doi: 10.1093/nar/14.22.8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Comparison of binding sites in DNA for berenil, netropsin and distamycin. A footprinting study. Eur J Biochem. 1987 Sep 1;167(2):281–289. doi: 10.1111/j.1432-1033.1987.tb13334.x. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Interaction of nucleosome core particles with distamycin and echinomycin: analysis of the effect of DNA sequences. Nucleic Acids Res. 1987 Feb 11;15(3):885–903. doi: 10.1093/nar/15.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullman A., Pullman B. Molecular electrostatic potential of the nucleic acids. Q Rev Biophys. 1981 Aug;14(3):289–380. doi: 10.1017/s0033583500002341. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Schurr J. M., Schurr R. L. DNA motions in the nucleosome core particle: a reanalysis. Biopolymers. 1985 Oct;24(10):1931–1940. doi: 10.1002/bip.360241007. [DOI] [PubMed] [Google Scholar]
- Waring M. J. DNA modification and cancer. Annu Rev Biochem. 1981;50:159–192. doi: 10.1146/annurev.bi.50.070181.001111. [DOI] [PubMed] [Google Scholar]
- White J. H., Cozzarelli N. R., Bauer W. R. Helical repeat and linking number of surface-wrapped DNA. Science. 1988 Jul 15;241(4863):323–327. doi: 10.1126/science.3388041. [DOI] [PubMed] [Google Scholar]
- White J. H., Gallo R., Bauer W. R. Dependence of the linking deficiency of supercoiled minichromosomes upon nucleosome distortion. Nucleic Acids Res. 1989 Jul 25;17(14):5827–5835. doi: 10.1093/nar/17.14.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Dattagupta N., Hogan M., Crothers D. M. Unfolding of nucleosomes by ethidium binding. Biochemistry. 1980 Feb 19;19(4):626–634. doi: 10.1021/bi00545a004. [DOI] [PubMed] [Google Scholar]


