Abstract
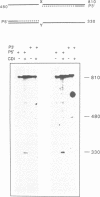
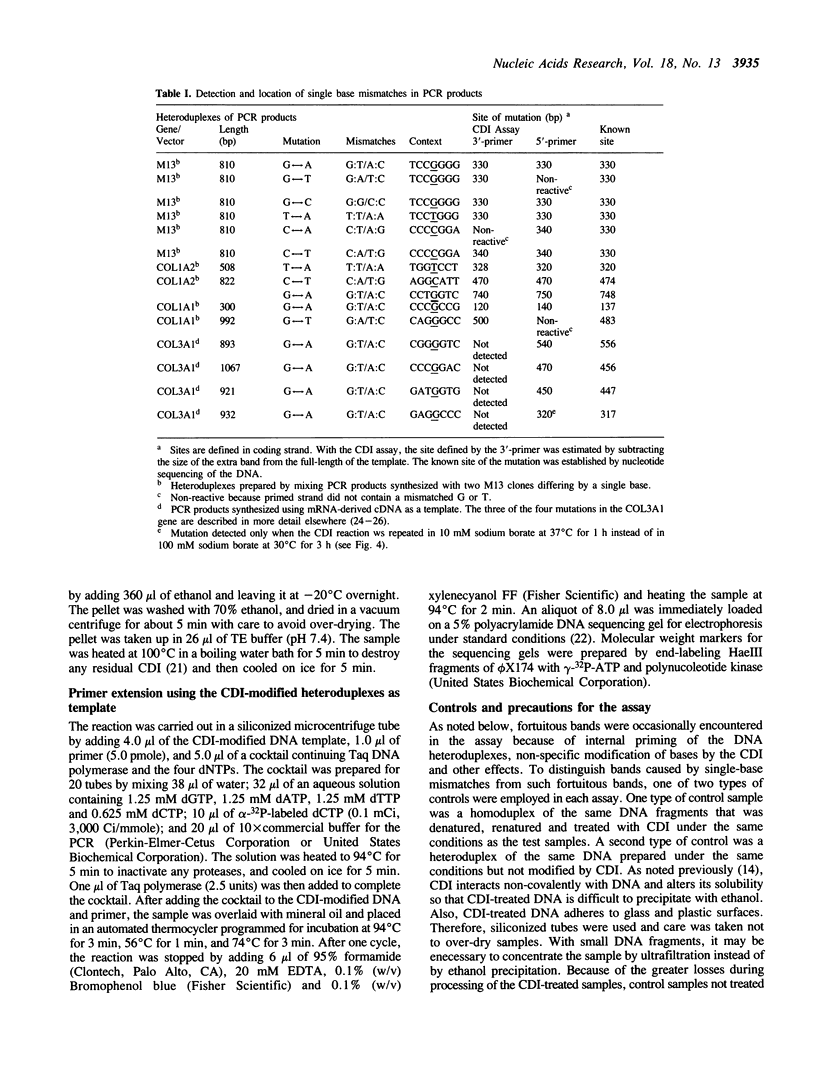
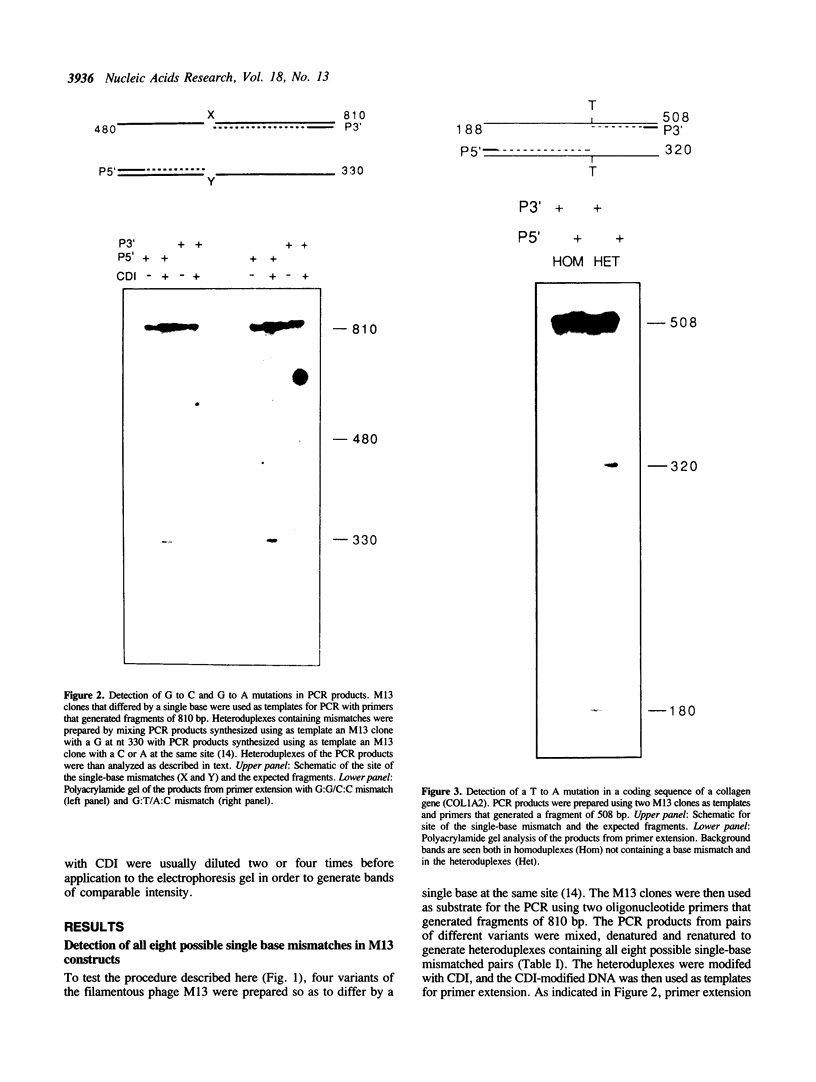
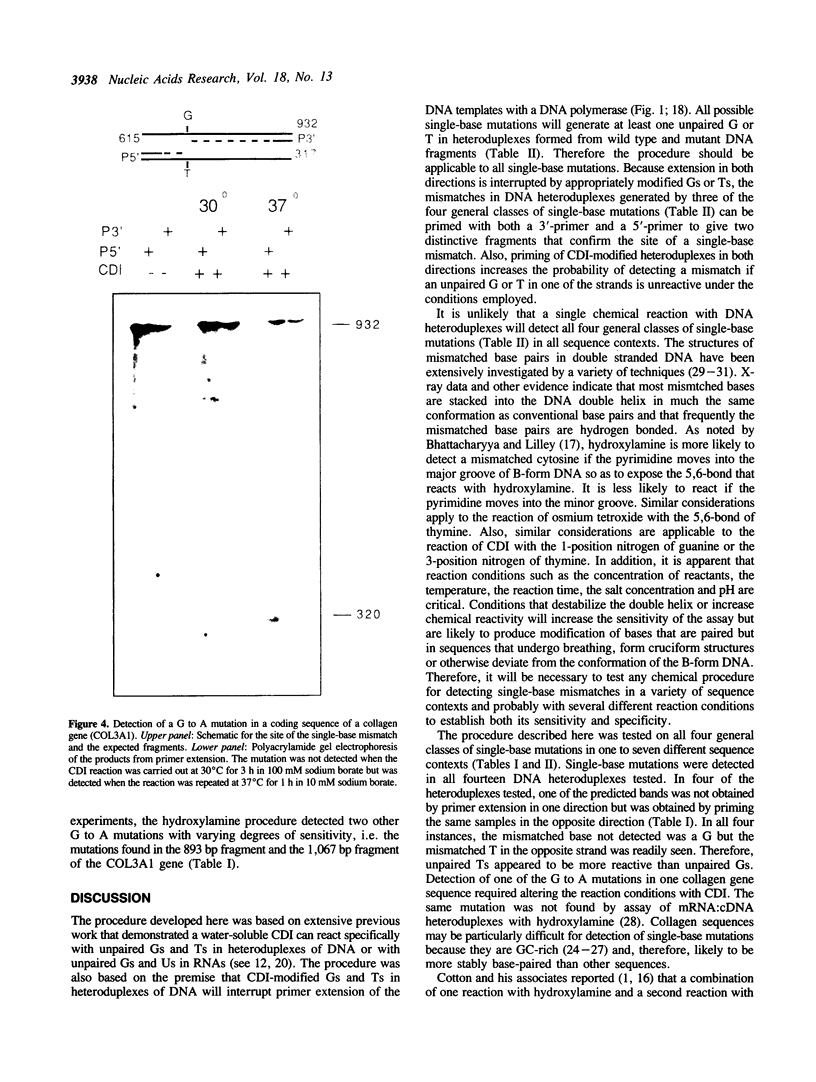
A new method was developed for the detection of single-base mutations in DNA. The polymerase chain reaction was used to prepare DNA fragments of up to 1 kb. Fragments that differed by a single-base were combined, denatured and renatured to generate heteroduplexes. The heteroduplexes were reacted with a water-soluble carbodiimide under conditions in which the carbodiimide modified Gs and Ts that were not base paired. The DNA was then used as a template for primer extension with Taq DNA polymerase under conditions in which extension terminated at the site of the carbodiimide-modified base and generated a 32P-labeled fragment that was identified by polyacrylamide gel electrophoresis as a fragment smaller than the full length product. The procedure detected all four general classes of single-base mutations in several different sequence contexts. The site of the mutation was located to within about 15 bp. Extension with both a 5'- and a 3'-primer made it possible to confirm the site of the mutation in most DNA samples or detect a mutation in heteroduplexes even if a G or T in one strand was unreactive because of its sequence context. The procedure appears to have several advantages over previously published techniques.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bateman J. F., Lamande S. R., Dahl H. H., Chan D., Mascara T., Cole W. G. A frameshift mutation results in a truncated nonfunctional carboxyl-terminal pro alpha 1(I) propeptide of type I collagen in osteogenesis imperfecta. J Biol Chem. 1989 Jul 5;264(19):10960–10964. [PubMed] [Google Scholar]
- Bhattacharyya A., Lilley D. M. Single base mismatches in DNA. Long- and short-range structure probed by analysis of axis trajectory and local chemical reactivity. J Mol Biol. 1989 Oct 20;209(4):583–597. doi: 10.1016/0022-2836(89)90596-2. [DOI] [PubMed] [Google Scholar]
- Brown T., Kneale G., Hunter W. N., Kennard O. Structural characterisation of the bromouracil.guanine base pair mismatch in a Z-DNA fragment. Nucleic Acids Res. 1986 Feb 25;14(4):1801–1809. doi: 10.1093/nar/14.4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G., Campbell R. D. Chemical reactivity of matched cytosine and thymine bases near mismatched and unmatched bases in a heteroduplex between DNA strands with multiple differences. Nucleic Acids Res. 1989 Jun 12;17(11):4223–4233. doi: 10.1093/nar/17.11.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G. Detection of single base changes in nucleic acids. Biochem J. 1989 Oct 1;263(1):1–10. doi: 10.1042/bj2630001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J. B., Wells R. D. Action of single-strand specific nucleases on model DNA heteroduplexes of defined size and sequence. Biochemistry. 1977 May 31;16(11):2374–2379. doi: 10.1021/bi00630a010. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Rooney J. E., Hosomi S., Zeiger A. R., Prockop D. J. Detection and location of single-base mutations in large DNA fragments by immunomicroscopy. Genomics. 1989 May;4(4):530–538. doi: 10.1016/0888-7543(89)90276-0. [DOI] [PubMed] [Google Scholar]
- Gibbs R. A., Caskey C. T. Identification and localization of mutations at the Lesch-Nyhan locus by ribonuclease A cleavage. Science. 1987 Apr 17;236(4799):303–305. doi: 10.1126/science.3563511. [DOI] [PubMed] [Google Scholar]
- Keohavong P., Thilly W. G. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz D. H., Brown G. L. The investigation of nucleic acid secondary structure by means of chemical modification with a carbodiimide reagent. I. The reaction between N-cyclohexyl-N'-beta-(4-methylmorpholinium)ethylcarbodiimide and model nucleotides. Biochemistry. 1969 Jun;8(6):2312–2328. doi: 10.1021/bi00834a012. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Maniatis T., Lerman L. S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Noll W. W., Collins M. Detection of human DNA polymorphisms with a simplified denaturing gradient gel electrophoresis technique. Proc Natl Acad Sci U S A. 1987 May;84(10):3339–3343. doi: 10.1073/pnas.84.10.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novack D. F., Casna N. J., Fischer S. G., Ford J. P. Detection of single base-pair mismatches in DNA by chemical modification followed by electrophoresis in 15% polyacrylamide gel. Proc Natl Acad Sci U S A. 1986 Feb;83(3):586–590. doi: 10.1073/pnas.83.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyadenosine-deoxycytidine pairing in the d(C-G-C-G-A-A-T-T-C-A-C-G) duplex: conformation and dynamics at and adjacent to the dA X dC mismatch site. Biochemistry. 1984 Jul 3;23(14):3218–3226. doi: 10.1021/bi00309a016. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Inagami S., Lovegren E., Chalkley R. DNA denatures upon drying after ethanol precipitation. Nucleic Acids Res. 1987 Nov 11;15(21):8739–8754. doi: 10.1093/nar/15.21.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. C., Kunkel T. A., Casna N. J., Ford J. P., Sancar A. Activities and incision patterns of ABC excinuclease on modified DNA containing single-base mismatches and extrahelical bases. J Biol Chem. 1986 Nov 5;261(31):14496–14505. [PubMed] [Google Scholar]
- Tromp G., Kuivaniemi H., Shikata H., Prockop D. J. A single base mutation that substitutes serine for glycine 790 of the alpha 1 (III) chain of type III procollagen exposes an arginine and causes Ehlers-Danlos syndrome IV. J Biol Chem. 1989 Jan 25;264(3):1349–1352. [PubMed] [Google Scholar]
- Tromp G., Kuivaniemi H., Stolle C., Pope F. M., Prockop D. J. Single base mutation in the type III procollagen gene that converts the codon for glycine 883 to aspartate in a mild variant of Ehlers-Danlos syndrome IV. J Biol Chem. 1989 Nov 15;264(32):19313–19317. [PubMed] [Google Scholar]
- Winter E., Yamamoto F., Almoguera C., Perucho M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7575–7579. doi: 10.1073/pnas.82.22.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]