Abstract
Pseudomonas aeruginosa is a Gram-negative bacterium that utilizes substrate-specific outer membrane (OM) proteins for the uptake of small, water-soluble nutrients employed in the growth and function of the cell. In this paper, we present for the first time a comprehensive single-channel examination of seven members of the OM carboxylate channel K (OccK) subfamily. Recent biochemical, functional and structural characterization of the OccK proteins revealed their common features, such as a closely related, monomeric, 18-stranded β-barrel conformation with a kidney-shaped transmembrane pore and the presence of a basic ladder within the channel lumen. Here, we report that the OccK proteins exhibited fairly distinct unitary conductance values, in a much broader range than earlier expectations, which includes low (~40–100 pS) and medium (~100–380 pS) conductance. These proteins showed diverse single-channel dynamics of current gating transitions, revealing one (OccK3)-, two (OccK4, OccK5 and OccK6)- and three (OccK1, OccK2 and OccK7)-open sub-state kinetics with functionally distinct conformations. Interestingly, we discovered that anion selectivity is a conserved trait among the members of the OccK subfamily, confirming the presence of a net pool of positively charged residues within their central constriction. Moreover, these results are in accord with an increased specificity and selectivity of these protein channels for negatively charged, carboxylate-containing substrates. Our findings might ignite future functional examinations and full-atomistic computational studies for unraveling a mechanistic understanding of the passage of small molecules across the lumen of substrate-specific, β-barrel OM proteins.
Keywords: Single-channel electrical recordings, Single-molecule biophysics, Channel gating, The kinetic rate theory, The OccK subfamily
Pseudomonas aeruginosa is a versatile human pathogen that exhibits a fundamental distinction from other Gram-negative organisms such as E. coli: its unusually low outer membrane (OM) permeability (1;2). The major reason for this dissimilarity is the absence of the large-conductance and nonspecific porins, such as the OM protein F (OmpF) and C (OmpC), which are otherwise abundantly present in the OM of E. coli. To overcome a poor permeability of their OMs, pseudomonads utilize a range of specialized transport pathways through which the water-soluble, low-molecular mass substrates navigate into the periplasm. OprF is an OM protein A (OmpA) analogue porin in P. aeruginosa (3). It has been shown by single-channel reconstitution into planar lipid bilayers that OprF forms low-conductance protein channels and has an electrical signature decorated by occasional openings (4). Moreover, P. aeruginosa has substrate-specific OM channels, such as OprP (5;6) and OprB (2;7), which exhibit high transport selectivity for phosphate (3;7) and sugar (3;7), respectively. High-resolution X-ray crystal structure of OprP revealed its trimeric assembly, an archetype of the OM porins (8), and a basic ladder formed by 9 arginines that are located from the extracellular surface through the constriction (9).
However, most small, water-soluble substrates are thought to be taken by the members of the OM carboxylate channel (Occ) family (2;3;10;11). This OM protein family from P. aeruginosa contains 19 substrate-specific channels that share significant amino acid similarity (40–50%) (12). Phylogenetic analysis indicated a division of the Occ family into the OccD and OccK subfamilies (12;13). The archetypes of these subfamilies are the OccD1 (12;14–17) and OccK1 (18;19) channels, which are believed to facilitate the uptake of basic amino acids (12;14;16), and vanillate (13;18) and benzoate (11), respectively. These sustained efforts to understand the major roles of the proteins of the Occ family were supplemented very recently by Eren and colleagues, who pursued biochemical, functional and structural analysis of the substrate specificity of three OccD and six OccK protein channels (11). This study confirmed that the OccD and OccK proteins share common structural features, such as the monomeric β-barrel conformation, which is quite different from what we have learned previously with trimeric porins of E. coli (20;21). Another hallmark structural trait of the Occ proteins is the presence of a basic ladder of arginines and lysines that are distributed from the extracellular surface to the constriction and from the constriction away to the periplasmic surface (11;17;18).
Therefore, it was proposed that this basic ladder forms an energetically favorable conduit for the small, carboxyl-containing substrates, which are pulled into the cell to sustain its growth and function (9;11;17;18). The OccK proteins show more variability in terms of shape, size and charge distribution along the channel longitudinal axis, suggesting distinctions in the functionality within this subfamily (11). Recent X-ray crystallography information on OccK proteins (11) ignited a detailed biophysical inspection of these subfamily members at the single-channel level. The sequence conservation among the residues facing the central constriction of the OccK proteins is generally quite low (Supplementary Materials, Fig. S1), and so different single-channel electrical signatures were expected.
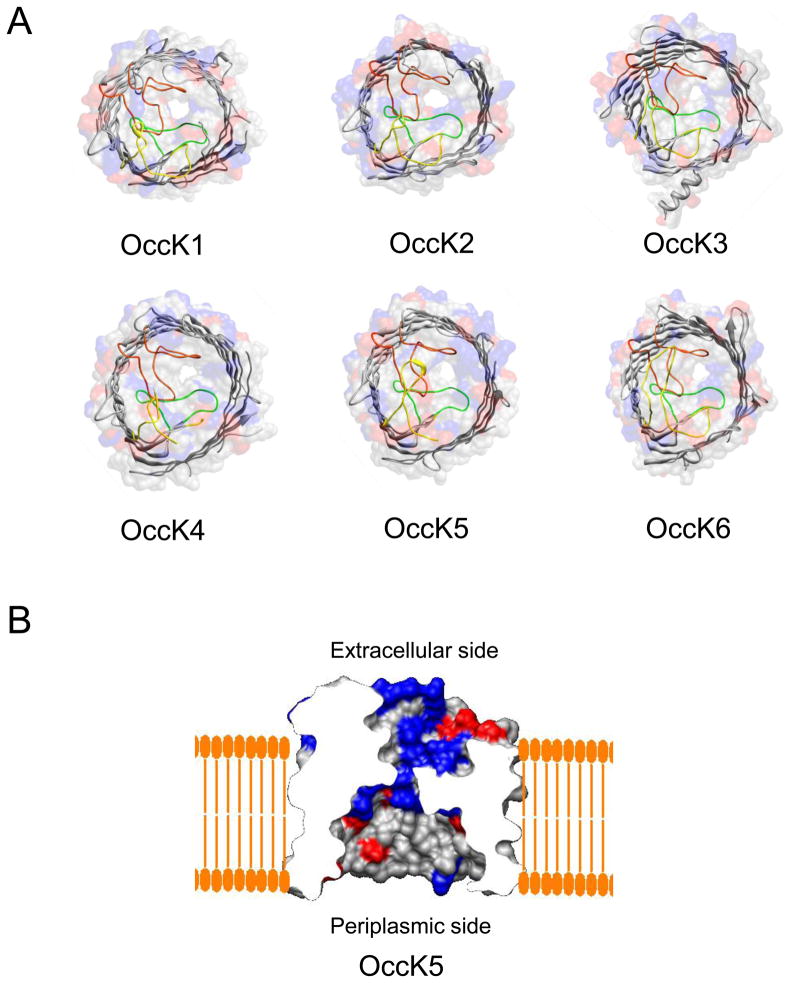
In this work, we pursued a systematical examination of seven members of the OccK subfamily using single-channel electrophysiology on planar lipid bilayers (22). The proteins inspected in this work were the following: OccK1, OccK2, OccK3, OccK4, OccK5, OccK6 and OccK7 (formerly called OpdK, OpdF, OpdO, OpdL, OpdH, OpdQ, and OpdD, respectively). Among these proteins, OccK7 does not have as yet an available crystal structure. Recently, we carried out a detailed biophysical analysis of the OccK1 channel (19). The results show that all these OccK proteins possess channel-forming activity in artificial planar lipid bilayers. We compared the OccK proteins based upon their unitary conductance, their kinetic and energetic data pertinent to single-channel discrete dynamics of their open sub-states, as well as their ionic selectivity. None of the OccK proteins forms a large-conductance channel, which is in accord with the recent high-resolution crystallographic information that revealed a narrow central constriction of these channels in the range of 3.5 through 5.0 Å diameter (Fig. 1) (11).
Figure 1. Structural representation of OccK1 family protein channels.
(A) The molecular surface and ribbon representations of six OccK subfamily members whose crystal structures have been solved recently (11), which include OccK1, OccK2, OccK3, OccK4, OccK5, and OccK6. On the molecular surface representation, the side chains of positively charged amino acids are marked blue, and those of negatively charged amino acids are marked red. On the ribbon representation, constriction-forming loops L3, L4, L7 are colored in green, yellow, and orange, respectively; (B) The side view of the OccK5 channel in a planar lipid bilayer. The charged amino acids are colored as described above. Note the positive surface resulting from the presence of the basic amino acid ladder within the channel constriction.
Materials and Methods
Cloning, expression, and purification of the OccK proteins
The mature parts of the occK genes from P. aeruginosa were amplified by PCR from genomic DNA and cloned into the E. coli expression vector pB22 (23;24), having the signal sequence of the E. coli OM protein YtfM and an N-terminal hexa-histidine tag for purification. DNA sequencing was performed at CFAR DNA sequencing facility (UMass Medical School, Worcester, MA). BL21(DE3) T1 phage-resistant cells (New England Biolabs, Ipswich, MA) were transformed with pB22-OccK constructs. The cells were grown to OD600 ~ 0.6 at 37°C, induced with 0.1% arabinose at 20°C overnight and harvested by centrifugation at 4500 rpm for 30 min (Beckman Coulter, J6-MC). Cell pellets were suspended in TSB (20 mM Tris, 300 mM NaCl, 10% glycerol, pH 8.0) and cells were lysed by sonication (3 × 40 s intervals) (Branson Digital Sonifier). Total membranes were obtained by centrifugation at 40,000 rpm for 40 min (45 Ti rotor, Beckman L8-70M ultracentrifuge). Membranes were homogenized in TSB and solubilized in 1% DM (n-Decyl-β-D-Maltopyranoside, Anatrace, Santa Clara, CA) and 1% LDAO (n-Dodecyl-N,N-Dimethylamine-N-Oxide, Anatrace) for 2 h at 4°C followed by centrifugation at 40,000 rpm for 30 min to remove precipitates and unsolubilized membranes. The membrane extract was applied to a 10 ml nickel column. The column was washed with 10 column volumes (CV) TSB containing 0.2% DM and 15 mM imidazole. The proteins were eluted with 3 CV TSB containing 0.2% DM and 200 mM imidazole, and further purified by gel filtration chromatography using 10 mM Tris, 50 mM NaCl and 0.12% DM, pH 8.0. For final polishing and detergent exchange, another gel filtration chromatography step was performed. The buffer used for this column varied depending on the protein; for most of the channels, 10 mM Tris, 50 mM NaCl and 0.3–0.35% C8E4 was used at pH 8.0. Typically, small amounts (0.02–0.1%) of another detergent (either LDAO, -OG, DM or diheptanoyl-phosphatidylcholine) were added to modify channel solubility. The purified proteins were concentrated to 5–15 mg/ml using 50 kDa molecular weight cutoff filters (Amicon) and directly flash-frozen in liquid nitrogen, without dialysis. The protein samples were aliquoted and kept at −80°C for several months. The purity of the OccK protein samples was assessed by SDS-PAGE gel electrophoresis (11).
Single-channel current recordings on planar lipid bilayers
Single-channel current measurements were carried out with planar lipid membranes (25;26)(Montal and Mueller, 1972; Braha et al., 1997; Howorka et al., 2000). Briefly, both chambers (1.5 ml each) of the bilayer apparatus were separated by a 25 μm thick teflon septum (Goodfellow Corporation, Malvern, PA). An aperture in the septum of ~60 μm diameter was pretreated with hexadecane (Aldrich Chemical Co., Milwaukee, WI) dissolved in highly purified n-pentane (Burdick & Jackson, Allied Signal Inc., Muskegon, MI) at a concentration of 10% (v/v). The standard electrolyte in both chambers was 1000 mM KCl, 10 mM potassium phosphate, pH 7.4, unless otherwise stated. The bilayer was formed with 1,2-diphytanoyl-sn-glycerophosphocholine (Avanti Polar Lipids Inc., Alabaster, AL, USA). OccK proteins were added to the cis chamber, which was grounded. A positive current is defined such that it represents a positive charge moving from the trans to cis chamber. Currents were recorded by using an Axopatch 200B patch-clamp amplifier (Axon Instruments, Foster City, CA) connected to the chambers by Ag/AgCl electrodes. An Optiplex Desktop Computer (Dell, Austin, TX) equipped with a Digitdata 1440 A/D converter (Axon) was used for data acquisition. The output from this amplifier was also filtered by an 8-pole low-pass Bessel filter (Model 900, Frequency Devices, Haverhill, MA) at a frequency of 10 kHz. The sampling rate was 50 kHz. Acquisition and analysis of single-channel data were performed using pClamp 10.2 software (Axon). The time resolution of the single-channel electrical recordings can be derived using the relationship for the rise time of the filter Tr=339/fc, where fc is the corner frequency of the low-pass Bessel filter (27;28). For example, for a value of fc=2 kHz, we obtain Tr= ~170 μs. This value would give us a dead time Td=0.54×Tr=92 μs. Events shorter than this value were missed. We used protein samples from one or more purification batches and found satisfactory reproducibility of the single-channel electrical signatures. We were able to consistently reproduce the single-channel electrical signatures of 12 members of the Occ family in the past few years (11;17–19). In this work, all averaged single-channel data resulted from at least three distinct single-channel reconstitutions.
Results
The members of the OccK subfamily feature a broad range of single-channel conductance values
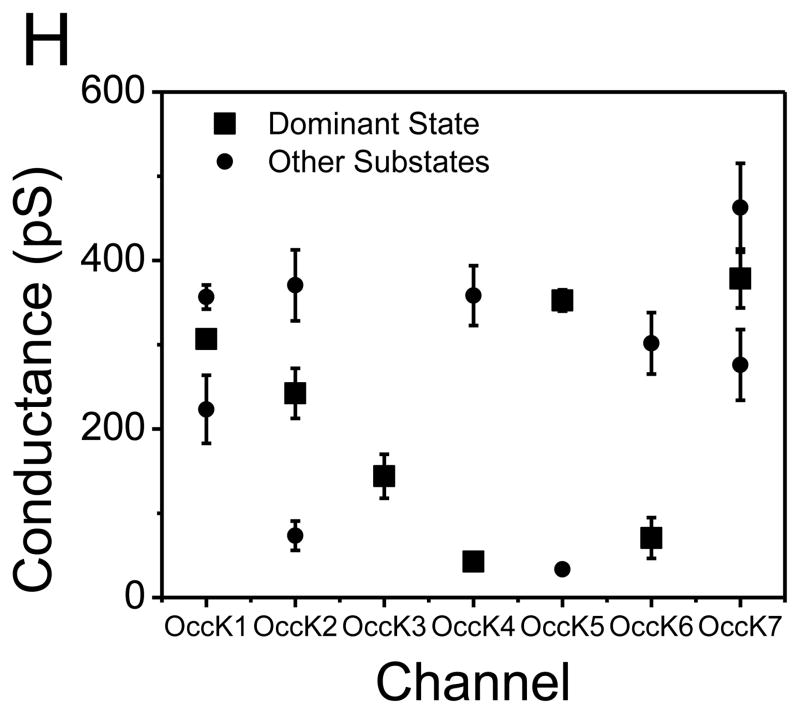
To obtain the single-channel electrical characteristics of the OccK proteins, our measurements were carried out in 1 M KCl, 10 mM potassium phosphate, pH 7.4. The unitary conductance values mentioned below are those corresponding to the most probable open sub-state of each channel. We determined that the unitary conductance of the OccK proteins covered a broad range, from ~40 pS to ~380 pS. Here, we define a low-, medium- and large-conductance channel as an OM protein whose single-channel conductance of the most probable open sub-state is in the range 0–100, 100–500 and 500–1000 pS, respectively. By comparing the unitary conductance among the seven members of the OccK subfamily, we divided them in two groups: (1) the medium-conductance channels, such as OccK1, OccK2, OccK3, OccK5, and OccK7, whose single-channel conductance values, obtained at +60 mV, were 307 ± 14 pS, 242 ± 40 pS, 144 ± 36 pS, 353 ± 22 pS, and 379 ± 45 pS (n=3 distinct single-channel experiments), respectively; (2) the low-conductance channels, which include OccK4 and OccK6, whose single-channel conductance values, under identical conditions, were 43 ± 11 pS and 71 ± 34 pS (n=3), respectively.
OccK proteins show multi-state, single-channel dynamics
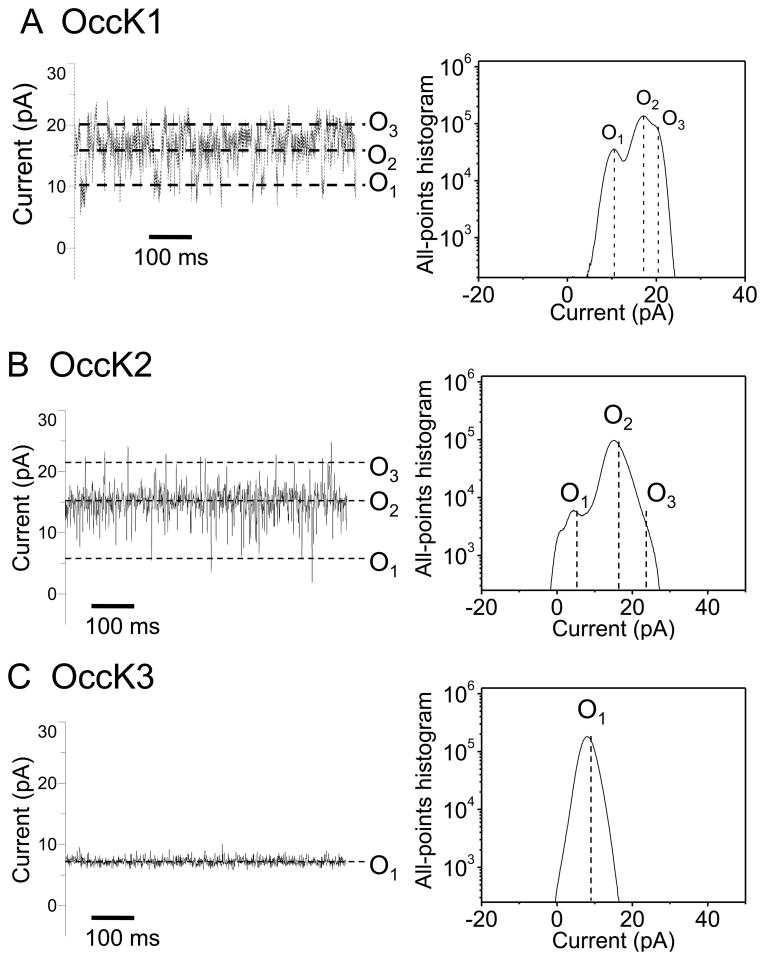
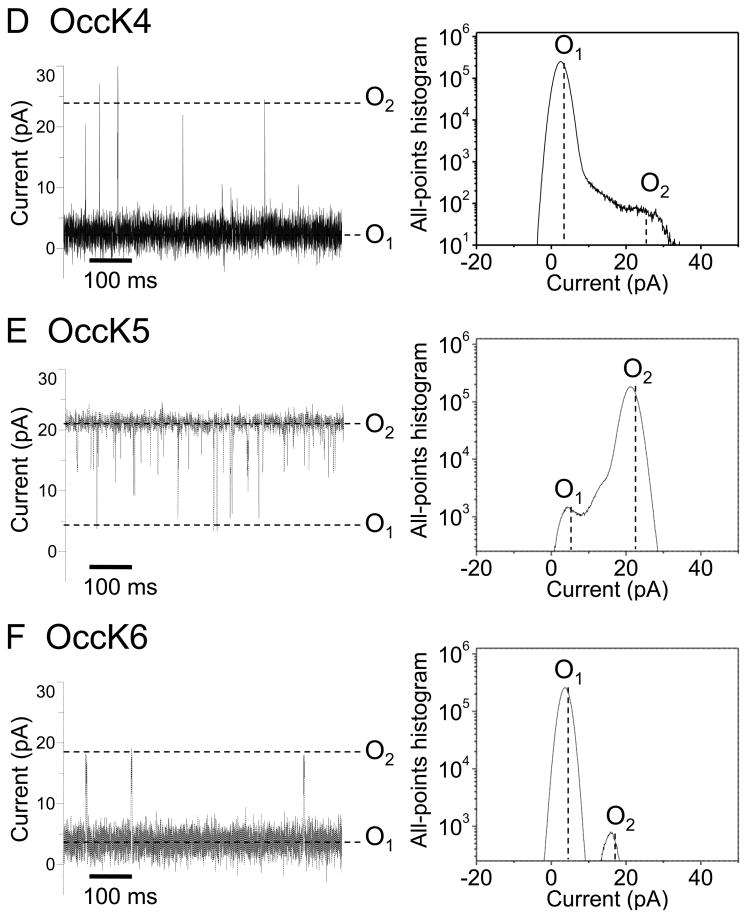
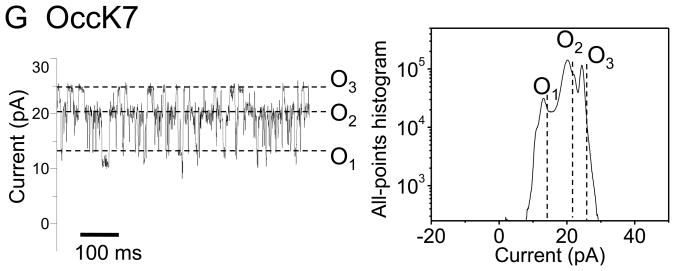
Recently, we showed that OccK1, the archetype of the OccK subfamily, has three major open sub-states (19). Fig. 2 presents typical single-channel electrical recordings of the seven reconstituted OccK subfamily members, which were collected at a transmembrane potential of +60 mV. Detailed single-channel data analysis permitted direct determination of the current amplitudes of the open sub-states (Fig. 2, the right-hand panels) as well as their average duration (Supplementary Materials, Fig. S2).
Figure 2. Typical single-channel electrical traces of the OccK subfamily members and the corresponding all-points current amplitude histograms.
Traces (the left panels) were recorded at a transmembrane potential of +60 mV, and in 1M KCl, 10 mM phosphate, pH 7.4. (A) OccK1; (B) OccK2; (C) OccK3; (D) OccK4; (E) OccK5; (F) OccK6; (G) OccK7. All-points current amplitude histograms (the right panels) included all acquired data points in a single-channel electrical trace. Current levels were marked on both the single-channel electrical traces and on the all-points current amplitude histograms. The sub-states O1, O2, and O3 (if applicable) were assigned to the current levels from the lowest to the highest conductance values, respectively. The all-points current amplitude histograms were determined from single experiments, whose electrical traces are shown on the left panels. In this figure, the single-channel electrical traces corresponding to OccK1, OccK2, OccK3, OccK5 and OccK7 were low-pass Bessel filtered at 2 kHz. Those single-channel electrical traces corresponding to OccK4 and OccK6 were low-pass Bessel filtered at 10 kHz.
Interestingly, all OccK proteins, except OccK3, have one dominant (or so called “most probable”) current level, which is decorated by other secondary short-lived open sub-states. We assigned the sub-states O1, O2, and O3 (if applicable) to the current levels from the lowest to the highest conductance values, respectively, as marked in Fig. 2. Table 1 summarizes the single-channel characteristics of the seven members of the OccK subfamily, including the most probable conductance as well as the channel conductance values associated with each open sub-state. In addition, Table 1 shows the average diameter and the charged residues of the channel eyelet of the OccK subfamily members. Based upon the single-channel electrical traces and the all-points current amplitude histograms, we distinguish two groups of channels. The first group includes the three-open sub-state OccK proteins, such as OccK1 (Fig. 2A), OccK2 (Fig. 2B) and OccK7 (Fig. 2G) (19). These proteins exhibited a most probable open sub-state O2, which was decorated by downward current transitions to the low-conductance O1 open sub-state and upward current transitions to the high–conductance O3 open sub-state. For example, OccK2 showed the most probable O2 open sub-state (gO2= 242 ± 40 pS) that fluctuated between the O1 open sub-state (gO1= 73 ± 27 pS) and the O3 open sub-state (gO3= 371 ± 52 pS). It is also worth mentioning that the OccK proteins that feature this three-open sub-state kinetics have medium unitary conductance in the range 240–380 pS.
Table 1. Biophysical properties of the OccK proteins.
The single-channel conductance values were measured in 1M KCl, 10 mM potassium phosphate, pH=7.4. The single-channel conductance that corresponds to most probable open sub-state is shown in bold. The g80/g−80 is the ratio of the conductance of the dominant state at +80 mV and −80mV. The diameter of the OccK proteins in the central constriction was determined using the HOLE software (59) and their crystal structures (11). The first column on the right-hand side indicates the pool of charged residues in the central constriction. This information was obtained by inspecting the lumen of the protein channels using the Chimera software (60).
| Protein | gO1 (pS)# | gO2 (pS)# | gO3 (pS)# | g80/g−80 | Diameter (Å) | Charges located within constriction |
|---|---|---|---|---|---|---|
| OccK1 | 223 ± 50 | 307 ± 14 | 357 ± 24 | 0.85 ± 0.09 | ~5.0 | (+) R22, R126, R158, R284 (−) D123, D289 |
| OccK2 | 73 ± 27 | 242 ± 40 | 371 ± 52 | 0.77 ± 0.04 | ~4.5 | (+) R26, R129, R161, R280, R327, R387 (−) D126, D292 |
| OccK3 | 144 ± 36 | * | * | 0.96 ± 0.26 | ~3.5 | (+) R123, R317, R358, R374 (−) D121, D276, D287 |
| OccK4 | 43 ± 11 | 358 ± 45 | * | 1.52 ± 0.47 | ~3.5 | (+) R13, R120, R124 (−) D121, D122 |
| OccK5 | 33 ± 12 | 353 ± 22 | * | 1.28 ± 0.06 | ~4.0 | (+)R31, R134, K179, R334, R374, R376, R392 (−) None |
| OccK6 | 71 ± 34 | 302 ± 47 | * | 0.58 ± 0.19 | ~4.0 | (+) R124, R156, R172, R384 (−) D285, E382 |
| OccK7 | 276 ± 52 | 379 ± 45 | 463 ± 63 | 0.98 ± 0.04 | N/A& | N/A& |
The conductance values for these open sub-states were calculated at a transmembrane potential of +60 mV.
This open sub-state is not present.
Crystal structure of the OccK7 protein is not available.
The second group includes the two-open sub-state OccK proteins, such as OccK4 (Fig. 2D), OccK5 (Fig. 2E) and OccK6 (Fig. 2F). OccK5 has a dominant O2 open sub-state with relatively high conductance (gO2 = 353 ± 22 pS) and typical gating transitions to the low-conductance O1 open sub-state (gO1= 33 ± 12 pS). The low-conductance protein channels OccK4 and OccK6 exhibited the most probable O1 open sub-state, which was accompanied by upward current spikes to the O2 open sub-state. Remarkably, the amplitude of the upward current spikes was large, making the single-channel conductance associated with the O2 open sub-state (gO2 (OccK4) = 358 ± 45 pS and gO2 (OccK6) = 302 ± 47 pS) comparable with the conductance range of the medium-conductance OccK proteins, except OccK3 (144 ± 36 pS). In contrast to the OccK proteins belonging to these two groups, again OccK3 (Fig. 2C) featured only one major open sub-state (O1), which was decorated by very low-amplitude and time-irresolvable current fluctuations.
Voltage dependence of the unitary conductance of the OccK proteins
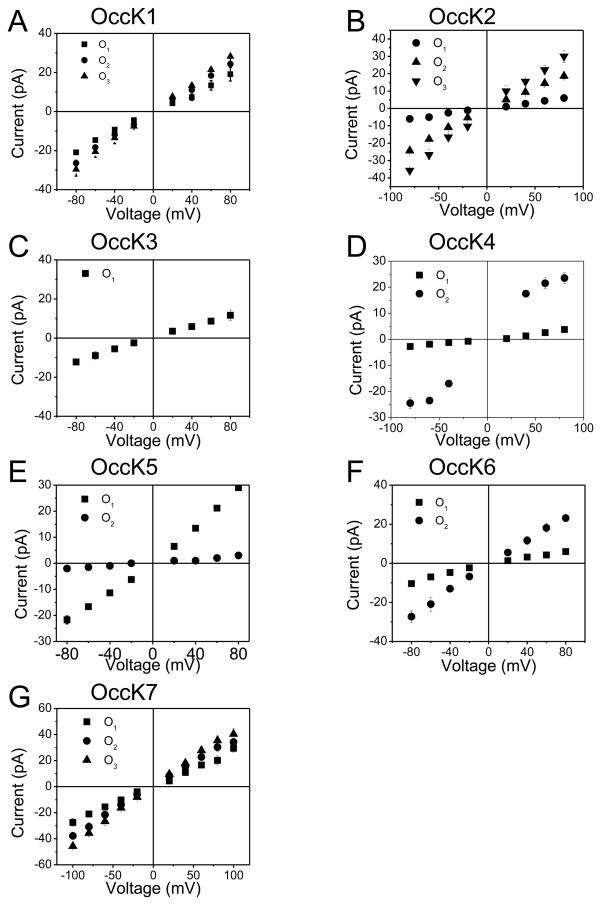
To better illustrate the multi-state behavior, we constructed I–V (current-voltage) profiles for all OccK proteins, revealing the current amplitude of individual open sub-states at each transmembrane potential examined in this work (Fig. 3). The range of the transmembrane potential used here was −80 through +80 mV, since at voltages greater than these values some OccK proteins became unstable. The I–V profiles demonstrate that most of these protein channels exhibited non-ohmic voltage dependence. Here, we introduce the asymmetry ratio (g80/g−80). This parameter is defined as the ratio between the unitary conductance at +80 mV and the unitary conductance at −80 mV. The values of g80/g−80 are listed in Table 1. All these ratio values corresponded to the most probable open sub-state of each channel. OccK3 and OccK7 showed ohmic voltage dependence of their unitary conductance. In contrast, all other OccK proteins featured asymmetric voltage dependence. The OccK6 channel exhibited the largest asymmetry with g80/g−80 of 0.58 ± 0.19. Based on their asymmetry ratio, we found that each channel has a preferred orientation for its insertion into the planar lipid bilayer, since this parameter was reproducible over ~80% of the executed single-channel experiments. Yet, in ~20% of single-channel experiments the orientation was flipped, resulting in a reciprocal value of the asymmetry ratio.
Figure 3. The I–V profiles of all open sub-states of the OccK subfamily members in 1M KCl, 10 mM potassium phosphate, pH 7.4.
(A) OccK1; (B) OccK2; (C) OccK3; (D) OccK4; (E) OccK5; (F) OccK6; (G) OccK7; (H) The plots represent the conductance levels of each open sub-state. The big squares indicate the conductance value corresponding to the most probable open sub-state of each channel. The small dots denote the conductance values corresponding to the low-probability open sub-states of each channel. These conductance values are also shown in Table 1.
Kinetics and energetics of current gating transitions in OccK proteins
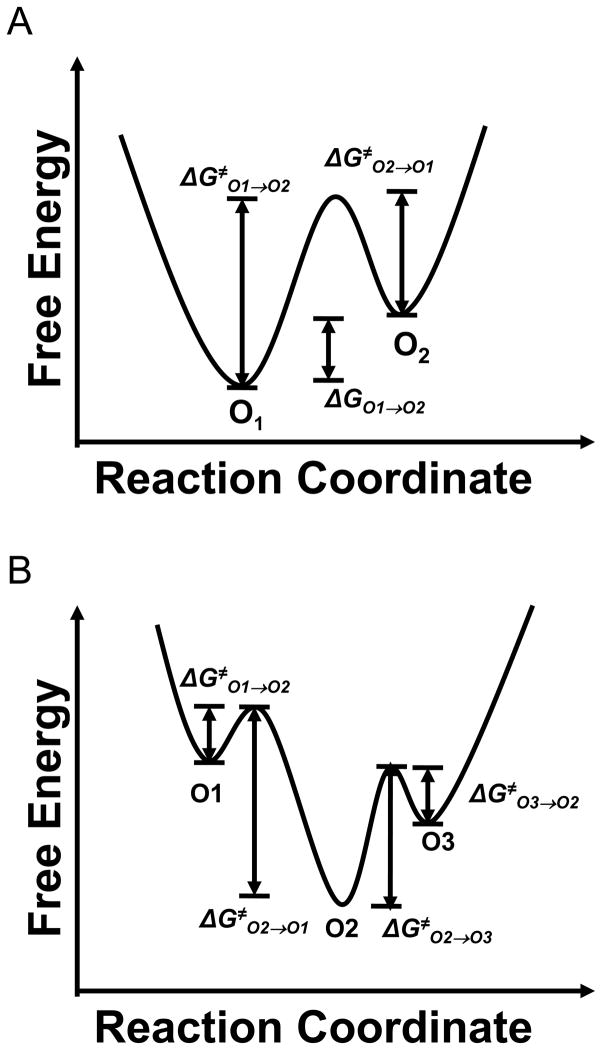
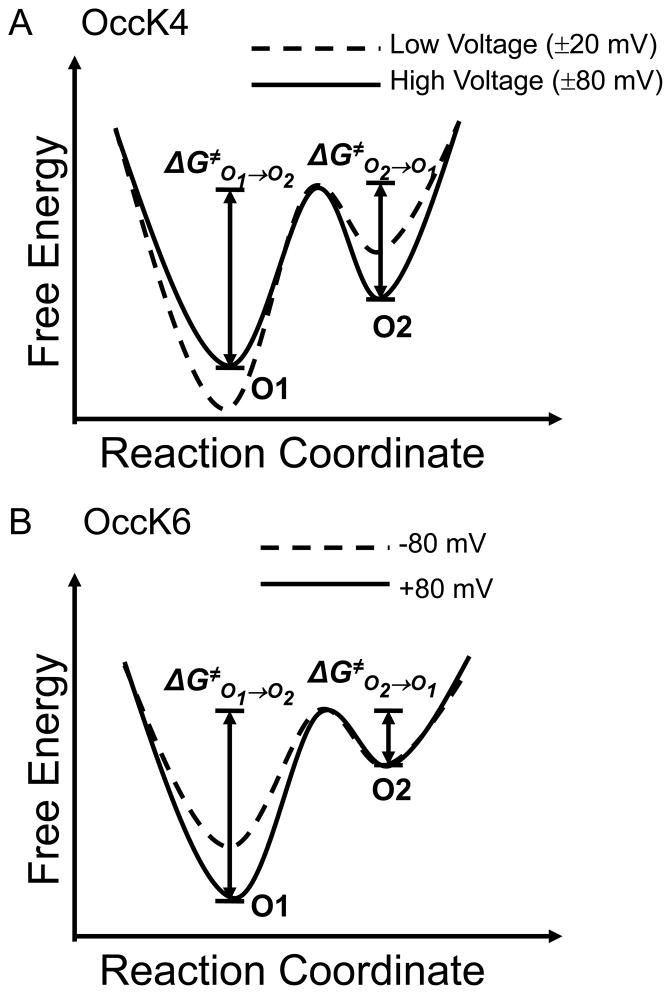
The current gating transitions in OccK proteins were further examined in detail using standard dwell time histograms (Supplementary Materials, Fig. S2) (19). All the dwell time histograms were fitted to single exponential distributions, as judged by log likelihood ratio (LLR) tests with a confidence level of 95% (29). The fitting results gave us the mean lifetime, which is the reciprocal of the transition rates. For the two-open sub-state channels (OccK4, OccK5, and OccK6), we employed a simple two-open sub-state kinetic model, which has two wells and one energetic barrier (Fig. 4A):
Figure 4. The free energy landscapes of the current gating transitions found in OccK proteins.
(A) The free energy landscape for the two-open sub-state kinetic model, in which the O1 level is assigned to the most probable open sub-state; (B) The free energy landscape for the three-open sub-state kinetic model, in which the O2 level is assigned to the most probable open sub-state.
This kinetic scheme has two kinetic rate constants kO1→O2 and kO2→O1.
For three-open sub-state channels (OccK1, OccK2 and OccK7), we constructed a linear model with three wells and two energetic barriers (Fig. 4B):
This kinetic scheme has four kinetic rate constants: kO1→O2, kO2→O1, kO3→O2 and kO2→O3. The details of this linear kinetic model are described in Supplementary Materials. Here, we show the kinetic data of six OccK proteins (Fig. 5). In addition to the transition rate constants, we estimated their corresponding activation free energies based on the standard Arrhenius equation:
| (1) |
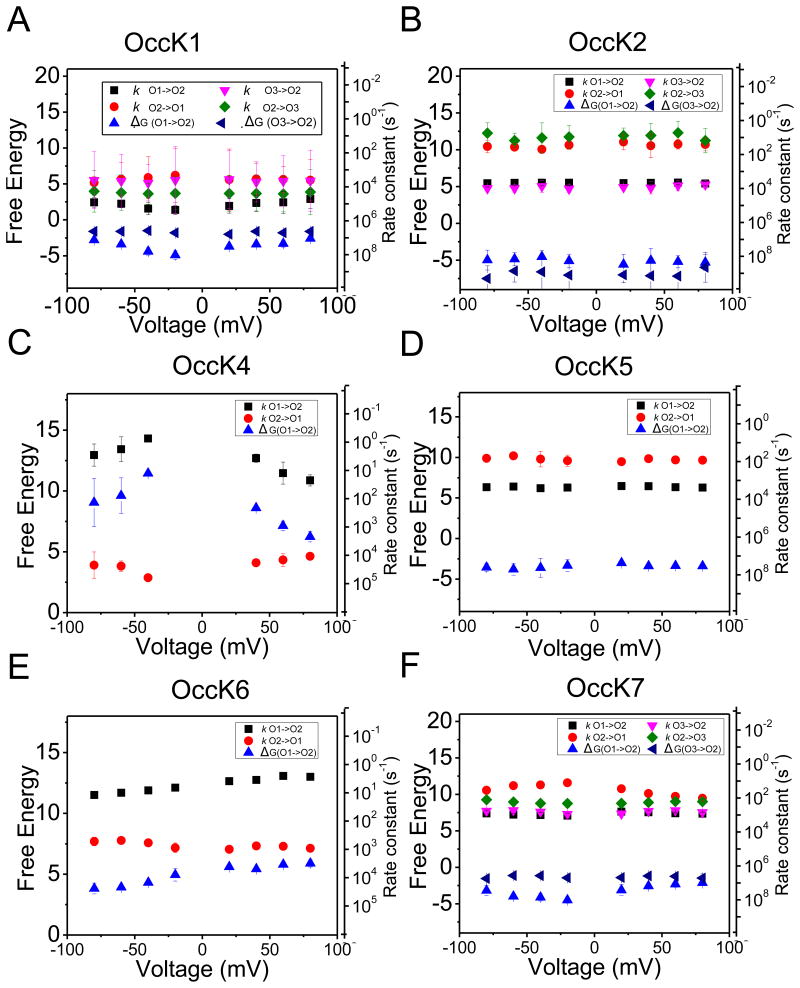
Figure 5. The kinetic rate constants of the current gating transitions and their corresponding free energy differences as well as estimated activation free energies.
(A) OccK2; (B) OccK4; (C) OccK5; (D) OccK6; (E) OccK7. The activation free energies were estimated using a frequency factor of 10−6 s−1 (30). The left y axes are the free energies in RT units, whereas the right y axes are the corresponding kinetic rate constants. Note that the kinetic rate constant axis is in a log scale, and the positive direction is downwards.
k is the kinetic rate constant. ff indicates the frequency factor, whereas ΔG≠ denotes the activation free energy. R and T are the gas constant and the absolute temperature, respectively.
The frequency factor in single-channel kinetics is usually taken as 106 s−1 (30). After calculating the activation free energy, we can determine the free energy difference (ΔG°) between two open sub-states. The free energy difference has a clear physical meaning, i.e., it reflects the probability ratio between different open sub-states:
| (2) |
In Fig. 5, we also show the scale of activation free energies. We present the kinetic rate constants on the right y-axis and the corresponding free energy expressed in RT units on the left y-axis to show the voltage-dependence of these transitions. Note that as the activation free energy increases the rate constant decreases. The rate constant axis is in logarithm scale and the positive direction is downwards. We were not able to find a voltage dependence of the kinetic rate constants for protein channels such as OccK2, OccK5 and OccK7. For instance, kO1→O2, kO2→O1, kO3→O2 and kO2→O3 of the OccK2 protein were always in the rage ~4200 ± 1500 s−1, ~35 ± 15 s−1, ~8000 ± 3000 s−1, and ~15 ± 10 s−1, respectively, regardless of the applied voltage (Fig. 5A). Similarly, kO1→O2 and kO2→O1 of the OccK5 protein were always ~1800 ± 300 s−1 and ~65 ± 15 s−1, respectively (Fig 5C). OccK7 exhibited a similar voltage independent kinetic profile (Fig. 5E). In contrast, the low-conductance OccK4 and OccK6 channels revealed a voltage dependence of their kinetic rate constants and of their free energy difference between the open sub-states (Fig. 5C and Fig. 5E).
Voltage dependence of the free energy landscapes of the low-conductance OccK4 and OccK6 channels
We constructed the voltage-dependent models of the free energy landscape of the low-conductance OccK protein channels (Fig. 6A and Fig. 6B). OccK4 displayed a greater kinetic rate constant from O1 to O2 at increased transmembrane potentials. For example, kO1→O2 was 3.2 ± 1.1 s−1 and 20.0 ± 8.9 s−1 at a transmembrane potential of +40 and +80 mV, respectively. In contrast, kO2→O1 decreased at increasing transmembrane potential. At +40 mV, kO2→O1 had a value of (16.9 ± 4.1) × 103 s−1, whereas at +80 mV, its value was (9.7 ± 0.3) × 103 s−1. These results were also obtained at a negative polarity of the applied transmembrane potential, revealing symmetry of the kinetic transitions in OccK4 with respect to voltage. We conclude that increasing the applied transmembrane potential decreases the activation free energy of the O1→O2 current transitions, but increases the activation free energy of the O2→O1 current transitions (Fig. 6A). On the other hand, OccK6 showed a monotonic decrease in the kO1→O2 kinetic rate constant from 10.1 ± 1.3 to 2.3 ± 0.3 s−1, when the transmembrane potential increased from −80 mV to +80 mV, whereas the kO2→O1 kinetic rate constant remained unaltered in the range ~650 ± 150 s−1. Therefore, the activation free energy of the O1→O2 current fluctuations increased when the voltage shifted towards more positive values, whereas the activation free energy of the O2→O1 current fluctuations did not undergo a statistically significant alteration (Fig. 6B).
Figure 6. The voltage-dependent free energy landscapes of the low-conductance OccK4 and OccK6 channels.
(A) Voltage dependence of the free energy landscape of the OccK4 channel; (B) Voltage dependence of the free energy landscape of the OccK6 channel.
Detailed inspection of the ion selectivity of the OccK1 channel, the archetype of the OccK subfamily
We explored the preferential permeability of OccK1 for cations versus anions in the form of permeability ratio (PK/PCl). The salt gradient assay was employed to determine the reversal potential (Vr). The calculation of the permeability ratio was achieved using the Goldman–Hodgkin–Katz formalism (31;32):
| (3) |
where the variable a represents the activity of either potassium or chloride in either the cis (subscript “c”) or trans (subscript “t”) chamber. Here, F, R and T are the Faraday constant, the gas constant and the absolute temperature, respectively.
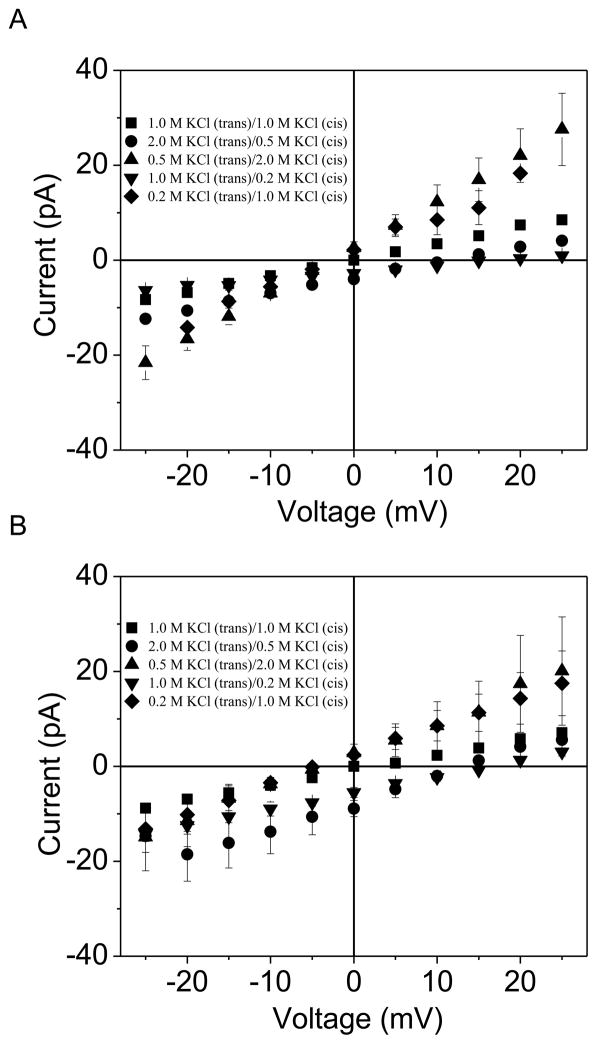
First, we comprehensively inspected the ion selectivity of the OccK1 channel under various experimental circumstances, including different salt concentration gradients and different pH values. The current-voltage curves, which were determined under asymmetric ionic concentrations, are shown in Fig. 7. The x-axis intercept of the I–V curve is the desired reversal potential, which is used to offset the resting potential, making the current zero. In Table 2, we present the reversal potential Vr and the calculated PK/PCl (eqn. (3)). Here, we define a weakly anion-selective, an anion-selective and a strongly anion selective channel as an OM protein whose permeability ratio PK/PCl is in the range 0.2–1.0 (PCl/PK is in the range 1–5), 0.1–0.2 (PCl/PK ≈ 5–10) and 0.01–0.1 (PCl/PK ≈ 10–100), respectively.
Figure 7. The I–V profiles of the OccK1 channel under asymmetric salt concentrations.
(A) pH 6.0; (B) pH 8.0. The corresponding numerical results pertinent to the ion selectivity features of OccK1 are shown in Table 2. The buffer solution contained 10 mM potassium phosphate.
Table 2. Permeability ratios and reversal potentials determined for the OccK1 protein under various experimental conditions.
(A) pH 6.0; (B) pH 8.0. The single-channel data were obtained under asymmetric conditions. The permeability ratio PK/PCl was calculated using reversal potential (Vr). The buffer solutions contained 10 mM potassium phosphate. The data represents averages ± SDs over a number of at least three distinct single-channel recordings.
| A. | |||
|---|---|---|---|
| cis cKCl (M) | trans cKCl (M) | Vr (mV) | PK/PCl |
| 1 | 0.2 | −3.2 ± 0.4 | 0.79 ± 0.04 |
| 0.2 | 1 | 13 ± 4 | 0.38 ± 0.07 |
| 2 | 0.5 | −3.1 ± 0.6 | 0.77 ± 0.06 |
| 0.5 | 2 | 12.1 ± 0.6 | 0.48 ± 0.08 |
| B. | |||
|---|---|---|---|
| cis cKCl (M) | trans cKCl (M) | Vr (mV) | PK/PCl |
| 1 | 0.2 | −6.4 ± 3.2 | 0.66 ± 0.12 |
| 0.2 | 1 | 15.9 ± 1.3 | 0.35 ± 0.03 |
| 2 | 0.5 | −3.8 ± 0.6 | 0.79 ± 0.03 |
| 0.5 | 2 | 14.1 ± 2.0 | 0.36 ± 0.05 |
In all asymmetric conditions, the OccK1 protein displayed a weak anion selectivity. For instance, under the experimental circumstances employed in this work, PK/PCl was in the range 0.35 – 0.79, so PCl/PK was in the range 1.3 – 2.9. Table 2A and Table 2B show the permeability ratios at pH 6.0 and 8.0, respectively, which give no statistically significant differences. The permeability ratios were greater when the KCl concentration was higher in the cis than in the trans chamber. The reversal potentials were positive when the KCl concentration was higher in the trans than in the cis chamber. For ccis = 0.2 M KCl and ctrans = 1 M KCl, the reversal potentials at pH 6.0 and 8.0 were 13 ± 4 mV and 16 ± 1 mV (n=3), respectively. On the other hand, for ccis = 1 M KCl and ctrans = 0.2 M KCl, the reversal potentials at pH 6.0 and 8.0 were −3.2 ± 0.4 mV and −6.4 ± 3.2 mV (n=3), respectively. Although different salt gradients resulted in slightly different values of the selectivity (from ~0.4 to ~0.8), all these results hold the sign of the preferred charge, i.e., OccK1 channel always preferably allows Cl− to penetrate more than K+, regardless of the experimental circumstances. The weak anion selectivity of the OccK1 channel is likely a result of the basic arginine ladder on the pore walls (18). This observation is also in accord with the anionic nature of the preferred substrate of this channel (e.g., benzoate) (11).
The anion selectivity is a conserved feature in the OccK subfamily
We also inspected the ion selectivity of all the other six OccK channels under one similar experimental condition, i.e., 0.2 M KCl in cis chamber and 1 M KCl in trans chamber, 10 mM potassium phosphate, pH=7.4. All the results are presented in Table 3. Strikingly, except for OccK3, all OccK proteins were anion-selective. OccK5 was strongly anion selective, with PK/PCl values of 0.01 ± 0.02. This finding was well correlated with the presence of a net pool of seven positively charged residues in the central constriction of the OccK5 channel (Table 1). OccK7 exhibited moderate anion selectivity (PK/PCl=0.18 ± 0.02), whereas OccK1, OccK2, OccK4 and OccK6 were weakly anion selective, with PK/PCl ranging from ~0.3 through ~0.7. Overall, these results are in line with the net negative pool of positively charged residues in the central eyelet of the channel.
Table 3.
Ion selectivity of the OccK proteins.The reversal potential (Vr) is the applied transmembrane potential which offsets the resting potential of the channel in salt gradient of 0.2 M KCl on the cis side and 1 M KCl on the trans side. The chamber also contained 10 mM potassium phosphate, pH=7.4. The reversal potentials (Vr) were provided as averages ± SD over at least three independent single-channel experiments.
| Protein | Vr (mV) | PK/PCl | Selectivity |
|---|---|---|---|
| OccK1 | 15.9 ± 1.3 | 0.35 ± 0.03 | Weakly anion-selective channel |
| OccK2 | 7.9 ± 1.0 | 0.63 ± 0.04 | Weakly anion-selective channel |
| OccK3 | ~0 | ~1 | Non-selective channel |
| OccK4 | 13.3 ± 1.5 | 0.45 ± 0.04 | Weakly anion-selective channel |
| OccK5 | 42.7 ± 3.1 | ~0.01 ± 0.02 | Strongly anion-selective channel |
| OccK6 | 6.7 ± 2.5 | 0.68 ± 0.10 | Weakly anion-selective |
| OccK7 | 25.7 ± 1.5 | 0.18 ± 0.02 | Anion-selective channel |
Discussion
In this work, we carried out an extensive single-channel examination of the members of the OccK subfamily from P. aeruginosa. This detailed study was prompted by recent biochemical, functional and structural analysis of the substrate specificity among the outer membrane carboxylate channels (Occ) (11). These OM protein channels exhibited diverse single-channel electrical signatures, including a broad range of unitary conductance, between 40 and 380 pS, a wide variety of transient current gating fluctuations, departing from the most probable open sub-state to other secondary open sub-states of shorter duration, and a spectrum of non-ohmic voltage dependence of their conductance. The OccK subfamily proteins displayed low- and medium-conductance open sub-states. For example, in the case of OccK1, OccK2, OccK5 and OccK7, the channel mostly remained on the medium-conductance O2 open sub-state (240–380 pS), from which either reversible fluctuations toward the low-conductance O1 open sub-state (OccK5) or multiple current transitions comprising a low-conductance O1 open sub-state and a medium-conductance O3 open sub-state (OccK1, OccK2, and OccK7) were observed.
For the three-open sub-state channels, we were not able to find direct transitions between the O1 and O3 open sub-states, suggesting that the O1 ↔ O2 and O2 ↔ O3 current transitions were made by distinct parts of the protein that fluctuated independently from each other. Another common feature among this group of channels is that the O2 ↔ O3 current transitions occurred with a greater kinetic rate constant than the O1 ↔ O2 current transitions. Moreover, in OccK1 (19), OccK2 and OccK7 channels, the high-frequency O2 ↔ O3 current transitions displayed a current amplitude lower than that value corresponding to the low-frequency O1 ↔ O2 current transitions (Fig. 2, Fig. 3 and Fig. 4). These striking similarities in the current amplitudes and event frequencies of the gating transitions observed in this work indicate that OccK1, OccK2 and OccK7 have similar mechanisms for the discrete current transitions among open sub-states O1, O2 and O3.
In contrast, low-conductance OccK4 and OccK6 proteins dwelled for long periods on the low-conductance O1 open sub-state (40–70 pS), and fluctuated in the form of infrequent and short-lived upward current transitions to a medium-conductance O2 open sub-state (302–358 pS). However, one distinction between the OccK4 and OccK6 protein channels is the voltage dependence of their current transitions (Fig. 6). This finding suggests that different mechanisms of the fluctuating parts of the low-conductance OccK channels are involved, resulting in upward large-amplitude current spikes that produce a more permeable pore. The members of the Occ family feature ~40~50% sequence similarity, and the OccK subfamily members share even a greater sequence similarity ~50%~55% (Supplementary Information, Fig. S1) (3;7;11). According to the recently determined, high-resolution crystal structures of the OccK proteins, the diameter of their constriction, including the molecular surface generated by the van der Waals radii, varies in a narrow range, from ~3.5 Å through ~5Å (Table 1). On the other hand, the X-ray crystal structures of the OccK proteins were determined at widely varying pH values, between 4 and 8.5 (11).
The unitary conductance should be a complex function that depends not only on the cross-sectional diameter of the central constriction, but also the average diameter along the longitudinal axis along the channel lumen, and the charge distribution throughout the inner surface of the protein. The most probable open sub-state in the OccK protein depends on the orientation of the large extracellular loops L3, L4 and L7 within the pore lumen. The single-channel electrical signatures of these OccK proteins are highly diverse (Fig. 2), suggesting that, despite of their sequence homology and closely similar cross-sectional diameter of the central constriction, minor alterations in the length, packing and structure of the extracellular loops located within the channel lumen (Supplementary Information, Fig. S2) have drastic implications on the unitary conductance as well as the functionality of these proteins (33;34).
Several experimental details of our single-channel electrical recordings, such as the lipid composition of the membranes, the applied transmembrane potential and the ion concentration in the chamber, depart from the physiological environment of P. aeruginosa (2;35). Given the duration of the fast and transient current fluctuations, it is conceivable that the single-channel current measured with the members of the OccK subfamily in 150 mM KCl and at a transmembrane potential that is specific to outer membrane proteins (~several mV) would not enable the detection of resolvable open sub-states. Therefore, a meaningful comparison among all members of the OccK subfamily required a substantial improvement in the signal-to-noise ratio, which was accomplished by using a buffer solution containing 1 M KCl and greater transmembrane potential values. One concern of using a higher salt concentration is the alteration of the sub-state dynamics. In a recent work, the change in the salt concentration featured a substantial impact on the single-channel electrical signature of the staphylococcal α-hemolysin (36). However, for the OccK1 protein, we performed single-channel electrical recordings using 500 mM KCl in the chamber, which is much closer to the physiological condition. In this case, we observed a conserved number of open sub-states (O1, O2 and O3) (19).
Interestingly, we found that the OccK proteins are anion selective, a feature conserved among the members of this subfamily. Our investigations also show a detailed analysis of the selectivity measurements on the OccK1 channel, which were performed under different KCl gradient and pH conditions. We were not able to find an impact of pH, in the range 6–8, on the ionic selectivity of the OccK1 channel. This result suggests a lack of chargeable groups within the channel lumen under these conditions. Indeed, a careful inspection of the X-ray crystal structure of this channel revealed that histidine residues (pKa = 6) are located outside the channel lumen. The highly conserved anion selectivity among the members of the OccK subfamily in P. aeruginosa is in accord with the net pool of positively charged residues, such as arginines and lysines, in the central constriction of the channel (Table 1; Supplementary Information, Fig. S1). The OccK proteins, whose crystal structure are available, show either one (OccK3, OccK4), two (OccK1, OccK6), four (OccK2) or seven (OccK5) net positive charges in the channel eyelet. In agreement with these structural distinctions among the members of the OccK subfamily, we determined that OccK5 is a strongly anion-selective channel (Table 3). This correlation also holds for OccK3 and OccK4, which are non-selective and poorly anion-selective channels, respectively. These ion selectivity results are also compatible with the substrate specificity of the OccK subfamily members. For example, carboxylate-containing benzoate, gluconorate and pyroglutamate are the preferred substrates for OccK1, OccK2 and OccK3, respectively (11). On the contrary, the OccD subfamily members do not transport these molecules. Most of the OccD channels transport arginine very well. Shall we anticipate that the OccD subfamily members are cation selective? If this is true, then this difference in ion selectivity among the members of the OccK and OccD subfamilies will add to the already known structural, phylogenetic and functional distinctions.
The positively charged residues within the central constriction of the OccK proteins might function as catalyzers for the navigation of carboxylate-containing substrates across the channel lumen. This mechanism is also present in other substrate-specific outer membrane proteins, such as the maltoporin of E. coli, which represents the pathway for the sugar uptake. In this case, the attractive sites are aromatic residues, thus reducing the activation free energies for sugar translocation from one side of the membrane to the other (37). In the past, we also demonstrated that attractive sites present within the channel lumen have major implications even for larger molecules, such as polypeptides traversing the channel (29;38), which is in accord with the theoretical predictions (39–42).
One question that emerges from this work is how the OccK channels compare with other well-studied substrate-specific or nonspecific outer membrane porins. The medium-conductance OccK channels have a greater conductance than that of the sugar-specific LamB porin from E. coli (20). LamB features a single-channel conductance of ~53 pS per each monomer in 1 KCl (43). This value is comparable with the single-channel conductance of the low-conductance OccK4 (~43 pS) and OccK6 (~71 pS) channels. This work also confirms that the OccK channels show an unitary conductance that is smaller than that value corresponding to nonspecific outer membrane porins from E. coli, such as the trimeric OmpF (~700 pS per monomer in 1 M KCl (44;45)) and OmpC (~900 pS per monomer in 1 M KCl (46)) channels. Each monomer of these outer membrane proteins forms a 16-stranded β barrel with a central constriction made by the inwardly folded L3 extracellular loop (47).
How do the OccK and OccD subfamily members compare each other? Recent work performed by our teams demonstrated that the observed most probable conductance values of the OccD and OccK subfamily members are generally in accord with the available crystal structures (11). For instance, the most probable conductance of OccD1 and OccD2 is about ~20 pS, which is very consistent with the very small diameter of these channels (less than 3 Ă) (17). These results are also in accordance with prior electrophysiological studies performed by Ishii and Nakae, who discovered a very small unitary conductance (~30 pS) in recordings with the OccD1 channel (48;49). The OccK subfamily includes a channel, OccK4, which also exhibits a very low conductance (~43 pS; Table 1). However, we determined that the most probable unitary conductance of the OccD3 channel is ~700 pS, the largest value among all examined OccD and OccK channels (11). This observation is not compatible with the small diameter of the channel eyelet of the OccD3 protein. Obviously, the length, conformation and location of each extracellular loop impact the single-channel conductance in a similar way in which a water-soluble polymer blocks the ionic flow (50;51). Huang and Hancock (1996) found that a deletion in loop L5 of the OccD1 channel produced a 33-fold increase in the single-channel conductance (52).
Very interestingly, Ishii and Nakae (1993) discovered that the OccD1 channel, the archetype of the OccD subfamily in P. aeruginosa, exhibited highly frequent, medium-conductance (~400 pS in 1 M KCl) openings in lipopolysacharide (LPS)-containing lipid bilayers (49). In LPS-free lipid bilayers, OccD1 showed a low-conductance (30 pS). However, in the LPS-containing lipid bilayers, the channel fluctuated between low- (~30 pS) and medium-conductance (~400 pS) open sub-states. Therefore, the lipid environment influenced the functional features of the OccD1 channel. This finding represents strong evidence that the composition of the lipid bilayer impacts the discrete dynamics of the open sub-states of the OccD1 channel. We speculate that LPS-containing bilayers might be an energetically favorable environment for transient openings of the Occ channels, promoting the transport of specific substrates or even antibiotics.
Molecular dynamics (MD) simulations show a great promise in obtaining a qualitative understanding of the stochastic motions of the extracellular long loops folded back into the channel lumen (53;54). For example, Bond and colleagues employed MD simulations to study the conformations of the extracellular loops in the outer membrane protein OpcA from Neisseria Meningitidis (55). They found that the lipid bilayer environment and physiological salt concentrations affected the conformation of the extracellular loops in OpcA. Therefore, it is conceivable that different crystallization conditions might influence the orientation of the extracellular loops folding back into the channel interior. Moreover, this experimental work might ignite future full-atomistic computational biophysics studies for obtaining a clarification on how small molecules, such as substrates and antibiotics traverse the complex environment of the channel lumen of the OccK subfamily members (56–58).
In summary, we show a detailed high-resolution, single-channel analysis of seven members of the OccK subfamily from P. aeruginosa. The electrical signature of the OccK proteins revealed diverse unitary conductance values with well-defined single-channel discrete dynamics, which is comprised of low- and medium-conductance open sub-states. The current gating transitions among various open sub-states were likely produced by stochastic fluctuations of the long extracellular loops folded back into the channel lumen. We found that the OccK proteins are anion selective, a feature that is explained at least in part by the presence of the basic ladder of arginine and lysine residues near the central constriction, and that is in accord with structural and in vitro substrate-transport reconstitution studies.
Supplementary Material
Acknowledgments
We also thank colleagues in the Movileanu and van den Berg research groups, who provided technical assistance at various stages of this work. This paper was funded in part by grants from the US National Science Foundation (DMR-1006332, L.M.) and the National Institutes of Health (R01 GM088403, L.M. and R01 GM085785, B.v.d.B).
ABBREVIATIONS AND SYMBOLS
- LamB
Sugar-specific porin of E. coli
- MD
Molecular dynamics
- OM
Outer membrane of Gram-negative bacteria
- Occ
Outer membrane carboxylate channel family of P. aeruginosa
- OccD
Outer membrane carboxylate channel D subfamily of P. aeruginosa
- OccK
Outer membrane carboxylate channel K subfamily of P. aeruginosa
- OmpC
Outer membrane protein C of E. coli
- OmpF
Outer membrane protein F of E. coli
- OprF
Outer membrane protein A analogue porin of P. aeruginosa
Footnotes
Supporting Materials and Methods Available: (i) amino acid sequence analysis of seven OccK subfamily members, (ii) analysis of the single-channel open sub-state transitions of the OccK proteins, (iii) a three-sub-state kinetic model for the current fluctuations in the OccK proteins, (iv) the standard free energies corresponding to various gating transitions in the OccK1 protein are available via the internet at http://www.acs.org
References
- 1.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991;266:770–779. [PubMed] [Google Scholar]
- 2.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock RE, Brinkman FS. Function of pseudomonas porins in uptake and efflux. Annu Rev Microbiol. 2002;56:17–38. doi: 10.1146/annurev.micro.56.012302.160310. [DOI] [PubMed] [Google Scholar]
- 4.Nestorovich EM, Sugawara E, Nikaido H, Bezrukov SM. Pseudomonas aeruginosa porin OprF: properties of the channel. J Biol Chem. 2006;281:16230–16237. doi: 10.1074/jbc.M600650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benz R, Hancock RE. Mechanism of ion transport through the anion-selective channel of the Pseudomonas aeruginosa outer membrane. J Gen Physiol. 1987;89:275–295. doi: 10.1085/jgp.89.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sukhan A, Hancock RE. The role of specific lysine residues in the passage of anions through the Pseudomonas aeruginosa porin OprP. J Biol Chem. 1996;271:21239–21242. doi: 10.1074/jbc.271.35.21239. [DOI] [PubMed] [Google Scholar]
- 7.Hancock REW, Tamber S. Porins of the Outer Membrane of Pseudomonas aeruginosa. In: Benz R, editor. Bacterial and Eukaryotic Porins: Structure, Function, Mechanism. Wiley-VCH; Weinheim: 2004. pp. 61–77. [Google Scholar]
- 8.Schirmer T. General and specific porins from bacterial outer membranes. J Struct Biol. 1998;121:101–109. doi: 10.1006/jsbi.1997.3946. [DOI] [PubMed] [Google Scholar]
- 9.Moraes TF, Bains M, Hancock RE, Strynadka NC. An arginine ladder in OprP mediates phosphate-specific transfer across the outer membrane. Nat Struct Mol Biol. 2007;14:85–87. doi: 10.1038/nsmb1189. [DOI] [PubMed] [Google Scholar]
- 10.Sampathkumar P, Lu F, Zhao X, Li Z, Gilmore J, Bain K, Rutter ME, Gheyi T, Schwinn KD, Bonanno JB, Pieper U, Fajardo JE, Fiser A, Almo SC, Swaminathan S, Chance MR, Baker D, Atwell S, Thompson DA, Emtage JS, Wasserman SR, Sali A, Sauder JM, Burley SK. Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution. Proteins. 2010;78:3056–3062. doi: 10.1002/prot.22829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eren E, Vijayaraghavan J, Liu J, Cheneke BR, Touw DS, Lepore BW, Indic M, Movileanu L, van den Berg B. Substrate specificity within a family of outer membrane carboxylate channels. PLoS Biology. 2012;10:e1001242. doi: 10.1371/journal.pbio.1001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamber S, Hancock RE. Involvement of two related porins, OprD and OpdP, in the uptake of arginine by Pseudomonas aeruginosa. FEMS Microbiol Lett. 2006;260:23–29. doi: 10.1111/j.1574-6968.2006.00293.x. [DOI] [PubMed] [Google Scholar]
- 13.Tamber S, Ochs MM, Hancock RE. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J Bacteriol. 2006;188:45–54. doi: 10.1128/JB.188.1.45-54.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochs MM, Lu CD, Hancock RE, Abdelal AT. Amino acid-mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa. J Bacteriol. 1999;181:5426–5432. doi: 10.1128/jb.181.17.5426-5432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochs MM, McCusker MP, Bains M, Hancock RE. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother. 1999;43:1085–1090. doi: 10.1128/aac.43.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 17.Biswas S, Mohammad MM, Patel DR, Movileanu L, van den Berg B. Structural insight into OprD substrate specificity. Nat Struct Mol Biol. 2007;14:1108–1109. doi: 10.1038/nsmb1304. [DOI] [PubMed] [Google Scholar]
- 18.Biswas S, Mohammad MM, Movileanu L, van den Berg B. Crystal structure of the outer membrane protein OpdK from Pseudomonas aeruginosa. Structure. 2008;16:1027–1035. doi: 10.1016/j.str.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Cheneke BR, van den Berg B, Movileanu L. Analysis of gating transitions among the three major open states of the OpdK channel. Biochemistry. 2011;50:4987–4997. doi: 10.1021/bi200454j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 2008;27:2171–2180. doi: 10.1038/emboj.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sackmann B, Neher E. Single-Channel Recording. Kluwer Academic/Plenum Publishers; New York: 1995. [Google Scholar]
- 23.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promotor. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den BB, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 25.Howorka S, Movileanu L, Lu XF, Magnon M, Cheley S, Braha O, Bayley H. A protein pore with a single polymer chain tethered within the lumen. J Am Chem Soc. 2000;122:2411–2416. [Google Scholar]
- 26.Goodrich CP, Kirmizialtin S, Huyghues-Despointes BM, Zhu AP, Scholtz JM, Makarov DE, Movileanu L. Single-molecule electrophoresis of beta-hairpin peptides by electrical recordings and Langevin dynamics simulations. J Phys Chem B. 2007;111:3332–3335. doi: 10.1021/jp071364h. [DOI] [PubMed] [Google Scholar]
- 27.Colquhoun D, Sigworth FJ. Fitting and statistical analysis of single-channel records. In: Sackmann BNE, editor. Single-channel recording. 2. Plenum Press; New York: 1995. pp. 483–587. [Google Scholar]
- 28.Movileanu L, Cheley S, Bayley H. Partitioning of individual flexible polymers into a nanoscopic protein pore. Biophys J. 2003;85:897–910. doi: 10.1016/S0006-3495(03)74529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammad MM, Movileanu L. Excursion of a single polypeptide into a protein pore: simple physics. but complicated biology. Eur Biophys J. 2008;37:913–925. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 30.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 31.Hille B. Ion Channels of Excitable Membranes. Sinauer Associates. Inc; Sunderland, Massachusetts, USA: 2001. [Google Scholar]
- 32.Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L. Catalyzing the translocation of polypeptides through attractive interactions. J Am Chem Soc. 2007;129:14034–14041. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 33.Jung Y, Bayley H, Movileanu L. Temperature-responsive protein pores. J Am Chem Soc. 2006;128:15332–15340. doi: 10.1021/ja065827t. [DOI] [PubMed] [Google Scholar]
- 34.Mohammad MM, Howard KR, Movileanu L. Redesign of a plugged beta-barrel membrane protein. J Biol Chem. 2011;286:8000–8013. doi: 10.1074/jbc.M110.197723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen K, Hellman J, Nikaido H. Porin channels in intact cells of Escherichia coli are not affected by Donnan potentials across the outer membrane. J Biol Chem. 1988;263:1182–1187. [PubMed] [Google Scholar]
- 36.Mohammad MM, Movileanu L. Impact of distant charge reversals within a robust beta-barrel protein pore. J Phys Chem B. 2010;114:8750–8759. doi: 10.1021/jp101311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilty C, Winterhalter M. Facilitated substrate transport through membrane proteins. Phys Rev Lett. 2001;86:5624–5627. doi: 10.1103/PhysRevLett.86.5624. [DOI] [PubMed] [Google Scholar]
- 38.Bikwemu R, Wolfe AJ, Xing X, Movileanu L. Facilitated translocation of polypeptides through a single nanopore. J Phys:Condens Matter. 2010;22:454117. doi: 10.1088/0953-8984/22/45/454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berezhkovskii AM, Pustovoit MA, Bezrukov SM. Channel-facilitated membrane transport: Transit probability and interaction with the channel. J Chem Phys. 2002;116:9952–9956. [Google Scholar]
- 40.Berezhkovskii AM, Bezrukov SM. Optimizing transport of metabolites through large channels: molecular sieves with and without binding. Biophys J. 2005;88:L17–L19. doi: 10.1529/biophysj.104.057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolomeisky AB, Kotsev S. Effect of interactions on molecular fluxes and fluctuations in the transport across membrane channels. J Chem Phys. 2008;128:085101. doi: 10.1063/1.2831801. [DOI] [PubMed] [Google Scholar]
- 42.Bauer WR, Nadler W. Molecular transport through channels and pores: effects of in-channel interactions and blocking. Proc Natl Acad Sci U S A. 2006;103:11446–11451. doi: 10.1073/pnas.0601769103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benz R, Schmid A, Nakae T, Vos-Scheperkeuter GH. Pore formation by LamB of Escherichia coli in lipid bilayer membranes. J Bacteriol. 1986;165:978–986. doi: 10.1128/jb.165.3.978-986.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saint N, Lou KL, Widmer C, Luckey M, Schirmer T, Rosenbusch JP. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II Functional characterization. J Biol Chem. 1996;271:20676–20680. [PubMed] [Google Scholar]
- 45.Chimerel C, Movileanu L, Pezeshki S, Winterhalter M, Kleinekathofer U. Transport at the nanoscale: Temperature dependance of ion conductance. Eur Biophys J. 2008;38:121–125. doi: 10.1007/s00249-008-0366-0. [DOI] [PubMed] [Google Scholar]
- 46.Biro I, Pezeshki S, Weingart H, Winterhalter M, Kleinekathofer U. Comparing the temperature-dependent conductance of the two structurally similar E. coli porins OmpC and OmpF. Biophys J. 2010;98:1830–1839. doi: 10.1016/j.bpj.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowan SW, Garavito RM, Jansonius JN, Jenkins JA, Karlsson R, Konig N, Pai EF, Pauptit RA, Rizkallah PJ, Rosenbusch JP. The structure of OmpF porin in a tetragonal crystal form. Structure. 1995;3:1041–1050. doi: 10.1016/s0969-2126(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 48.Ishii J, Nakae T. Specific interaction of the protein-D2 porin of Pseudomonas aeruginosa with antibiotics. FEMS Microbiol Lett. 1996;136:85–90. doi: 10.1016/0378-1097(95)00497-1. [DOI] [PubMed] [Google Scholar]
- 49.Ishii J, Nakae T. Lipopolysaccharide promoted opening of the porin channel. FEBS Lett. 1993;320:251–255. doi: 10.1016/0014-5793(93)80597-n. [DOI] [PubMed] [Google Scholar]
- 50.Movileanu L, Bayley H. Partitioning of a polymer into a nanoscopic protein pore obeys a simple scaling law. Proc Natl Acad Sci U S A. 2001;98:10137–10141. doi: 10.1073/pnas.181089798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Movileanu L, Cheley S, Howorka S, Braha O, Bayley H. Location of a constriction in the lumen of a transmembrane pore by targeted covalent attachment of polymer molecules. J Gen Physiol. 2001;117:239–251. doi: 10.1085/jgp.117.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H, Hancock RE. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol. 1996;178:3085–3090. doi: 10.1128/jb.178.11.3085-3090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luan B, Caffrey M, Aksimentiev A. Structure refinement of the OpcA adhesin using molecular dynamics. Biophys J. 2007;93:3058–3069. doi: 10.1529/biophysj.107.106724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luan B, Carr R, Caffrey M, Aksimentiev A. The effect of calcium on the conformation of cobalamin transporter BtuB. Proteins. 2010;78:1153–1162. doi: 10.1002/prot.22635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bond PJ, Derrick JP, Sansom MS. Membrane simulations of OpcA: gating in the loops? Biophys J. 2007;92:L23–L25. doi: 10.1529/biophysj.106.097311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hajjar E, Bessonov A, Molitor A, Kumar A, Mahendran KR, Winterhalter M, Pages JM, Ruggerone P, Ceccarelli M. Toward screening for antibiotics with enhanced permeation properties through bacterial porins. Biochemistry. 2010;49:6928–6935. doi: 10.1021/bi100845x. [DOI] [PubMed] [Google Scholar]
- 57.Hajjar E, Mahendran KR, Kumar A, Bessonov A, Petrescu M, Weingart H, Ruggerone P, Winterhalter M, Ceccarelli M. Bridging timescales and length scales: from macroscopic flux to the molecular mechanism of antibiotic diffusion through porins. Biophys J. 2010;98:569–575. doi: 10.1016/j.bpj.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar A, Hajjar E, Ruggerone P, Ceccarelli M. Molecular simulations reveal the mechanism and the determinants for ampicillin translocation through OmpF. J Phys Chem B. 2010;114:9608–9616. doi: 10.1021/jp9110579. [DOI] [PubMed] [Google Scholar]
- 59.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–60. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 60.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.