Abstract
Hydroxyproline (Hyp) metabolism is a key source of glyoxylate production in the body and may be a major contributor to excessive oxalate production in the primary hyperoxalurias where glyoxylate metabolism is impaired. Important gaps in our knowledge include identification of the tissues with the capacity to degrade Hyp and the development of model systems to study this metabolism and how to suppress it. The expression of mRNA for enzymes in the pathway was examined in 15 different human tissues. Expression of the complete pathway was identified in liver, kidney, pancreas, and small intestine. HepG2 cells also expressed these mRNAs and enzymes and were shown to metabolize Hyp in the culture medium to glycolate, glycine, and oxalate. [18O]- and [13C5]Hyp were synthesized and evaluated for their use with in vitro and in vivo models. [18O]Hyp was not suitable because of an apparent tautomerism of [18O]glyoxylate between enol and hydrated forms, which resulted in a loss of isotope. [13C5]Hyp, however, was metabolized to [13C2]glycolate, [13C2]glycine, and [13C2]oxalate in vitro in HepG2 cells and in vivo in mice infused with [13C5]Hyp. These model systems should be valuable tools for exploring various aspects of Hyp metabolism and will be useful in determining whether blocking Hyp catabolism is an effective therapy in the treatment of primary hyperoxaluria.
Keywords: oxalate, glycolate, glyoxylate
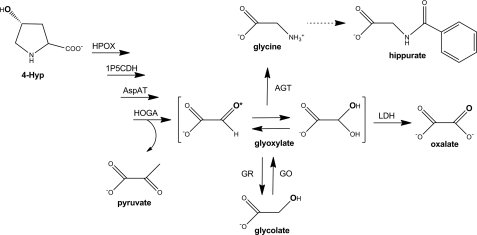
degradation of the amino acid 4-hydroxyproline (Hyp) results in the formation of glyoxylate and is the largest contributor to the glyoxylate pool in mammals yet identified (12). Glyoxylate synthesis has clinical significance, as it is the only known precursor of oxalate. An overproduction of oxalate can lead to the formation of kidney stones and the failure of organs due to crystal deposition. The importance of pathways involving glyoxylate is evident in the primary hyperoxalurias (PH), rare diseases where oxalate is overproduced. In PH type 1, mutations in alanine:glyoxylate aminotransferase (AGT) reduce the transamination of glyoxylate to glycine (10). In PH type 2, mutations in glyoxylate reductase (GR) decrease the conversion of glyoxylate to glycolate. A novel form of PH, type 3, which is caused by mutations in 4-hydroxy-2-oxoglutarate aldolase (HOGA) of the Hyp catabolism pathway, was recently identified (6, 22, 26).
Collagen is the main Hyp-containing protein (12–14% Hyp) in the body. Turnover of collagen is estimated to be 2–3 g/day (24). The resulting release of 300–450 mg of Hyp must be degraded or excreted, as it cannot be reutilized for protein synthesis. The urinary excretion of <30 mg of Hyp within peptides and <5 mg of free Hyp indicates that most of the Hyp is metabolized (15). The diet can also be a source of Hyp from collagen in meat products and foods containing gelatin. We recently determined that high-protein diets containing chicken, beef, and pork as protein sources contributed ∼240 mg Hyp/day (17). Thus, significant levels of Hyp must be degraded on a daily basis, which may amount to >500 mg Hyp/day when moderate levels of animal protein are consumed. The degradation of 500 mg of Hyp would result in the formation of ∼280 mg glyoxylate/day. On the basis of our current understanding of glyoxylate metabolism, the glyoxylate from Hyp catabolism should be predominantly converted to glycine and glycolate by AGT and GR, respectively (Fig. 1). In the liver, glycolate oxidase (GO) could convert much of the created glycolate back to glyoxylate in the peroxisome, where it could be transaminated to glycine by AGT (23).
Fig. 1.
Degradation of 4-hydroxyproline (Hyp) in mammals and its influence on glyoxylate metabolism. Conversion of Hyp to glyoxylate occurs through consecutive action of 4 mitochondrial enzymes: Hyp oxidase (HPOX), Δ1-pyrroline-5-carboxylate dehydrogenase (1P5CDH), aspartate aminotransferase (AspAT), and 4-hydroxy-2-oxoglutarate adolase (HOGA). A variety of enzymes, including alanine:glyoxylate aminotransferase (AGT), lactate dehydrogenase (LDH), and glyoxylate reductase (GR), can act on glyoxylate produced from 4-hydroxy-2-oxoglutarate. Glyoxylate primarily exists as its gem-diol hydrate. As such, the 18O label can be lost from the position indicated by the asterisk. Glycolate can be converted back to glyoxylate by glycolate oxidase (GO). Glycine can be further metabolized through several steps to yield hippurate. Sites of hydroxylation and 18O labeling by Phy1 are shown in bold.
The liver and kidney are known to be major sites of Hyp degradation (1, 15). Additional tissues active in Hyp metabolism are uncertain in humans, but this metabolism has been suggested to occur in the small intestine and heart (3, 8). The enzymes involved in the conversion of Hyp to glyoxylate are known to be localized in mitochondria (1, 24). This degradation process (Fig. 1) involves the step-wise action of hydroxyproline oxidase (HPOX), Δ1-pyrroline-5-carboxylate dehydrogenase (1P5CDH), aspartate aminotransferase (AspAT), and HOGA (1, 24, 31). Of the enzymes relevant to the metabolism of glyoxylate, GR is located in mitochondria and cytoplasm of all cells, whereas AGT and GO, expressed in the liver, are peroxisomal enzymes. The studies described here were designed to determine which tissues possess the enzymatic machinery to metabolize Hyp, to identify a suitable cell model to study this metabolism, and to determine whether isotopically labeled forms of Hyp, [13C5]Hyp and [18O]Hyp, would be suitable tools for studying the metabolism in vitro and in vivo. Such a model system would be useful for identifying functional roles of this pathway and possible ways of suppressing Hyp metabolism to glyoxylate.
METHODS
Analysis of Hyp catabolic pathway mRNA tissue distribution.
The human multiple-tissue cDNA panels MTC1 and MTC2 (Clonetech, Mountain View, CA) were screened using PCR and the primers designed to specifically target and amplify a 500- to 1,000-bp fragment of each target gene and one housekeeping gene, GAPDH, according to the instructions of the manufacturer. Each 15-μl reaction contained 0.5 μl of cDNA panel stock from one tissue, each primer at 430 nM, and the Stratagene Herculase polymerase (Agilent Technologies, Santa Clara, CA). The parameters were as follows: 95°C for 1 min followed by 24 cycles (GAPDH and actin) and 34 cycles (target genes) of 95°C for 30 s, 55°C for 30 s, and 68°C for 3 min. Densitometric analyses were performed using Quantity One (version 4.5.1, Bio-Rad, Hercules, CA).
A similar PCR-based analysis was performed for all the enzymes in the Hyp pathway in HepG2 cells and human liver. For these experiments, RNA was purified using the TRIzol procedure (Invitrogen, Carlsbad, CA), and cDNA was generated using the avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI); 5 μg of cDNA were used for the subsequent PCRs.
Western analysis of HPOX and HOGA expression.
Liver tissue was homogenized in hypotonic lysis buffer [25 mM HEPES (pH 7.1) and 0.1% Triton X-100] prior to analysis. Mitochondria from bovine kidney and HepG2 cells were isolated as previously described (18). After SDS-PAGE separation, the samples were electroblotted onto Hybond-C nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ). Membranes were incubated for 1 h in 5% (wt/vol) nonfat dry milk dissolved in PBS (pH 7.4) containing 1% (vol/vol) Tween 20 and for 1.5 h at room temperature with affinity-purified rabbit antibodies to human recombinant HOGA (1 μg/ml) or human recombinant HPOX (1 μg/ml), both of which were generated by Proteintech Laboratories (Chicago, IL). The membrane was then washed and incubated for 1 h at room temperature with peroxidase-conjugated goat anti-rabbit immunoglobulin G (0.2 μg/ml; Dako, Carpinteria, CA). Enhanced chemiluminescence (Amersham Biosciences) was used to visualize immunoreactive bands.
Synthesis of Hyp metabolic tracers.
13C and 18O isotopes of Hyp were synthesized using the recombinant Dactylsporangium proline 4-hydroxylase (Phy1), as described below. This enzyme stereospecifically adds a hydroxyl group to the 4 position of Pro to produce Hyp (29, 30). For [13C]Hyp, universally labeled Pro ([13C5]Pro), i.e., all five carbon atoms containing the 13C isotope, was used as the starting material (Cambridge Isotope Laboratories, Andover, MA). All restriction enzymes, chemicals, media, column materials, and 18O2 were purchased from New England Biolabs, Bio-Rad, and Sigma.
The gene for Phy1 (phy1) from Dactylsporangium sp. RH1 was synthesized by Blue Heron Biotechnology (Bothell, WA) using optimized codons for expression in Escherichia coli and cloned into the Nde I and BamH I sites of a modified pET19 vector (Novagen) with ampicillin resistance. In this construct, Phy1 contained an NH2-terminal poly-His tag and an intervening, engineered PreScission protease site (GE Healthcare, Piscataway, NJ). A 10-liter fermentation culture was inoculated with a 200-ml overnight culture of C41(DE3) E. coli cells containing the Phy1 expression plasmid and grown at 37°C and 200 rpm until the optical density at 600 nm reached 1.0. The temperature was reduced to 16°C, isopropyl β-d-1-thiogalactopyranoside was added to 0.5 mM, and the cells were incubated overnight.
The harvested cells (30–50 g) were resuspended in 100 ml of buffer A [50 mM Tris (pH 8.5), 500 mM KCl, 5 mM imidazole, 0.1% Triton X-100, and 10% glycerol] containing 1 mM MgCl2, 4 mg of DNase, and two Complete EDTA-free protease inhibitor tablets (Roche Applied Science, Indianapolis, IN). The cells were lysed by two passes through a cell disruptor (EmulsiFlex-C5, Avestin, Ottawa, ON, Canada). The cell lysate was centrifuged twice at 18,000 g. The resulting supernatant was loaded onto a Ni-NTA column (Qiagen, Valencia, CA) equilibrated with buffer A. The column was washed sequentially with buffer A and buffer B (buffer A without Triton X-100 and glycerol). Phy1 was eluted from the column with a 5-250 mM linear imidazole gradient in buffer B. The appropriate fractions, as judged by SDS-PAGE, were pooled and treated with PreScission protease (1:25 ratio) at 16°C for 3 days. Removal of the His tag was verified by SDS-PAGE. The protein was then concentrated to 5 ml and loaded onto a gel filtration column (HiLoad Superdex 75) equilibrated with 25 mM Tris (pH 7.5) and 100 mM NaCl. The appropriate fractions were pooled, concentrated to 52.4 mg/ml, divided into aliquots, and frozen with liquid N2 for storage at −80°C. The protein concentration was determined using the theoretical extinction coefficient (0.966 M−1·cm−1) determined by the PROTPARAM program on the EXPASY web server.
The initial conditions used to produce Hyp from Pro via the Phy1 reaction were based on the work by Shibasaki et al. (30). The reaction mixture contained 80 mM buffer, 4 mM Pro, 8 mM 2-oxoglutarate (2-OG), 2 mM ferrous sulfate, 4 mM l-ascorbic acid, and 0.08, 0.4, and 1.2 mg Phy1/ml. The efficiency of the reaction was tested using several temperatures (25–40°C) and buffers [citric acid (pH 5–5.5), MES (pH 6–6.5), HEPES (pH 7–7.5), and Tris (pH 8.5)]. The optimal conditions for the large-scale production of Hyp were 480 mM MES (pH 6.5), 24 mM l-Pro, 48 mM 2-OG, 12 mM ferrous sulfate, 24 mM l-ascorbic acid, and 0.4 mg Phy1/ml for [18O]Hyp production or 1.2 mg Phy1/ml for [13C5]Hyp production. For [18O]Hyp synthesis, a 10-ml reaction was performed in a multichamber tonometer connected to an oxygen-scavenged anaerobic train. Phy1 was kept separate from all the other reaction components in a sidearm until the entire vessel was made anaerobic by ≥12 cycles of vacuum administration and argon replacement. The vessel was then pressurized to 0.4 psi with 97% 18O2, and the reaction components were mixed. The solution was incubated at 35°C overnight. [13C5]Hyp was produced in a 40-ml reaction and did not require anaerobic conditions.
Purification of Hyp metabolic tracers.
The large-scale Phy1 reactions were quenched by incubation at 100°C for 2 min. Hyp was separated from the unreacted Pro by loading the supernatant onto a 1.3 × 50 cm H+-form AG 50W-X8 column (Bio-Rad) prewashed consecutively with 1 M HCl and water until the pH of the eluent was >4.5 (20). After the sample was loaded, the column was washed with 100 ml of water and then 50 ml of 40 mM HCl to remove the other reaction components. Hyp and Pro were differentially eluted with 0.5 M HCl at a flow rate of 0.5 ml/min. An aliquot of every fifth fraction was analyzed as described below to determine the location of the Hyp and Pro peaks. The majority of the HCl was removed from the Hyp by lyophilization. The remaining chloride was exchanged with formate by dissolution of the Hyp in 4 ml of deionized water and passage through a 1 × 30 cm formate-form AG1 X8 column (Bio-Rad) with deionized water as the mobile phase. The sodium in the Hyp was exchanged for NH4+ by adjustment of the pH to >12 with 12.7 M ultrapure NaOH and passage through a 1 × 30 cm NH4+-form AG 50W-X8 column with deionized water used as the mobile phase.
The final Hyp powder obtained after lyophilization was redissolved in water to make a 100 mM Hyp stock solution and quantified using the μHPLC assay. The purity of [18O]- and [13C]Hyp was determined by ion chromatography (IC) coupled with electrospray mass spectrometry (IC-MS; Dionex, Sunnyvale, CA). The IC step utilized a 2 × 250 mm Amino Pac PA10 column with guard column and a KOH gradient from 80 to 90 mM over 10 min at 0.4 ml/min. Removal of sodium and chloride ions was verified by the following IC methods: a 2 × 250 mm AS11-HC column (formate and chloride) with a 0.5–3 mM KOH gradient and a 2 × 250 mm CS-12 column (sodium and ammonium) with 18 mM methanesulfonic acid as mobile phase.
Cell culture.
HepG2 cells were obtained from the American Type Culture Collection (Rockville, MD) and used for only 30 passages. The cells were routinely grown at 37°C in DMEM containing 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, and 25 mM glucose (Invitrogen, Carlsbad, CA) in a humidified atmosphere containing 5% CO2. For experiments, tissue culture-treated polystyrene 35-mm dishes (Corning, Lowell, MA) were seeded with 2 × 106 cells and grown to confluency in DMEM.
Analytic methods.
Hyp was measured by the AccQ·Tag method (Waters, Milford, MA), as previously described (25). The protein content of HepG2 cell monolayers was measured using a Coomassie Plus assay kit (Pierce, Rockford, IL), with bovine serum albumin as the standard, after dissolution of the cells with 0.1 M NaOH. Total oxalate was determined in urine and cell culture media by IC, as previously described (16). IC-MS was used to measure total glycolate, total hippurate, and 13C enrichment in oxalate, glycolate, and hippurate, as previously described (16). The enrichment of [13C2]glycine in cell culture media was measured by gas chromatography-mass spectroscopy by Metabolic Solutions (Nashua, NH).
Animals.
Three-month-old male mice, in a mixed genetic background (C57BL6/129), were fed a purified casein/lactalbumin-based test diet, the nutrient composition of which is described elsewhere (2). They were maintained in a barrier facility with a 12:12-h light-dark cycle and an ambient temperature of 23 ± 1°C. All animals had free access to food and water. All animal studies were approved by the Institutional Animal Use and Care Committee.
[13C5]Hyp intravenous infusion metabolic cage experiments.
Wild-type mice and mice deficient in GR activity [Grhpr knockout (KO)] were housed in metabolic cages (Nalgene, Rochester, MN). The mice, under isoflurane anesthesia, were catheterized via the jugular vein with a funnel catheter (Instech Lab), which was attached to a swivel (Instech Lab) and mounted to the metabolic cage. Saline containing 7.8 mM [13C5]Hyp was infused immediately after surgery at a rate of 0.3 μl/min or 0.14 μmol/h for the duration of the experiment. After a 3-day recovery period, three 24-h urine samples were collected from each animal.
RESULTS
Tissue expression of mRNA for Hyp catabolic enzymes.
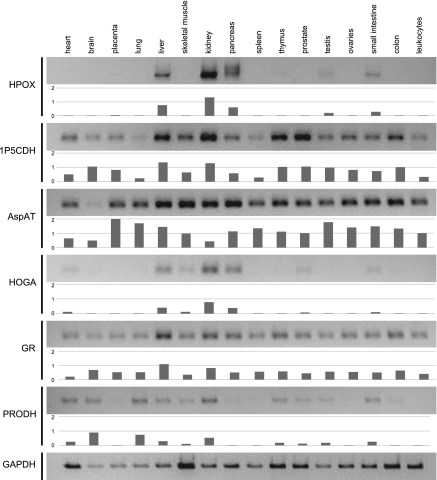
The expression of mRNA for the enzymes involved in Hyp catabolism (HPOX, 1P5CDH, AspAT, and HOGA; Fig. 1) and for GR was compared with the expression of proline dehydrogenase and the housekeeping gene GAPDH in 16 different human tissues (Fig. 2). All enzymes of the Hyp catabolic pathway were expressed in liver, kidney, pancreas, and small intestine. Some expression of HPOX was evident in testis, and HOGA was detected in heart, skeletal muscle, and prostate. 1P5CDH, AspAT, and GR were ubiquitously expressed. Expression of proline dehydrogenase was detected in all tissues except placenta, pancreas, spleen, ovaries, and leukocytes.
Fig. 2.
Analysis of human cDNA tissue panel by PCR. Housekeeping gene GAPDH and proline dehydrogenase (PRODH) were also analyzed. Relative intensity for each band compared with GAPDH is shown below each blot to account for the different level of GAPDH expression in the tissues. See Fig. 1 legend for abbreviations for all the enzymes of the Hyp pathway and GR.
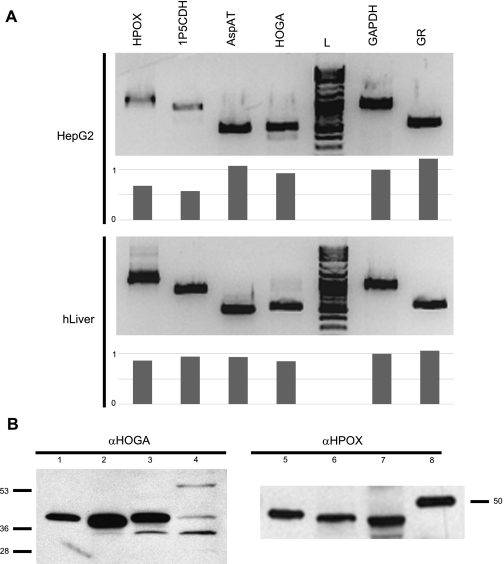
HepG2 cells retain many of the functional properties of liver cells, including the capacity to synthesize oxalate and glycolate (5). Figure 3A shows that these cells express mRNA for all enzymes in the pathway at levels similar to those in human liver, except for a lower expression of HPOX and 1P5CDH. Expression of the HPOX and HOGA proteins in HepG2 cells, human liver, and bovine kidney was also verified by Western blotting (Fig. 3B).
Fig. 3.
Assessment of mRNA and protein levels of Hyp pathway enzymes in HepG2 cells and various tissue lysates. A: PCR analysis of mRNA levels within HepG2 cells and human liver. Relative intensity for each band compared with GAPDH is shown below each blot. B: Western blot analysis of HPOX and HOGA expression in cells and tissues. Lane 1, recombinant HOGA (0.5 ng protein); lane 2, bovine kidney mitochondria (1 μg protein); lane 3, human liver (5 μg protein); lane 4, HepG2 mitochondria (1 μg protein); lane 5, bovine kidney mitochondria (1 μg protein); lane 6, human liver (5 μg protein); lane 7, HepG2 mitochondria (1 μg protein); lane 8, Precision Plus Western C Standard, 50 kDa.
Uptake and metabolism of Hyp in cultured cells.
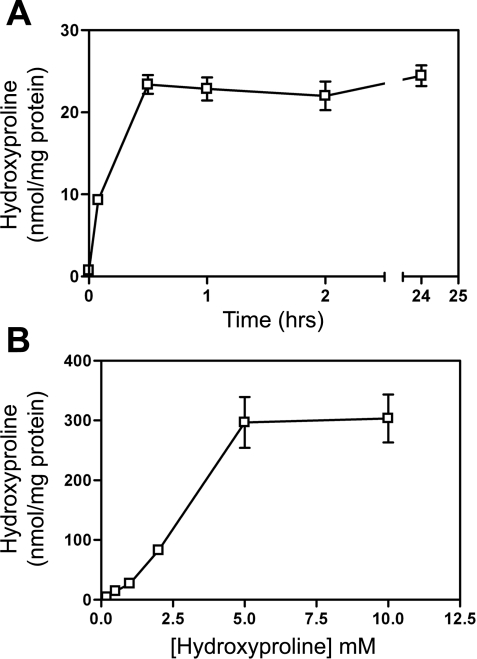
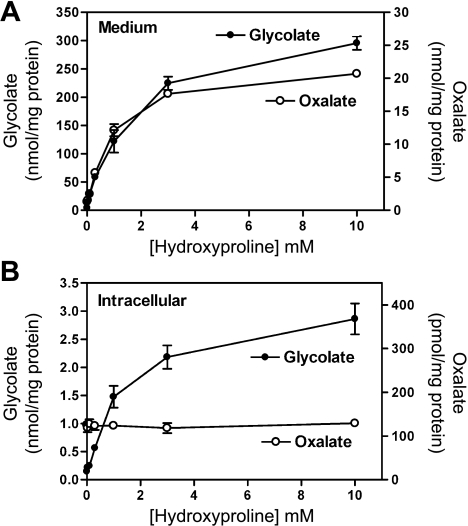
HepG2 cells took up Hyp from the culture medium, and equilibration was reached in 30 min (Fig. 4A). The intracellular pool was saturated when the extracellular concentration reached 5 mM (Fig. 4B). The metabolism of Hyp to glycolate and oxalate began to plateau with 3 mM Hyp in the medium (Fig. 5A). Approximately 10 times as much glycolate as oxalate was produced at all Hyp concentrations tested. The intracellular glycolate concentration (Fig. 5B) increased as the Hyp increased, whereas the oxalate concentration remained constant. These metabolic activities and the mRNA and Western analyses for HPOX and HOGA support the idea that HepG2 cells retain the complete pathway for the catabolism of Hyp.
Fig. 4.
Uptake of Hyp by HepG2 cells. A: time dependence of uptake of Hyp by cells incubated in medium containing 1 mM Hyp. B: uptake of 0–12.5 mM Hyp in 1 h.
Fig. 5.
Metabolism of Hyp by HepG2 cells. A: amount of glycolate and oxalate in the medium of cells incubated with 0–10 mM Hyp for 24 h. B: intracellular concentrations of glycolate and oxalate in cells analyzed in A following extraction of saline-washed cells with 10% trichloroacetic acid.
Synthesis and evaluation of stable isotopes of Hyp in vitro.
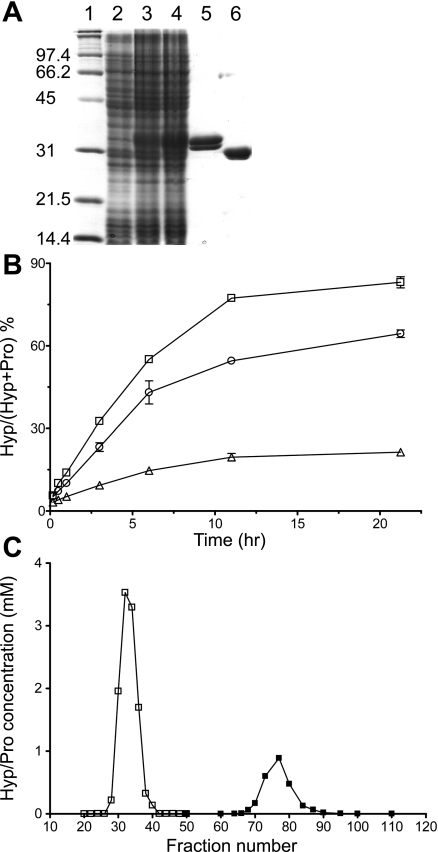
To specifically follow the metabolism of Hyp to glycolate, oxalate, and glycine in vitro and in vivo, 13C5 and 18O isotopes of Hyp were synthesized using the recombinant proline hydroxylase Phy1 from Dactylsporangium sp. RH1. The protein was efficiently overexpressed and purified from E. coli utilizing a synthetic gene optimized for codon usage and an NH2-terminal fusion to a poly-His tag (Fig. 6A). The His tag was removed by proteolytic digestion and verified by electrospray mass spectrometry. On the basis of the observation that the in vivo production of Hyp was maximal in media at pH 6.5, we first determined the level of Hyp production with the following conditions in a 100-μl reaction: 0.1 mg/ml Phy1, 80 mM MES (pH 6.5), 4 mM Pro, 8 mM 2-OG, 2 mM ferrous sulfate, and 4 mM l-ascorbic acid (30). Subsequent experiments testing pH 5–8.5 buffers at 25–40°C (data not shown) indicated that pH 6.5 and 35°C were optimal. In an effort to increase the production scale of Hyp, the volume of the reaction and the concentrations of the reagents were increased sixfold except for Phy1. For the three concentrations of Phy1 tested, 0.1, 0.4, and 1.2 mg/ml (Fig. 6B), the amount of Hyp produced leveled off after 10–12 h. The use of 0.4 mg Phy1/ml was found to be the most economical, with ∼64% of Pro converted to Hyp.
Fig. 6.
Purification of recombinant proline-4-hydroxylase (Phy1) and optimization of Hyp production. A: SDS-PAGE analysis of Phy1 purification. Lane 1, molecular mass standards (kDa); lane 2, Escherichia coli cell extract at 0-h induction; lane 3, cell extract after 3-h induction with isopropyl β-d-1-thiogalactopyranoside; lane 4, cell extract after 6-h induction; lane 5, purified Phy1 containing the NH2-terminal His tag; lane 6, purified Phy1 after removal of the His tag. B: reaction curves for Phy1 at 0.1 mg/ml (▵), 0.4 mg/ml (○), and 1.2 mg/ml (□). C: purification of Hyp from Pro via AG 50W-X8 column chromatography. □, Hyp-containing peaks; ■, Pro- containing peaks.
For the generation of [18O]- and [13C5]Hyp, the reaction volumes were increased to 10 and 40 ml, respectively. An anaerobic tonometer with 18O2 and 0.4 mg Phy1/ml were used for the synthesis of [18O]Hyp. However, 1.2 mg Phy1/ml was used to produce [13C5]Hyp aerobically to limit loss of [13C5]Pro due to its high cost. The labeled Hyp species were purified from the unreacted Pro via an H+-form AG 50W-X8 ion-exchange column (Fig. 6C). Typical conversion efficiencies were as follows: 38% and 83% Pro converted to [18O]Hyp and [13C5]Hyp, respectively. Yields following all purification and deionization steps ranged from 76 to 100%. The mass of the ionized form of Pro increased correctly from 114 Da to 132 and 135 Da for [18O]Hyp and [13C5]Hyp, respectively. This analysis also showed that 18O isotope incorporation was 95.4%, consistent with the use of 97% 18O2, and 13C isotope incorporation was 100%.
Confluent HepG2 cells were incubated in media containing 0.5 mM unlabeled Hyp ([12C5]Hyp), [13C5]Hyp, and [18O]Hyp. The levels of glycolate, glycine, and oxalate in the culture media after a 24-h incubation are shown in Table 1. Cells incubated with unlabeled Hyp produced a significant increase in the glycolate and oxalate excreted into the culture medium. Supplementation with [18O]- and [13C5]Hyp resulted in the formation of 18O- and 13C-labeled glycolate and oxalate. Less 18O than 13C label was incorporated, suggesting that some of the 18O label in [18O]Hyp was lost following its conversion to glyoxylate because of its tautomerism (Fig. 1) between hydrated and enol forms (21). The detection of [13C2]glycine in the media most likely indicates that glyoxylate and/or glycolate produced from Hyp metabolism enters the peroxisomal compartment, where it is converted to glycine by AGT. The bulk of the consumed [13C5]Hyp not accounted for most likely represents [13C2]glycine that has been further metabolized. For example, glycine could have been converted to 13CO2 by the activity of the glycine cleavage system or to serine via the methyltransferase pathway (13). Labeled hippurate was not detected in the media, suggesting that glycine conjugation to benzoic acid is not a significant pathway in these cells (11). These observations indicate that [13C5]Hyp is a superior tracer to [18O]Hyp and that [13C5]Hyp can be used to monitor Hyp breakdown and the metabolic fate of Hyp-derived glyoxylate in HepG2 cells.
Table 1.
Metabolism of isotopically labeled Hyp by HepG2 cells
| Treatment |
||||
|---|---|---|---|---|
| Metabolite | Control | [12C5]Hyp | [18O]Hyp | [13C5]Hyp |
| Hyp consumed | 122 (9) | 151 (38) | 152 (9) | |
| [12C2]glycolate | 5.6 (0.7) | 41.3 (2.4) | 18.7 (3.9) | 7.4 (1.2) |
| [18O]glycolate | 25.7 (7.9) | |||
| [13C2]glycolate | 38.3 (0.5) | |||
| [13C2]glycine | 5.6 (0.5) | |||
| [12C2]oxalate | 1.9 (0.5) | 4.8 (0.7) | 4.0 (0.4) | 1.9 (0.8) |
| [18O]oxalate | 1.6 (0.3) | |||
| [13C2]oxalate | 3.2 (0.3) | |||
Values are means (SD) in nmol/mg protein. Cells were incubated for 24 h in medium containing no additional hydroxyproline (Hyp) or 0.5 mM [12C5]Hyp, [18O]Hyp, or [13C5]Hyp. Hyp consumed was determined by subtracting the amount of Hyp remaining in the medium following incubation.
In vivo infusion of [13C5]Hyp.
Wild-type and Grhpr KO mice were infused with tracer levels of [13C5]Hyp while housed in metabolic cages. Plasma Hyp levels increased from 9.7 ± 2.1 to 11.8 ± 3.3 μM following infusion, which was not significant (P = 0.28). The data suggest that these infusion conditions did not significantly alter normal Hyp turnover. Urinary measures following [13C5]Hyp infusion are shown in Table 2. In both mouse strains, the synthesis of 13C-labeled oxalate and hippurate was detected. Animals deficient in GR activity excreted 12.4 times more [13C2]oxalate than their wild-type littermates, indicating that GR plays an important role in metabolizing Hyp-derived glyoxylate and limiting its conversion to oxalate. The labeling of hippuric acid, which is synthesized predominantly in the liver, indicates that [13C2]glycine was formed from Hyp metabolism and AGT action and did not appear to differ between strains. Grhpr KO animals excreted no detectable [13C2]glycolate, and wild-type animals excreted only small amounts.
Table 2.
Urinary excretions after [13C5]Hyp infusion
| Animals |
|||
|---|---|---|---|
| Wild-type | Grhpr KO | P Value | |
| Urine volume, ml | 1.2 (0.2) | 1.2 (0.4) | 0.96 |
| Urine creatinine, mg | 0.38 (0.03) | 0.45 (0.1) | 0.30 |
| Urine oxalate, μM | 340 (28) | 900 (281) | 0.027 |
| Urine [13C2]oxalate, μM | 5.7 (2.7) | 70.7 (22.5) | 0.008 |
| Urine glycolate, μM | 239 (51) | 276 (99) | 0.597 |
| Urine [13C2]glycolate, μM | 2.4 (0.9) | <0.5 | |
| Urine hippuric acid, μM | 411 (153) | 334 (36) | 0.443 |
| Estimated [13C2]hippurate, μM | 2.1 (0.7) | 1.5 (0.1) | 0.175 |
Values are means (SD); n = 3 animals. Three 24-h urine collections were obtained after intravenous infusion of [13C5]Hyp (0.14 μmol/h). Urinary [13C2]glycolate levels in Grhpr knockout (KO) animals were below the limit of detection of 0.5 μM. A t-test was used to determine significance of difference between wild-type and Grhpr KO animals; P < 0.05 was considered significant.
DISCUSSION
The liver and kidney (Figs. 2 and 3) were confirmed to be major sites of Hyp catabolism in humans, consistent with previous reports in other mammals (1). The small intestine and pancreas were also shown to express these genes. These tissues have not previously been identified as capable of degrading Hyp. Cooper et al. (8) reported that HPOX is expressed in cell lines derived from colon tumors and suggested that its activity in these cells may play a role in growth regulation, oxidative stress, and activation of apoptosis. Thus the functional role of Hyp catabolism in the whole intestinal tract and pancreas warrants further examination. Low levels of HOGA expression in the absence of detectable HPOX expression were observed in heart, skeletal muscle, and prostate. It is possible that the method we used lacked sufficient sensitivity to detect low levels of HPOX expression in some of these tissues or that there are differences in mRNA stability. Rat heart mitochondria, for instance, have been reported to metabolize Hyp, with a resultant reduction of NAD+, indicating that, in this species, heart tissue most likely expresses HPOX activity as well as 1P5CDH (4).
HepG2 cells have previously been shown to retain several hepatic functions, including the ability to synthesize glycolate and oxalate (5). The present results indicate that these cells express all the genes in the Hyp catabolic pathway (Fig. 3) and that they retain the capacity to take up and degrade Hyp to glycolate and oxalate (Fig. 4, Table 1). It is noteworthy that, over a wide range of Hyp concentrations in the culture medium, the amount of glycolate was consistently ∼10 times greater than the amount of oxalate secreted into the growth medium (Fig. 5). Cells appeared to release oxalate to maintain a constant low intracellular concentration, whereas glycolate intracellular concentration increased. An elevation in intracellular oxalate could potentially be detrimental, as it could inhibit glycolysis (7). Therefore, HepG2 cells should be well suited for evaluating ways to modify Hyp metabolism for use as a potential therapy to reduce oxalate production in individuals with PH. Whether HPOX activity is induced in these cells by p53 and is associated with the production of reactive oxygen species, as reported in tumor-derived intestinal cells, will also be of interest (8).
After infusion of [13C5]Hyp, wild-type and Grhpr KO animals excreted [13C2]oxalate into their urine (Table 2). The amount of oxalate excreted by Grhpr KO animals was ∼12 times higher than that excreted by wild-type animals, indicating that GR plays a significant role in metabolizing the glyoxylate derived from Hyp breakdown and limiting its conversion to oxalate. In wild-type animals, the amount of urinary [13C2]glycolate was half that of urinary [13C2]oxalate. This is in contrast to formation of 10 times more glycolate than oxalate from Hyp in HepG2 cells (Fig. 5) and human subjects consuming Hyp (19). This excretion pattern appears to be unique to male mice, as female mice excrete more glycolate than oxalate from dietary Hyp (Knight and Holmes, unpublished results). As only tracer levels of [13C5]Hyp were infused, the results suggest that, even in wild-type animals, the breakdown of Hyp contributes to endogenous oxalate synthesis. In future experiments, the exact contribution of this metabolism to urinary oxalate excretion will be determined in humans and experimental animals with use of [13C5]Hyp that is custom-synthesized for infusion experiments.
The glyoxylate that is derived each day from Hyp catabolism is normally metabolized by AGT and GR. This study provides the first evidence that glyoxylate produced within the mitochondria may be converted to glycine within the peroxisome by AGT. In individuals with PH types 1 and 2, where these enzymes are deficient, Hyp breakdown may make a substantial contribution to their excessive oxalate synthesis (9). The enzymes HPOX and HOGA are unique to the Hyp catabolic pathway. Mutations in HOGA have recently been reported to result in an excessive oxalate synthesis for reasons not yet ascertained (6, 22, 26). However, HPOX may be a suitable target, as an inherited deficiency in this enzyme that results in an elevated Hyp excretion, but no apparent other abnormal physiology, has been detected in some individuals (14, 27, 28).
In summary, we have identified the liver, kidney, pancreas, and possibly regions of the intestine as potential sites of Hyp degradation. HepG2 cells retain the capacity to break down Hyp. Metabolism of Hyp in these cells and in mice can be monitored using the stable 13C isotope of Hyp. These model systems will allow us to investigate the metabolism of Hyp, understand changes that occur in mouse models of PH, and identify therapeutic strategies that decrease Hyp catabolism and oxalate synthesis in PH.
GRANTS
This work was supported in part by the Oxalosis and Hyperoxaluria Foundation and National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-54468 and DK-74945.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.J., L.C.J., J.K., M.F.C., R.P.H., and W.T.L. are responsible for conception and design of the research; J.J., L.C.J., J.K., and M.F.C. performed the experiments; J.J., L.C.J., J.K., T.J.R., R.P.H., and W.T.L. analyzed the data; J.J., L.C.J., J.K., T.J.R., R.P.H., and W.T.L. interpreted the results of the experiments; J.J., L.C.J., J.K., T.J.R., and W.T.L. prepared the figures; J.J., L.C.J., J.K., R.P.H., and W.T.L. drafted the manuscript; J.J., L.C.J., J.K., M.F.C., T.J.R., R.P.H., and W.T.L. edited and revised the manuscript; J.J., L.C.J., J.K., M.F.C., T.J.R., R.P.H., and W.T.L. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank Kendrah Kidd for technical assistance.
Present address of J. Jiang: Department of Microbiology and Biotechnology, Northeast Agricultural University, Harbin City, People's Republic of China.
REFERENCES
- 1. Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem 49: 1005–1061, 1980 [DOI] [PubMed] [Google Scholar]
- 2. Adams MR, Golden DL, Franke AA, Potter SM, Smith HS, Anthony MS. Dietary soy β-conglycinin (7S globulin) inhibits atherosclerosis in mice. J Nutr 134: 511–516, 2004 [DOI] [PubMed] [Google Scholar]
- 3. Atlante A, Passarella S, Quagliariello E. Spectroscopic study of hydroxyproline transport in rat kidney mitochondria. Biochem Biophys Res Commun 202: 58–64, 1994 [DOI] [PubMed] [Google Scholar]
- 4. Atlante A, Seccia TM, Marra E, Minervini GM, Vulpis V, Pirrelli A, Passarella S. Carrier-mediated transport controls hydroxyproline catabolism in heart mitochondria from spontaneously hypertensive rat. FEBS Lett 396: 279–284, 1996 [DOI] [PubMed] [Google Scholar]
- 5. Baker PR, Cramer SD, Kennedy M, Assimos DG, Holmes RP. Glycolate and glyoxylate metabolism in HepG2 cells. Am J Physiol Cell Physiol 287: C1359–C1365, 2004 [DOI] [PubMed] [Google Scholar]
- 6. Belostotsky R, Seboun E, Idelson GH, Milliner DS, Becker-Cohen R, Rinat C, Monico CG, Feinstein S, Ben-Shalom E, Magen D, Weissman I, Charon C, Frishberg Y. Mutations in DHDPSL are responsible for primary hyperoxaluria type III. Am J Hum Genet 87: 392–399, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beutler E, Forman L, West C. Effect of oxalate and malonate on red cell metabolism. Blood 70: 1389–1393, 1987 [PubMed] [Google Scholar]
- 8. Cooper SK, Pandhare J, Donald SP, Phang JM. A novel function for hydroxyproline oxidase in apoptosis through generation of reactive oxygen species. J Biol Chem 283: 10485–10492, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coulter-Mackie MB. 4-Hydroxyproline metabolism and glyoxylate production: a target for substrate depletion in primary hyperoxaluria? Kidney Int 70: 1891–1893, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Danpure CJ. Primary hyperoxaluria. In: The Metabolic and Molecular Bases of Inherited Disease, edited by Scriver CR, Beaudet AL, Sly WS, Vallee D, Childs B, Kinzler KW, Vogelstein B. New York: McGraw-Hill, 2001, p. 3323–3367 [Google Scholar]
- 11. Gatley SJ, Sherratt HS. The synthesis of hippurate from benzoate and glycine by rat liver mitochondria. Submitochondrial localization and kinetics. Biochem J 166: 39–47, 1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holmes RP, Assimos DG. The impact of dietary oxalate on kidney stone formation. Urol Res 32: 311–316, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem 1: 169–187, 1973 [DOI] [PubMed] [Google Scholar]
- 14. Kim SZ, Varvogli L, Waisbren SE, Levy HL. Hydroxyprolinemia: comparison of a patient and her unaffected twin sister. J Pediatr 130: 437–441, 1997 [DOI] [PubMed] [Google Scholar]
- 15. Kivirikko KI. Urinary excretion of hydroxyproline in health and disease. Int Rev Connect Tissue Res 5: 93–163, 1970 [DOI] [PubMed] [Google Scholar]
- 16. Knight J, Assimos DG, Easter L, Holmes RP. Metabolism of fructose to oxalate and glycolate. Horm Metab Res 42: 868–873, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knight J, Easter LH, Neiberg R, Assimos DG, Holmes RP. Increased protein intake on controlled oxalate diets does not increase urinary oxalate excretion. Urol Res 37: 63–68, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knight J, Holmes RP. Mitochondrial hydroxyproline metabolism: implications for primary hyperoxaluria. Am J Nephrol 25: 171–175, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knight J, Jiang J, Assimos DG, Holmes RP. Hydroxyproline ingestion and urinary oxalate and glycolate excretion. Kidney Int 70: 1929–1934, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levine M. A new method for isolation of hydroxy-l-proline and l-proline from gelatin. J Biol Chem 234: 1731–1732, 1959 [PubMed] [Google Scholar]
- 21. Meany JE, Pocker Y. The dehydration of glyoxylate hydrate: general-acid, general-base, metal ion, and enzymatic catalysis. J Am Chem Soc 113: 6155–6161, 1991 [Google Scholar]
- 22. Monico CG, Rossetti S, Belostotsky R, Cogal AG, Herges R, Olson JB, Bergstrahl E, Williams H, J, Haley WE, Frishberg Y, Milliner DS. Primary hyperoxaluria type III gene HOGA1 (formerly DHDPSL) as a possible risk factor for idiopathic calcium oxalate urolithiasis. Clin J Am Soc Nephrol 6: 2289–2295, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murray MS, Holmes RP, Lowther WT. Active site and loop 4 movements within human glycolate oxidase: implications for substrate specificity and drug design. Biochemistry 47: 2439–2449, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: The Metabolic and Molecular Bases of Inherited Disease, edited by Scriver CR, Beaudet AL, Sly WS, Vallee D, Childs B, Kinzler KW, Vogelstein B. New York: McGraw-Hill, 2001, p. 1821–1838 [Google Scholar]
- 25. Reverter M, Lundh T, Lindberg JE. Determination of free amino acids in pig plasma by precolumn derivatization with 6-N-aminoquinolyl-N-hydroxysuccinimidyl carbamate and high-performance liquid chromatography. J Chromatogr B Biomed Sci Appl 696: 1–8, 1997 [DOI] [PubMed] [Google Scholar]
- 26. Riedel TJ, Johnson LC, Knight J, Hantgan RR, Holmes RP, Lowther WT. Structural and biochemical studies of human 4-hydroxy-2-oxoglutarate aldolase: implications for hydroxyproline metabolism in primary hyperoxaluria. PLos One 6: e26021, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson MJ, Menzies IS, Sloan I. Hydroxyprolinaemia with normal development. Arch Dis Child 55: 484–486, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roesel RA, Blankenship PR, Lynch WR, Coryell ME, Thevaos TG, Hall WK. Hydroxyproline metabolism in two sisters with hydroxyprolinemia. Hum Hered 29: 364–370, 1979 [DOI] [PubMed] [Google Scholar]
- 29. Shibasaki T, Mori H, Chiba S, Ozaki A. Microbial proline 4-hydroxylase screening and gene cloning. Appl Environ Microbiol 65: 4028–4031, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shibasaki T, Mori H, Ozaki A. Enzymatic production of trans-4-hydroxy-l-proline by regio- and stereospecific hydroxylation of l-proline. Biosci Biotechnol Biochem 64: 746–750, 2000 [DOI] [PubMed] [Google Scholar]
- 31. Valle D, Goodman SI, Harris SC, Phang JM. Genetic evidence for a common enzyme catalyzing the second step in the degradation of proline and hydroxyproline. J Clin Invest 64: 1365–1370, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]