Abstract
Although the Austronesian expansion had a major impact on the languages of Island Southeast Asia, controversy still exists over the genetic impact of this expansion. The coexistence of both Asian and Papuan genetic ancestry in Eastern Indonesia provides a unique opportunity to address this issue. Here, we estimate recombination breakpoints in admixed genomes based on genome-wide SNP data and date the genetic admixture between populations of Asian vs. Papuan ancestry in Eastern Indonesia. Analyses of two genome-wide datasets indicate an eastward progression of the Asian admixture signal in Eastern Indonesia beginning about 4,000–3,000 y ago, which is in excellent agreement with inferences based on Austronesian languages. The average rate of spread of Asian genes in Eastern Indonesia was about 0.9 km/y. Our results indicate that the Austronesian expansion had a strong genetic as well as linguistic impact on Island Southeast Asia, and they significantly advance our understanding of the biological origins of human populations in the Asia–Pacific region.
Keywords: expansion time, admixture time, migration rate, single nucleotide polymorphism
The spread of Austronesian-speaking peoples ultimately extended well over halfway around the world from Madagascar in the west to Easter Island in the east. This process is often termed the Austronesian expansion, which represents a complex demographic process of interaction between migrating Neolithic farmers and indigenous Mesolithic hunter–gatherer communities, a frequent phenomenon in world prehistory. However, our knowledge about the historical origin and spread of Austronesian-speaking peoples has been overwhelmingly from linguistic, archaeological, and anthropological studies (1). The genetic impact of the Austronesian expansion has been contested, with some studies claiming that the genetic ancestry of Island Southeast Asia mostly reflects pre-Austronesian migrations (2, 3) and that the Austronesian expansion did not involve large-scale population movements (4).
Eastern Indonesia (consisting of the Nusa Tenggaras and Moluccas) (Figs. 1 and 2) provides a unique opportunity to investigate the extent to which the genetic ancestry in this region reflects Austronesian or pre-Austronesian migration(s). Both Asian and Papuan (related to New Guinea) genetic ancestry is evident in East Indonesia based on mtDNA and Y-chromosome studies (5, 6) as well as a recent study (7) of multiple autosomal and X-chromosomal SNPs, and there is a decline in Asian ancestry (and corresponding increase in Papuan ancestry) going from west to east. Because the time of the entry of the Austronesians into Indonesia has been dated by both archaeological and linguistic evidence to about 4,000 y ago (8, 9), estimating the time of the admixture between Asian and Papuan ancestry in East Indonesian groups could, therefore, distinguish between a pre-Austronesian vs. Austronesian explanation for the Asian ancestry. However, none of the previous genetic studies of East Indonesia analyzed sufficiently dense genome-wide data to estimate the admixture time, which relies on recombination information extracted from throughout the genome. In this study, we use high-density genome-wide SNP data to estimate the amount and time of admixture in East Indonesian groups to determine if the admixture time corresponds to Austronesian or pre-Austronesian expansions.
Fig. 1.
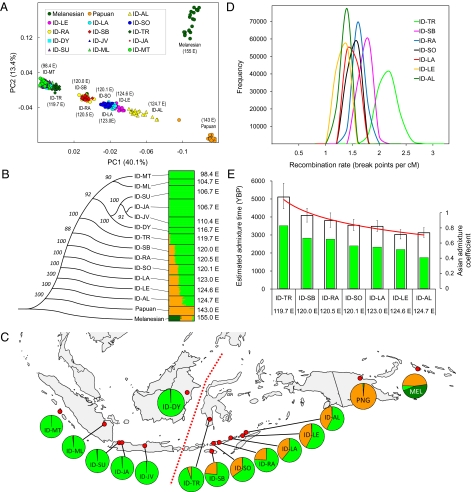
Analyses of the Pan-Asian SNP dataset. (A) PCA of individuals representing 15 populations. A plot for the first two principle components is shown. Individuals are shaded by different colors according to their population affiliation. Numbers in parentheses indicate the longitude of each population sample. (B) Phylogenetic analysis of 15 populations and estimated population structure. (Left) An unrooted phylogenetic tree shows genetic relationship of the 15 populations using the neighbor-joining method on pairwise FST matrices. Bootstrap values based on 1,000 replicates are also shown. (Right) A plot shows the estimated population structure, with each colored horizontal line representing an individual assigned proportionally to one of the K = 3 clusters with the proportions represented by the relative lengths of the three different colors. Black lines separate individuals from different populations. Populations are labeled on the left by lines connecting to the nodes of the tree. The longitude of each population sample is displayed on the right of the figure. The results shown were based on the highest probability run of 10 STRUCTURE runs at K = 3. (C) Geographical distribution of genetic components. Red dots on the map are sampling locations. Each circle graph represents a population sample, with the frequency of the three genetic components inferred by STRUCTURE analysis (B) indicated by the colored sectors. Red dashed line denotes Wallace's biogeographic line. (D) Posterior distribution of the recombination parameters for seven Eastern Indonesian populations showing admixture. Posterior distribution of the recombination parameter r (breakpoints per centimorgan) estimated by STRUCTURE analysis is indicated with colors for populations as shown in the legend. The mean and 90% CI of r for each population are shown in Table 3. (E) Estimated admixture time in seven Eastern Indonesian populations. White bars indicate estimated admixture times with 90% CIs (scale on the left y axis); color bars denote the admixture proportion (fraction of Asian ancestry) in each population as indicated by the y axis on the right.
Fig. 2.
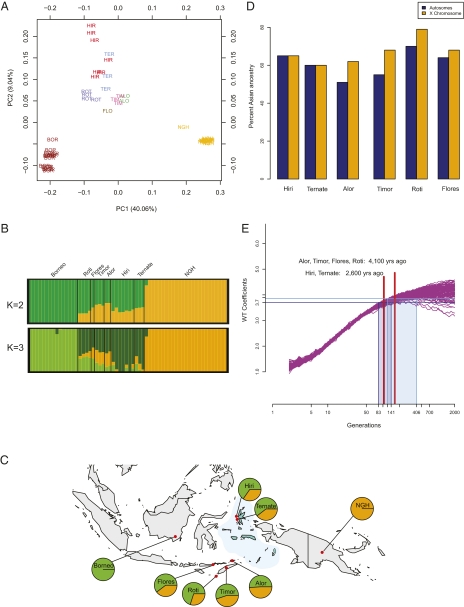
Analyses of the Affymetrix 6.0 dataset. (A) PCA. A plot of the first two principle components is shown. Population labels are abbreviated as the first three letters of the population name. (B) frappe results for K = 2 and K = 3. Each color indicates a different ancestry component. (C) Geographical distribution of genetic components. Red dots on the map are sampling locations. Each circle represents a population sample with the frequency of two ancestry components inferred by StepPCO and frappe analyses. The Moluccas are shaded in light blue. (D) Admixture estimate for autosomes vs. X chromosome. (E) Admixture time estimate. Simulated data from 100 simulations with a 40% migration rate are presented. Each curve represents a single admixed population. Average WT coefficients calculated for 100 chromosomes drawn at random from each population at exponentially growing time points are plotted as a function of time. Measurements obtained for the Molucca and the Nusa Tenggara populations are shown by blue horizontal lines. Red vertical lines indicate the time estimate, and shaded boxes define the CIs.
Results
We analyzed dense genome-wide SNP data from two studies (10, 11). The first study consists of about 50,000 SNPs analyzed in 288 individuals from 13 Austronesian-speaking populations and two Papuan-speaking populations across Indonesia and Papua New Guinea (Table 1), whereas the second study consists of about 680,000 SNPs analyzed in 36 individuals from 7 populations from Indonesia and 25 individuals from Papua New Guinea (Table 2). Apart from a general description of population structure and its relationship with geography, language, and demographic history, we focus on estimating the amount and time of admixture involving Asian and Papuan ancestry across East Indonesia. Because genetic recombination breaks down parental genomes into segments of different sizes, the genome of a descendant of an admixture event is composed of different combinations of these ancestral segments or blocks (12). Admixture time can be estimated from the information based on the distribution of ancestral segments and the recombination breakpoints in an admixed genome (12–16).
Table 1.
Information on population samples in the Pan-Asian dataset
| Sample ID | Sample size | Latitude | Longitude | Ethnicity | Language | Language family |
| ID-MT | 15 | 0.3 S | 98.4 E | Mentawai | Mentawai | Austronesian |
| ID-ML | 12 | 3.0 S | 104.7 E | Malay | Malay | Austronesian |
| ID-SU | 25 | 6.2 S | 106.7 E | Sundanese | Sunda | Austronesian |
| ID-JA | 34 | 6.2 S | 106.7 E | Javanese | Javanese | Austronesian |
| ID-JV | 19 | 7.3 S | 110.4 E | Javanese | Javanese | Austronesian |
| ID-DY | 12 | 1.2 S | 116.7 E | Dayak | Benuak | Austronesian |
| ID-TR | 20 | 4.7 S | 119.7 E | Toraja | Toraja | Austronesian |
| ID-SB | 20 | 9.8 S | 120.0 E | Kambera | Kambera | Austronesian |
| ID-RA | 17 | 8.7 S | 120.5 E | Manggarai | Manggarai | Austronesian |
| ID-SO | 19 | 8.6 S | 120.1 E | Manggarai | Manggarai | Austronesian |
| ID-LA | 20 | 8.3 S | 123.0 E | Lamaholot | Lamaholot | Austronesian |
| ID-LE | 19 | 8.3 S | 124.6 E | Lembata | Lembata | Austronesian |
| ID-AL | 19 | 8.3 S | 124.7 E | Alorese | Alor | Austronesian |
| Papuan | 17 | 4.0 S | 143.0 E | Papuan | East Papuan | Papuan |
| Melanesian | 20 | 6.0 S | 155.0 E | Melanesian | Nasioi | Papuan |
Table 2.
Information on population samples in the Affymetrix 6.0 dataset
| Sample ID | Sample size | Location | Latitude | Longitude | Language family |
| BOR | 16 | Borneo | 3.3 S | 114.6 E | Austronesian |
| FLO | 1 | Flores | 8.6 S | 121.1 E | Austronesian |
| ROT | 4 | Roti | 10.7 S | 123.2 E | Austronesian |
| TIM | 3 | Timor | 10.2 S | 123.6 E | Austronesian |
| ALO | 2 | Alor | 8.3 S | 124.6 E | Unknown |
| HIR | 7 | Hiri | 0.9 N | 127.3 E | Papuan |
| TER | 3 | Ternate | 0.8 N | 127.4 E | Papuan |
| NGH | 25 | New Guinea Highlands | 6.2 S | 143.6 E | Papuan |
Genetic Relationship of Populations, Correlation of Genetic Ancestry and Linguistic Affiliations, and Geography.
We first analyzed relationships among individuals in the Pan-Asian SNP dataset using principle component (PC) analysis based on genome-wide data. Fig. 1A is a 2D plot displaying the first two PCs for all individuals. Individuals tend to cluster with other members of their linguistic or ethnic affiliations, especially for the Papuan [New Guinea Human Genome Diversity Project–Centre d'Etude du Polymorphisme Humain (HGDP-CEPH) group] and Melanesian (Bougainville HGDP-CEPH group) samples. The first PC axis shows an obvious separation of the Papuan and West Indonesian (ID-MT, ID-ML, ID-SU, ID-JA, ID-JV, and ID-DY) groups, with all East Indonesian populations (ID-TR, ID-SB, ID-SO, ID-RA, ID-LE, and ID-AL) lying in between to form a genetic cline (ID, Indonesian; MT, Mentawai; ML, Malay; SU, Sundanese; JA, Javanese; JV, Javanese; DY, Dayak; TR, Toraja; SB, Kambera; RA, Manggarai; SO, Manggarai; LA, Lamaholot; LE, Lembata; and AL, Alorese). There is also clinal variation in the genetic (FST) distances (Fig. S1 and Table S1) and a significant correlation between geographic and genetic distances (R2 = 0.69, Mantel test, P < 0.00005) (Fig. S2). A neighbor-joining tree constructed from the FST distances (Fig. 1B) is also consistent with the geographical distribution of populations. Interestingly, lexical distances, which were obtained by calculating patristic distances between each language along the maximum clade credibility tree in the work by Gray et al. (9), also showed good correlation with genetic distances (r = 0.46, Mantel test, P = 0.02).
We also performed Bayesian cluster analysis as implemented in the STRUCTURE algorithm (17) to examine the ancestry of each person. This analysis considers each person's genome as having originated from K ancestral but unobserved populations, and the contributions are described by K coefficients that sum to one for each individual. Individuals are posited to derive from an arbitrary number of ancestral populations, denoted by K. We ran STRUCTURE from K = 2 to K = 15, with results at K = 3 showing the greatest posterior probability. Estimated individual membership fractions in K = 3 genetic clusters are shown in Fig. 1 B and C, with the three clusters corresponding to Melanesian, Papuan, and Asian ancestry, respectively. Similar analyses were also performed using the program frappe (18), which implements a maximum likelihood method, and the results showed a good concordance with the results of STRUCTURE. The results showed that individuals from the same population often shared membership coefficients in the inferred cluster, with individuals from the seven Eastern Indonesian populations (ID-TR, ID-SB, ID-RA, ID-SO, ID-LA, ID-LE, and ID-AL) displaying significant amounts of both Papuan and Asian ancestry (Fig. 1B and Table S2). Among the seven East Indonesian populations, the proportion of Asian ancestry decreases as the longitude increases (R2 = 0.705, P < 0.02), whereas the proportion of Papuan ancestry goes in the other direction (R2 = 0.701, P < 0.02), which is consistent with the results of the PC analysis (Fig. 1A) and the tree analysis (Fig. 1B). The admixture proportion of populations under the assumption of K = 3 is shown in Table S2; ID-AL, the closest population to Papuan, has the highest proportion (55.4%) of Papuan ancestry, whereas ID-TR, which is the East Indonesian population most distant from the Papuans, has the lowest proportion (5.1%) of Papuan ancestry.
Admixture Time Estimation.
To estimate admixture time, we first selected 2,807 ancestry informative markers according to the allele frequencies of the Papuan and West Indonesian populations and ran the STRUCTURE program (15) under the linkage model with correlated allele frequencies. The geographical distribution of admixture proportions obtained by this analysis is in excellent agreement with the distribution for the entire dataset (Fig. 1 B and C and Table S2). We then used the linkage model to estimate the population recombination parameter r (break points per centimorgan), the posterior distribution of which is shown in Fig. 1D and Table 3. Again, we observed a cline in the distribution of r according to longitude; the population (ID-TR) located farthest to the west showed the highest recombination rate of 2.04 break points per cM [90% confidence interval (CI) = 1.79, 2.35], whereas the population (ID-LE) located farthest to the east showed the lowest recombination rate of 1.21 break points per cM (90% CI = 1.11, 1.32). Under the assumption of a hybrid isolation model, the admixture events of Eastern Indonesian populations were estimated to have occurred about 121–204 generations or 3,026–5,109 y ago (Table 3), assuming 25 y per generation.
Table 3.
Estimated recombination parameters r (breakpoints per centimorgan) and time since admixture [years before present YBP)]
| Recombination parameter (r) |
Admixture time (YBP) |
|||
| Sample ID | Mean | 90% CI | Mean | 90% CI |
| ID-TR | 2.04 | (1.79, 2.35) | 5,109 | (4,463, 5,868) |
| ID-SB | 1.63 | (1.49, 1.79) | 4,085 | (3,716, 4,484) |
| ID-RA | 1.52 | (1.37, 1.69) | 3,803 | (3,431, 4,225) |
| ID-SO | 1.42 | (1.30, 1.55) | 3,540 | (3,248, 3,871) |
| ID-LA | 1.39 | (1.28, 1.52) | 3,481 | (3,200, 3,798) |
| ID-LE | 1.21 | (1.11, 1.32) | 3,026 | (2,780, 3,296) |
| ID-AL | 1.25 | (1.15, 1.36) | 3,123 | (2,870, 3,405) |
Replicated Analysis with a Different Dataset and Method.
To substantiate these results and gain additional insights, we also analyzed a different genome-wide dataset (11) consisting of about 680,000 SNPs analyzed in 36 individuals from seven populations from Indonesia (including two populations from the Moluccas, four populations from the Nusa Tenggaras, and one population from Borneo) and 25 individuals from the highlands of Papua New Guinea (Methods and Table 2). In agreement with the Pan-Asia data analysis, principle component analysis (PCA) for these data shows a cline in the genetic ancestry of East Indonesian populations between Borneo and New Guinea, with Asian ancestry gradually decreasing from west to east across Indonesia (Fig. 2A). Interestingly, we observe that PC2 is largely driven by differences between the two Moluccas groups (who speak Papuan languages) and the other groups (who speak Austronesian languages). Next, we estimated ancestry components in each population using the maximum likelihood-based algorithm implemented in the program frappe (18). Our results show good concordance with the results of STRUCTURE (Fig. 2 B and C). At K = 3, the third ancestry component is seen predominantly in the Moluccas and to a lesser degree, in the Nusa Tenggaras, again suggesting that there might be something different about the history of these two regions of Indonesia (Fig. 2B). This notion is also supported by the next analysis, where we compared the admixture rates on the autosomes and the X chromosome in these groups. In agreement with previous studies (7), we observe a higher frequency of Asian ancestry on the X chromosome relative to the genome-wide estimates, which suggests that the admixture in Eastern Indonesia was sex-biased, with greater contribution from Asian women. However, this sex-biased pattern of admixture is only seen in the Nusa Tenggaras and not in the Moluccas (Fig. 2D). Because our samples from the Moluccas come from Papuan-speaking groups, whereas the Nusa Tenggara samples are from Austronesian-speaking groups, this result suggests that sex-related differences in the admixture between Papuan and Austronesian groups are consistent with the hypothesis that the Austronesian groups were matrilocal (19, 20). We, therefore, estimated the time of Austronesian–Papuan admixture separately for the Nusa Tenggaras and the Moluccas using the wavelet transform method described previously (12). The admixture time estimated for the Moluccas corresponds to 104 generations (95% CI = 83–141 generations) or about 2,600 y ago. The estimate for the Nusa Tenggaras corresponds to an admixture time of 164 generations (95% CI = 121–406 generations) or about 4,100 y ago (Fig. 2E). These results are in excellent agreement with the results reported for the Pan-Asia SNP dataset (Fig. 1E).
Implications for Austronesian Expansion.
Two important points arise from the admixture time analysis. First, both datasets indicate a cline of decreasing time of admixture across East Indonesia, with oldest time in the west and youngest time in the east. These results strongly suggest that the admixture cline in East Indonesia reflects the spread of individuals of Asian ancestry coming from the west and admixing with resident groups of Papuan ancestry. Second, the time of admixture is highly consistent for the two different datasets, which were estimated by two different methods, and suggest that the admixture began about 4,000 y ago (Figs. 1E and 2E and Table 3). This time is in excellent agreement with estimates from archaeological and linguistic data for the arrival of Austronesian speakers in East Indonesia about 3,500–4,000 y ago (9, 21). For example, the oldest pottery found in East Indonesia, associated with the Austronesian culture, dates back to 3,500 y ago (22). Moreover, a Bayesian analysis of Austronesian languages dates the Austronesian expansion into Indonesia to about 4,000 y ago and also suggests a west to east spread across East Indonesia (9). In addition, as mentioned before, there is a significant correlation between the patristic distances from the language tree (9) and the genetic distances for the groups in the Pan-Asian SNP dataset (r = 0.46, Mantel test, P = 0.02). However, other linguistic analyses, which are not quantitative but instead, are based on inferences regarding the center of diversification of the relevant languages, instead suggest an east to west spread of Austronesian languages across East Indonesia (23). Overall, our admixture analyses indicate that the Asian ancestry in East Indonesia is associated with the Austronesian expansion.
A potential caveat to this interpretation of the results is that recent gene flow in the direction from New Guinea to East Indonesia could artificially reduce the admixture time estimate in those populations closest to New Guinea, which is because such recent gene flow would introduce new, larger blocks of New Guinea ancestry on top of the older, smaller blocks of Papuan ancestry in East Indonesia. An instructive example is the case of Fiji, which is inferred to have received recent migration from New Guinea that did not extend farther to the east in Remote Oceania (24). Dating of the Asia–New Guinea admixture signal in Fiji indeed produces a more recent time of about 1,100 y compared with about 2,700 y in Polynesia (12). In principle, recent gene flow to East Indonesia from New Guinea could similarly reduce the admixture time in the affected populations. However, the spectrum of wavelet coefficients derived from the admixture signal is more bimodal in Fiji, suggestive of multiple admixture events, whereas in East Indonesia, the spectrum is unimodal (Fig. S3). Thus, there is no indication in the data of recent gene flow from New Guinea.
Migration Rate of Austronesian Genes.
In addition, from the admixture cline and its geographical distribution (Fig. S4), we estimated an average rate of spread of Austronesian genes in East Indonesia of about 0.9 (± 1.3) km/y or 22 (± 32.3) km per 25-y generation, which is somewhat slower than the estimation based on archaeological evidence of about 75 km per 25-y generation (1). These results would be expected considering the fact that culture transmission and language dispersal are generally easier and thus, faster than gene introgression. Although the real rate of coastal colonization was much slower than the rate across sea, these average rates still give some perspective on the relative speed of the Austronesian expansion in these particular areas.
Discussion
In summary, the admixture time analysis of two different genome-wide datasets provides compelling evidence that people of Asian ancestry began moving through East Indonesia about 4,000 y ago from west to east and admixed with resident groups of Papuan ancestry. Furthermore, this scenario is in excellent agreement with linguistic, archaeological, and genetic evidence for a pre-Austronesian presence of Papuans in East Indonesia (4, 25, 26) and an eastward spread of Austronesian-speaking farmers across East Indonesia (9, 21). Our results showed that the major direction of Asian gene flow has been from west to east across East Indonesia, but they do not rule out other proposed migration events that would have had a lesser genetic impact. Our analyses, thus, refute suggestions that the Asian ancestry observed in Indonesia largely predates the Austronesian expansion (2, 27), or that the Austronesian expansion was not accompanied by large-scale population movement (4). To be sure, other migrations (both before and after the Austronesian expansion) have undoubtedly left a genetic legacy in Indonesia. However, our analyses of genome-wide data do indicate that there was a strong and significant genetic impact associated with the Austronesian expansion in Indonesia, just as similar analyses have pointed to a genetic impact associated with the Austronesian expansion through Near and Remote Oceania (24, 28). Given that admixture among human populations (and between modern and archaic humans) is increasingly being recognized as a significant aspect of modern human biology (29), estimates of the time of admixture should provide important insights into the history of our species.
Materials and Methods
Additional details are in SI Materials and Methods.
Population Samples and Data.
Two datasets were analyzed. The first dataset consists of 288 unrelated samples from 13 Indonesian populations with genotype data generated using the Affymetrix Genechip Human Mapping 50K Xba array and was obtained from the Pan-Asia SNP Project (10). Samples of one Papuan population from Papua New Guinea and one Melanesian population from Bougainville, genotyped with the Illumina Genechip Human Mapping 650K Xba array, were collected from the database of the HGDP-CEPH (30). Detailed information for these groups is described in Table 1. With data integration, we obtained 19,934 SNPs shared by 15 populations. This dataset is referred to as the Pan-Asian SNP dataset.
The second dataset consists of 61 individuals from eight populations from Indonesia and New Guinea that were all genotyped on the Affymetrix 6.0 platform as described previously (11, 24). Detailed information on these populations is presented in Table 2. After data integration, there were 685,582 SNPs included in the analysis. This dataset is referred to as the Affymetrix 6.0 dataset.
Statistical Analysis.
In the Pan-Asian dataset, PCA was performed at the individual level using EIGENSOFT version 3.0 (31). Unbiased estimates of FST were calculated using the work by Weir and Hill (32) with PEAS V1.0 (33). Great circle distance calculations followed the approach of Ramachandran et al. (34). The tree of populations was reconstructed based on FST distances and the neighbor-joining algorithm (35) implemented in the Molecular Evolutionary Genetics Analysis software package (MEGA version 4.0) (36). The ancestry of each person was inferred using a Bayesian cluster analysis as implemented in the STRUCTURE program (15) and a maximum likelihood method as implemented in the frappe program (18). In estimating the admixture time of Eastern Indonesian populations showing admixture, we selected a panel of 2,807 ancestry informative markers with large allele frequency difference (FST > 0.3) between ID-MT and Papuan, and we ran STRUCTURE with linkage model and followed the procedure and parameter settings described in previous studies (13, 14). In the Affymetrix 6.0 dataset, PCA, admixture proportions, and time of admixture estimation were performed using the StepPCO software (12).
Supplementary Material
Acknowledgments
We thank R. Gray and S. Greenhill for supplying the linguistic distances and R. Blust and R. Gray for valuable discussion. This research was supported in part by the Ministry of Science and Technology International Cooperation Base of China. These studies were supported by National Science Foundation of China Grants 31171218 (to S.X.), 30971577 (to S.X.), and 30890034 (to L.J.), Shanghai Rising-Star Program 11QA1407600 (S.X.), and Science Foundation of the Chinese Academy of Sciences (CAS) Grants KSCX2-EW-Q-1-11 (to S.X.), KSCX2-EW-R-01-05 (to S.X.), and KSCX2-EW-J-15-05 (to S.X.). S.X. is Max-Planck Independent Research Group Leader and member of the CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges the support of the K. C. Wong Education Foundation, Hong Kong. I.P. and M.S. acknowledge support from the Max Planck Society. M.K. acknowledges support from the German Research Council Sonderforschungsbereich 680 “Molecular Basis of Evolutionary Innovations.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
2A complete list of The HUGO Pan-Asian SNP Consortium can be found in SI Text.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118892109/-/DCSupplemental.
References
- 1.Bellwood PS, Fox JJ, Tryon DT. The Austronesians: Historical and Comparative Perspectives. Canberra, Australia: ANU E Press; 1995. [Google Scholar]
- 2.Hill C, et al. A mitochondrial stratigraphy for island southeast Asia. Am J Hum Genet. 2007;80:29–43. doi: 10.1086/510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soares P, et al. Climate change and postglacial human dispersals in southeast Asia. Mol Biol Evol. 2008;25:1209–1218. doi: 10.1093/molbev/msn068. [DOI] [PubMed] [Google Scholar]
- 4.Donohue M, Denham T. Farming and language in Island Southeast Asia: Reframing Austronesian history. Curr Anthropol. 2010;51:223–256. [Google Scholar]
- 5.Karafet TM, et al. Major east-west division underlies Y chromosome stratification across Indonesia. Mol Biol Evol. 2010;27:1833–1844. doi: 10.1093/molbev/msq063. [DOI] [PubMed] [Google Scholar]
- 6.Mona S, et al. Genetic admixture history of Eastern Indonesia as revealed by Y-chromosome and mitochondrial DNA analysis. Mol Biol Evol. 2009;26:1865–1877. doi: 10.1093/molbev/msp097. [DOI] [PubMed] [Google Scholar]
- 7.Cox MP, Karafet TM, Lansing JS, Sudoyo H, Hammer MF. Autosomal and X-linked single nucleotide polymorphisms reveal a steep Asian-Melanesian ancestry cline in eastern Indonesia and a sex bias in admixture rates. Proc Biol Sci. 2010;277:1589–1596. doi: 10.1098/rspb.2009.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellwood P. The First Farmers: The Origins of Agricultural Societies. Oxford: Blackwell; 2005. [Google Scholar]
- 9.Gray RD, Drummond AJ, Greenhill SJ. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science. 2009;323:479–483. doi: 10.1126/science.1166858. [DOI] [PubMed] [Google Scholar]
- 10.The HUGO Pan-Asian SNP Consortium Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 11.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pugach I, Matveyev R, Wollstein A, Kayser M, Stoneking M. Dating the age of admixture via wavelet transform analysis of genome-wide data. Genome Biol. 2011;12:R19. doi: 10.1186/gb-2011-12-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Jin L. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am J Hum Genet. 2008;83:322–336. doi: 10.1016/j.ajhg.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S, Huang W, Qian J, Jin L. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet. 2008;82:883–894. doi: 10.1016/j.ajhg.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 19.Jordan FM, Gray RD, Greenhill SJ, Mace R. Matrilocal residence is ancestral in Austronesian societies. Proc Biol Sci. 2009;276:1957–1964. doi: 10.1098/rspb.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hage P, Marck J. Matrilineality and the Melanesian origin of Polynesian Y chromosomes. Curr Anthropol. 2003;44:S121–S127. [Google Scholar]
- 21.Lewis MP. Ethnologue: Languages of the World, 16th ed. 2009. Available at http://www.ethnologue.com/. Accessed May 5, 2011.
- 22.Bellwood P, et al. 35,000 Years of Prehistory in the Northern Moluccas. Rotterdam: Balkema; 1998. [Google Scholar]
- 23.Blust R. Central and Central-Eastern Malayo-Polynesian. Oceanic Linguistics. 1993;32:241–293. [Google Scholar]
- 24.Wollstein A, et al. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 25.Denham T, Donohue M. Pre-Austronesian dispersal of banana cultivars West from New Guinea: Linguistic relics from Eastern Indonesia. Archaeol Oceania. 2009;44:18–28. [Google Scholar]
- 26.Ross M. Pronouns as a preliminary diagnostic for grouping Papuan languages. In: Pawley A, Attenborough R, Golson J, Hide R, editors. Papuan Pasts: Cultural, Linguistic, and Biological Histories of Papuan-Speaking Peoples. Canberra, Australia: Pacific Linguistics; 2005. pp. 15–65. [Google Scholar]
- 27.Soares P, et al. Climate change and postglacial human dispersals in southeast Asia. Mol Biol Evol. 2008;25:1209–1218. doi: 10.1093/molbev/msn068. [DOI] [PubMed] [Google Scholar]
- 28.Friedlaender JS, et al. The genetic structure of Pacific Islanders. PLoS Genet. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat Rev Genet. 2011;12:603–614. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 30.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 31.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- 33.Xu S, Gupta S, Jin L. PEAS V1.0: A package for elementary analysis of SNP data. Mol Ecol Resour. 2010;10:1085–1088. doi: 10.1111/j.1755-0998.2010.02862.x. [DOI] [PubMed] [Google Scholar]
- 34.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.