Abstract
Algorithms derived from measurements of short-peptide (8–10 mers) binding to class I MHC proteins suggest that the binding groove of a class I MHC protein, such as Kb, can bind well over 1 million different peptides with significant affinity (<500 nM), a level of ligand-binding promiscuity approaching the level of heat shock protein binding of unfolded proteins. MHC proteins can, nevertheless, discriminate between similar peptides and bind many of them with high (nanomolar) affinity. Some insights into this high-promiscuity/high-affinity behavior and its impact on immunodominant peptides in T-cell responses to some infections and vaccination are suggested by results obtained here from testing a model developed to predict the number of cell surface peptide–MHC complexes that form on cells exposed to extracellular (exogenous) peptides.
The heterodimeric αβ antigen-binding receptors on T cells (TCR) and the antigen-binding Fab fragments of antibodies are similar structurally and in the great diversity of their ligand-binding site. Their diversity is also generated by similar stochastic gene segment rearrangements in developing B and T cells. But antibodies can be raised against virtually any existing or imaginable organic structures (antigens), if sufficiently stable, while the antigens recognized by T cells’ TCRs are remarkably limited to complexes formed by peptides bound noncovalently to proteins encoded by genes in the major histocompatibility complex (SI Text, R1 and R2).
Although the peptides are bound with 1:1 stoichiometry, well over 1 million different peptides are estimated to bind with significant affinity to an MHC protein's binding site (see below). This extreme binding promiscuity, or degeneracy, is probably matched only by the binding of amino acid sequences in unfolded proteins by some heat shock proteins (1, 2) (SI Text, R3). It is not unlimited, however, because each MHC type (encoded by an MHC allele) usually exercises preferences for peptides having particular motifs (e.g., one or two strategically placed anchor amino acid residues) (3). The MHC binding site can also distinguish between peptides that differ by single amino acid mutations or substitutions, apparently binding them with different affinities (4, 5). Moreover, in the in vivo responses to foreign proteins such as to those proteins of an invading virus, only one (immunodominant) or a few other subdominant peptides out of the great many potential peptides in the pathogen's proteome are recognized by most of a host's responding T cells (6). These disparate aspects of peptide/MHC interactions, some seemingly contradictory, are considered here from the viewpoint of a peptide–MHC interaction model described below.
In the peptide–class I MHC complexes (pMHC-I) that elicit responses by CD8+ T cells, the peptides are usually 8–10 aa in length (SI Text, R4), and they are mostly generated intracellularly by proteolytic cleavage of ubiquinated cytosolic proteins in proteasomes (SI Text, R5). The peptides of cell surface pMHC-I complexes can also arise from extracellular proteins that enter cells by endocytosis, are processed proteolytically, and are then loaded on MHC molecules and ultimately cell-surface–displayed (cross-presented) (7) (SI Text, R6).
A third way to generate cell surface pMHC, the way with which we are here concerned, is to simply expose cells to extracellular peptides. Although generally thought not to occur in vivo, this last process (often termed peptide pulsing or loading) is critically important for efforts to identify peptides that are recognized by particular TCR and stimulate T cells with peptide-pulsed dendritic cells, such as in some peptide-based vaccines (SI Text, R7 and R8). That it can also occur under physiological conditions is suggested by the finding of peptides in lymphatic fluid (8) and by evidence for extracellular loading of cells in the pancreas with an insulin-derived peptide (9).
The dynamics of intracellular (endogenous) peptide production and their association with MHC have been extensively examined (10, 11, 12) (SI Text, R9), and the thermodynamics and kinetics of some pMHC interactions have been characterized in considerable detail (13–15). Still, although peptide pulsing is indispensable for efforts to identify peptides recognized by T cells, the binding of extracellular peptides to cell surface MHC is not sufficiently well-understood to predict the number of pMHCs formed on cells after exposing them to an extracellular peptide. Our aims here are to evaluate a model that may serve as the basis for making such estimates and predictions and to see if the model leads to any insights into promiscuous peptide binding by MHC proteins and some of its ramifications. Our results show good agreement between predicted and measured numbers of cell surface complexes formed by extracellular peptides, and they support evidence that promiscuous binding arises from conformational flexibility of MHC proteins and peptides. They also support and extend evidence that (i) a class I MHC protein can bind millions of different short peptides and (ii) the very slow dissociation of some pMHC complexes accounts for their high (nanomolar) affinity; (iii) they also suggest how MHC binding promiscuity might support the focusing of T-cell responses to some pathogens and vaccines on only a few immunodominant peptides.
Model and the Strategy for Testing
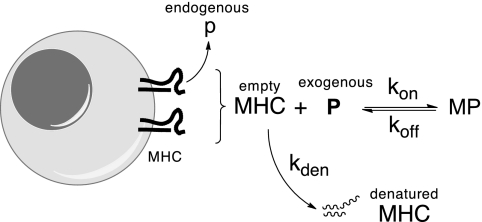
The model (Fig. 1) stems from evidence that extracellular (exogenous) peptides can bind to those cell surface MHC molecules that have vacant peptide-binding sites (16) (SI Text, R10). These empty MHC molecules (M) might have been loaded with an endogenous peptide that subsequently dissociated to leave a vacant groove, or they might have appeared on the cell surface as newly synthesized MHC molecules that escaped being loaded in the endoplasmic reticulum with an endogenous peptide. Whatever their origin, the empty MHCs are unstable: they either bind the extracellular peptide (P), forming an MP complex, or undergo denaturation (17).
Fig. 1.
Diagram outlining the model. Empty MHC molecules on the cell surface bind extracellular (exogenous) peptide (P) or undergo irreversible denaturation.
The model focuses on changes over time in the number of cell surface empty MHC that bind a P and form MP complexes. As is shown below, the number of these complexes is determined by the rates at which empty MHC appear on the cell surface (dM0/dt) and undergo denaturation (kden) and the rates at which they associate with the peptide (kon) and the MP complexes dissociate (koff) (Materials and Methods). To determine koff, cells were pulsed with a peptide, and loss of the resulting MP complexes was then followed over time. Values for the other three parameters were obtained by measuring the numbers of MP on cells that had been incubated with a peptide at diverse concentrations for various times. All of the measured MP values, together with the independently derived koff, were then subjected to a multidimensional search program (MATLAB fminsearch) to obtain kon, kden, and dM0/dt values that best fit the measured MP.
We examined the binding of two synthetic octapeptides to Kb, a mouse class I MHC molecule. The Kb-expressing cells were two cell lines, RMA-S and DC2.4, and bone marrow-derived dendritic cells (1° DCs) from C57BL/6 mice. Most experiments were performed with RMA-S cells, which lack the transporter (TAP) activity that translocates proteasome-generated peptides into the endoplasmic reticulum; such TAP-deficient cells are thought to have (at steady state) a relatively large number of empty class I MHCs on the cell surface. DC2.4, a dendritic cell line, and the 1° DCs are TAP-competent.
The peptides tested were SIINFEKL (often referred to as OVA257–264 but here referred to simply as OVA) and SIYRYYGL (termed SIY). OVA is produced by cells that express chicken ovalbumin; the OVA–Kb complex is a potent agonist for the widely studied TCR on CD8+ T cells from OT-1 TCR transgenic mice (5). The SIY peptide was identified in a combinatorial library (4); the SIY–Kb complex is a potent agonist for another widely studied TCR on cloned CD8+ T cells (2C) that are maintained as cultured cell lines or obtained from 2C TCR+ transgenic mice (18) (SI Text, R11).
Results
Peptide–MHC Dissociation Rate Constants (koff).
When fluorescein-labeled OVA was previously added to live cells under conditions similar to those used in the present work, some peptide was endocytosed and bound to Kb in the endoplasmic reticulum (19). In another earlier study, some cell surface pMHC complexes formed from an exogenous peptide with Ld were rapidly endocytosed and later displayed on the cell surface after a lag of many hours (20). To minimize endocytosis, cytochalasin D was added to cells before and during loading of the exogenous peptides. Under these conditions, peptide dissociation from MP followed single exponential kinetics (Fig. S1). For OVA, koff was 0.0495/h (t1/2 ∼ 14 h), and for SIY, koff was 0.119/h (t1/2 ∼ 5 h) (Table 1). Previously reported koff values for dissociation of various peptides from pMHC complexes have been found to span a wide range, from a few minutes (21) to days, for a class II MHC (SI Text, R12). For OVA–Kb, the finding of a t1/2 of 14 h at 37 °C (Table 1) is consistent with the 21-h t1/2 value found at 25 °C by surface plasmon resonance with recombinant Kb, and the OVA peptide modified covalently for immobilization on a chip (22).
Table 1.
Affinity and rate constants for SIINFEKL (OVA) and SIYRYYGL (SIY) binding to cell surface Kb
| Rate constants | SIINFEKL | SIYRYYGL |
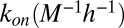 |
1.627 × 107 | 7.889 × 105 |
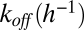 |
0.0495 | 0.1191 |
 |
3.042* | 151 |
 |
1.0872 | 0.4683 |
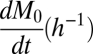 |
[1.00 − 3.19] × 104† | [1.36 − 1.88] × 104† |
| [2.44 − 5.69] × 104‡ | ||
| 1.23 × 104§ |
*In other studies of OVA binding to Kb, KD values ranged from 1.5 to 7 nM (21, 22) (SI Text, R25).
†RMA-S cells.
‡DC2.4 cells.
§Primary dendritic cells.
Binding of OVA and SIY to Kb (kon, kden, dM0/dt).
The numbers of OVA–Kb complexes that formed on RMA-S and DC2.4 cells after incubating them for 20 min to 4 h with OVA at concentrations that ranged from 0.01 to 10 μM were measured with the 25D1.16 antibody. The data (Fig. 2A) were all combined and analyzed according to the equations shown in Materials and Methods using the fminsearch program (MATLAB) and the independently determined koff value (Fig. S1). The best-fitting values for kon, kden, and dM0/dt are shown in Table 1.
Fig. 2.
Binding of peptides to cell surface Kb (37 °C). The curves are based on Eq. 4 and values for kon, koff, kden, and dM0/dt in Table 1. (A) Binding of OVA peptide to Kb on RMA-S (a–f), DC2.4 (g and h), and primary dendritic cells (i). Peptide concentrations are indicated by color in order of orange, green, purple, and (when applicable) blue. (a) 7.85E-009, 7.85E-008, 7.85E-007; (b) 6.30E-008, 6.30E-007, 6.30E-006; (c) 3.00E-007, 3.00E-006, 3.00E-005; (d) 3.00E-007, 3.00E-006, 3.00E-005; (e) 1.00E-008, 1.00E-007, 1.00E-006; (f) 1.00E-008, 1.00E-007, 3.00E-007, 1.00E-006; (g) 1.00E-006, 3.00E-006, 3.00E-005; (h) 3.00E-007, 3.00E-006, 3.00E-005; (i) 1.00E-007, 1.00E-006, 1.00E-005. (B) Binding of SIY peptide to Kb on RMA-S cells. SIY concentrations are indicated by color in the order of orange, green, purple, and (only for a) blue. (a) 1.00E-008, 1.00E-007, 1.00E-006, 1.00E-005; (b) 8.30E-008, 8.30E-007, 8.30E-006.
Antibodies that are highly specific for a particular pMHC complex—like mAb 25D1.16 for the OVA–Kb complex—are rare, perhaps because they are more stable than most other pMHCs and thus exceptionally immunogenic. It was of interest, therefore, to test the model with more commonplace anti-MHC antibodies, which react with allele-specific elements of MHC proteins regardless of the associated peptide. mAb Y3 recognizes such elements in the α1 and α2 domains of Kb (23). Hence, fluorescein-labeled Y3 was used to follow the binding of the SIY peptide to Kb on RMA-S cells. Under these conditions, it was necessary to subtract from the amount of Y3 that bound to SIY-pulsed cells the amount of Y3 bound to cells that were not peptide-pulsed. After incubating the cells with SIY at various concentrations and different times, the numbers of SIY–Kb complexes formed were measured (Fig. 2B). The SIY–Kb dissociation constant (koff) was determined as shown in Fig. S1, Lower, and the values found for kon, kden, and dM0/dt for SIY binding to Kb, shown in Table 1, were obtained as described above for the OVA–Kb interaction.
Unlike kon and kden, which reflect inherent properties of OVA and SIY interactions with empty Kb and of empty Kb alone, it was expected that dM0/dt might vary considerably from experiment to experiment, because the experiments were carried out over a period of many months with cells maintained more or less continuously in culture. The results of each experiment were, therefore, analyzed separately for dM0/dt using the koff, kon, and kden values from Table 1. dM0/dt was seen to vary only from about 10,000 to 20,000 cells−1 h−1 for RMA-S cells and from about 20,000 to 60,000 cells−1 h−1 for DC2.4 cells.
The experimentally determined and predicted numbers of MP per cell are compared in Fig. 3. Agreement was equally evident over short and long pulsing times (20 min to 4 h) with low- or high-peptide concentrations and both the TAP− RMA-S and the TAP+ DC2.4 cells.
Fig. 3.
Scatter plot comparing predicted and measured number of MP complexes per cell (y and x axes, respectively) from data in Fig. 2. For OVA peptide binding, red dots are from RMA-S cells, blue dots are from DC2.4 cells, and green dots are from primary bone marrow-derived dendritic cells. For SIY peptide binding, purple dots are from RMA-S cells. Linear regression values (95% confidence intervals in parentheses) are slope = 0.9757 (0.9291–1.022), y-intercept = −405, and r2 = 0.922. Dashed line corresponds to perfect match between predicted and measured values.
Sensitivity of Calculated Numbers of MP per Cell to Changes in Parameter Values.
To examine the impact of parameter value variations on calculated MP values, each parameter was allowed to vary, in turn, and the other three parameters were kept unchanged. The effects on the calculated values after 1 and 2 h simulated incubation with extracellular peptides are shown in Fig. 4. Over the range of values examined, the largest effects were seen with varying dM0/dt and, notably, varying kden, where lower values, corresponding to increased stability of empty Kb, led to accumulation of more MP per cell.
Fig. 4.
Sensitivity of predicted MP complexes per cell to variations in parameter values; y axes are predicted MP per cell with OVA peptide after 1- and 2-h incubation times (dark and light lines, respectively) at various kon, koff, kden, and dM0/dt values. Vertical dashed lines refer to parameters for OVA peptide in Table 1.
Limitations of the Model.
The pMHC values on dendritic cells are of particular interest, because under physiological conditions, these cells are especially effective, perhaps uniquely so, in activating naïve T cells. For the dendritic cell line DC2.4, the predicted and observed pMHC values were in agreement. With 1° DCs, however, the results were only partially in agreement. The 1° DCs obtained after 7 d of culture of bone marrow cells from B6 mice with growth factor granulocyte macrophage colony-stimulating factor (GMCSF) were highly variable in size and likely in extent of maturation (SI Text, R13). Preliminary trials with these cells, regardless of whether they were activated with LPS, showed that the number of MP formed from extracellular peptide (OVA) increased with incubation times from 20 min to 4 h as with the cultured cell lines (Fig. 3, green symbols). However, after longer incubation times, MP values on the dendritic cells decreased (e.g., after 24 h, they were substantially lower than at 4 h). The decrease was faster than expected from the OVA–Kb koff (Table 1) and was likely caused by the marked endocytotic activity of 1° DC, especially immature DCs (24). The present model does not explicitly take into account the effects of endocytic activity or changes in MP complexes over longer times as cells grow and divide.
Frequency of Short Peptides That Bind to Class I MHC (Kb).
The affinities of various class I MHC proteins for thousands of peptides, measured as IC50 values, have served as the basis for predictive algorithms that sort peptides with considerable accuracy into strong and moderate binders (IC50 < 500 nM) and weak or nonbinders (IC50 > 500 nM) to various MHC-I proteins (25). Using the 500-nM affinity cutoff, it has recently been estimated that about 1% of the ∼10 million unique nonamers in the human proteome bind to human MHC-I: 0.7% bind to what may be the least promiscuous class I MHC protein, HLA-B57, and 1.8% bind to a more conventional protein, HLA-B7 (26). Using a more stringent 50-nM cutoff to identify strong binders, Istrail et al. (27) found a lower frequency of short peptides that bind (0.2–0.5% depending on the MHC-I allele), but importantly, the frequency was the same for predicted proteins from a wide variety of proteomes: human, mouse, Drosophila, Caenorhabditis elegans, many viruses, bacterial species (including archeobacteria from the ocean floor), and even the randomly permuted (shuffled) amino acid sequences of various proteomes.
To further examine this issue, we considered the frequency of Kb-binding octamers from two proteins that are evolutionarily remote from mammals and their pathogens. The selected proteins were from the set of putatively plant-specific proteins that have been identified in a plant genome (Arabidopsis thaliana; i.e., proteins with similar sequences are found only in genomes of plant species and not in genomes from Eukaryota, Bacteria, and Archea) (28). In one of these plant proteins (a β-amylase, 77 kDa; accession no. AT2G45880, UniProt 90/Swiss KB database) with 684 unique 8-mers, nine (1.3%) octamers are predicted by NetMHCpan (version 2.4) (25) to bind to Kb (with higher affinity than IC50 < 500 nM). In another plant-specific protein (a methyltransferase, 77 kDa; accession no. AT1G78240, UniProt 90/Swiss KB database) with 677 unique 8-mers, seven (1.0%) octamers are predicted to bind to Kb with affinity above the cutoff (one of them, SSFAYSRL, with a predicted 2-nM affinity). Thus, it seems that, in general, a representative MHC-I protein, such as Kb, can bind with significant affinity about 1% of all potential octamers. It can be estimated that the number of unique octamers in all 1.3 million proteins in the UniProt 90/Swiss KB database (as of 2005) is about 354 million. Thus, although Kb, a typical MHC-I protein, binds only around 1% of octamers, this small fraction amounts to over 3 million different peptides. And this number is an underestimate because (i) the 500-nM affinity cutoff is somewhat arbitrary (Table 2), (ii) there are limitations to the algorithm's accuracy—estimated in the work by Kosmryl et al. (26) for a similar algorithm to be about 80% accurate, and (iii) excluded from the total are peptides with covalent modifications, such as phosphoryl groups, and other posttranslational modifications, haptenated peptides [like 2,4-dinitrophenyl (DNP) and the great many other low molecular-mass substances that cause allergic contact dermatitis and drug hypersensitivities], and peptides generated in combinatorial libraries, like the one in which the SIY peptide was identified (4). Indeed, the SIY peptide does not exist in known proteomes (as of 2005).
Table 2.
Predicted relative affinity (IC50) of some class I-MHC proteins for the indicated peptides
| Peptide | MHC-I | Affinity (IC50; nM)* |
| 1. SIYRYYGL | Kb | 13.27 |
| 2. SIINFEKL | Kb | 215.07 |
| 3. EQYKFYSV | Kb | 236.63 |
| 4. LSPFPEQL | Kb | 716.55 |
| 5. LSPFPEQL | Ld | 17,349 |
| 6. QLSPFPEQL | Ld | 28,392 |
*Values are from the netMHCpan predictive algorithm, v2.4 (23).
Table 2 shows that, of several extensively studied peptides, NetMHCpan correctly identified three binders to Kb; it also shows, correctly, that peptide #3 (dEV8) is a better binder than peptide #4 (p2Ca) to Kb, consistent with Kb having been successfully crystallized in association with dEV8 but not with p2Ca (SI Text, R14). Still, p2Ca (LSPFPEQL), with affinity for Kb that is below the 500-nM cutoff, actually forms complexes with Kb (SI Text, R15) that are recognized by a TCR (2C), albeit very weakly (29), and elicits a specific cytolytic response by cloned CD8+ 2C T cells (30). Likewise, the two peptides (#4 and #5) that are predicted not to be bind significantly to Ld actually form complexes with Ld that may be the most potent natural T-cell agonists known (29, 31). It may be that predicted values for peptide-Ld complexes are erratic because the training set is limited.
Discussion
According to the model illustrated in Fig. 1, four parameters (kon, koff, kden, and dM0/dt) determine how many pMHC complexes are formed on cells that are exposed to extracellular peptides. How well do the parameters values found here match the corresponding values reported previously, and what bearing do they have on binding promiscuity? The koff and KD values found here are consistent with those reported previously for various pMHC complexes, including OVA–Kb (21, 22) (Table 1, SI Text, R25, and Fig. S1).
The association (kon) and denaturation rate constants (kden), however, are of particular interest for promiscuity. The procedures used to measure kon for peptide binding to ostensibly empty MHC molecules have varied widely, and the reported association rate constants have spanned a wide range, roughly from 102 to 106 M−1s−1. Many reported values are at the low end of this range (21) (SI Text, R16 and R17), similar to those values found here and also for peptide binding to a class II MHC (32). When expressed in conventional units (M−1s−1) rather than in the units (M−1h−1) used in Table 1, we found 4,900 M−1s−1 for OVA–Kb and 217 M−1s−1 for SIY–Kb (Table 1). In contrast, the highest kon reported was about 106 M−1s−1 for a nonamer binding to Db (33). The latter, however, is still 10–100 times slower than the value found for some antibody–hapten reactions; for instance, the kon for binding an ε-2,4-DNP-lysine nonamer [DNP-(lysine)9] by a homogeneous antibody (myeloma protein) was 107 M−1s−1, and the same protein's binding of the smaller hapten, ε-2,4-DNP-lysine, was 108 M−1s−1, which was almost diffusion limited (SI Text, R18 and R19).
A basis for these disparate kon values is suggested by studies in which the binding of dansyl- or fluorescein-labeled peptides to recombinant MHC proteins was monitored continuously (13–15, 33). Under these conditions, the association kinetics were biphasic: one rate, reflecting a slow unimolecular step, was attributed to changes of MHC conformation from peptide-unreceptive to -receptive conformations, and the other rate was attributed to peptide binding to the MHC's receptive conformation. Because there is also evidence that peptides are flexible and can adopt different configurations (9, 34, 35) (SI Text, R26), the very slow overall on-rate values could reflect the time required to achieve sufficient mutual configurational complementarity for flexible MHC and flexible peptides to form an initial complex, which might then undergo an induced fit process. The resulting very slow dissociation of some pMHC could account for their high affinity (Table 1, OVA–Kb). The binding of flexible MHC and peptides is expected to have an entropic cost. To assess the cost, the entropy change for a peptide–MHC interaction was determined and actually found to be favorable, probably because of the hydrophobicity of the peptide and the MHC's binding site (13). Whether other peptide–MHC reactions are also entropy-driven remains to be seen.
Although conformational variability underlies an MHC molecule's ability to bind many different peptides (13), a structural basis for this promiscuity is evident in X-ray crystallographic findings. Most of the hydrogen bonds between bound peptide and an MHC's binding site residues involve the peptide's backbone main chain atoms, a common feature of peptides bound in extended conformation (SI Text, R20–R22). In contrast, the preferential binding of some peptides (determinant selection) arises from interactions of side chains of peptide residues at the anchor and some other positions of the peptide with MHC residues in depressions or pockets in the binding site.
If slow association rates and MHC conformational variability are the keys to promiscuous peptide binding, the lifetime of empty MHC molecules (indicated by kden) is significant. The kden found here correspond to t1/2 values of 38–88 min (at 37 °C). These values (∼50 min on average) (Table S1) are similar to the only other t1/2 (55 min) for an empty MHC of which we are aware; the latter was found for a human class II MHC, HLA-DR [calculated by Kevin Fowler from data in the work by Grotenbreg et al. (32) using a model that differed considerably from the model used here]. Whether these unexpectedly long-lived empty MHCs found for one mouse class I and one human class II MHC are representative of MHC proteins in general remains to be seen. For effective peptide pulsing of cells, the lifetimes of empty MHCs should exceed peptide–MHC association rates (kon > kden). This inference is supported by the simulated effect of reducing kden on the number of predicted pMHC complexes formed (Fig. 4).
In contrast to promiscuous peptide binding to MHC, only one or very few of the short peptides that can be potentially derived from a virus's proteins are actually recognized in vivo by the most of the T cells that respond to some virus infections (6). An extreme example of such immunodominant peptides is seen in C57/BL6 (B6) mice, which have two MHC-I proteins, Kb and Db. In these mice, the majority of CD8+ T cells produced in response to influenza virus infection of the respiratory tract recognize only 1 octamer of viral origin (NP366–374 in association with Db) of the over 4,000 potential octamers in the virus's proteome (about 4,400-aa residues encoded in 11 proteins). Similar immunodominance of a few peptides is also evident in humans infected with influenza virus, but the dominant peptides differ among various individuals, even those individuals sharing the same restricting MHC (36). Altering the route of vaccinia virus infection in mice also results in changes in the vaccinia virus-derived dominant peptides that elicit T-cell responses (37, 38).
Various mechanisms could contribute to peptide immunodominance (6). It could also be favored by promiscuous peptide binding if, as is likely, binding encounters between unselected pMHC and naïve, unselected T cells only rarely leads to activation of a T cell. MHC binding promiscuity allows the generation of large pMHC libraries from antigenic proteins, and the larger and more diverse the library, the greater the chance that one or a few of its pMHC will be bound strongly enough to the TCR on a T cell to stimulate the cell to proliferate and thereby identify an immunodominant peptide. Because somewhat different pMHC libraries may be generated by differences in antigen-presenting cells at various anatomic sites, the immunodominant peptides arising from a pathogen may well vary and depend on the route of infection (37, 38).
It is of interest to note similarities between promiscuous peptide binding by MHC proteins and by some heat shock proteins (Hsp 70 family). DnaK, the bacterial homolog of ubiquitous Hsp70 proteins, binds (with 5-nM to 5-μM affinity) many short amino acid sequences in unfolded proteins, the consensus sequence of bound peptides consisting of four to five residues enriched in hydrophobic amino acids flanked by cationic residues. These sequences are typically separated in unfolded proteins by around 50–100 intervening residues (1, 2), a frequency not unlike that of octamers that bind to class I-MHC proteins.
The enormous peptide binding promiscuity of MHC is to be expected, of course, because the proteins encoded by the few class I MHC genes in each individual (e.g., HLA-A, -B, and -C in humans and HLA-K, -L, and -D in mice) have to be able to effectively present to an individual's T cells short peptides generated from a great multitude of proteins, including those from exotic microbial pathogens never encountered in a species' evolutionary history. Although many of the T cells produced in immune responses to an antigenic protein may be elicited by only one or a few of the many peptides that are potentially generated from that protein, it is ironic that the adaptive immune system, which is noted for its great specificity, should depend on an initiating ligand–protein reaction that is among the most promiscuous known, ranking with the binding of unfolded proteins by some heat shock proteins.
Materials and Methods
Model.
The processes shown in Fig. 1 can be represented by (Eq. 1)
 |
where M refers to empty MHC molecules and dM0/dt refers to the rate at which empty MHC molecules appear on the cell surface. kon is the rate constant for P binding to empty MHC to form MP, koff is the rate constant for P dissociation from MP, and kden is the rate constant for denaturation of the empty MHC molecules (i.e., the rate at which they lose, essentially irreversibly, the ability to bind peptides).
The rate at which the pMHC complexes of interest (MP) are formed is (Eq. 2)
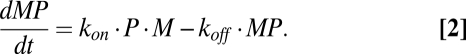 |
Eqs. 1 and 2 combine to (Eq. 3)
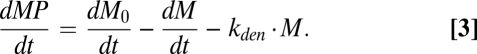 |
The solution to Eq. 3, given in SI Text, yields the following formula (Eq. 4) for MP(t), the number of cell surface MP per cell formed by incubating cells for a specified time with the peptide at a known concentration. The derivation of Eq. 4 assumes there are no significant changes in peptide concentration or rate of appearance of empty MHC during the period of observation (Eq. 4):
 |
where 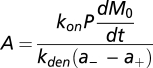 ,
, 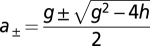 ,
, 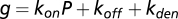 , and
, and 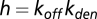 .
.
Cells.
RMA-S cells were from American Type Cell Culture. DC2.4 cells, a gift from K. L. Rock (University of Massachusetts, Worcester, MA), are c-myc–transfected (immortalized) dendritic cells from B6 mice. Primary dendritic cells were derived from bone marrow of B6 mice by standard procedures (SI Text, R23).
Monoclonal Antibodies.
Hybridoma cells producing the 25D1.16 antibody (39) were a gift from Ron Germain (National Institutes of Health, Bethesda, MD). The hybridoma that produces antibody Y3 (SI Text, R24) was from American Type Cell Culture. Both antibodies were purified from hybridoma cell culture supernatants on a protein A column. Antibodies were labeled as described in SI Text with 125I, AlexaFluor 680 (AF), or fluorescein (F), or they were doubly labeled with both 125I and AF. The number of AF or F groups per antibody molecule was determined by UV absorption using a molar extinction coefficient for fluorescein = 77,000 (pH 7.3, 494 nm) and a molar extinction coefficient for AF = 187,400 cm−1 (679 nm). Protein concentrations were determined by bicinchoninic acid (BCA) assay or UV absorption at 280 nm and corrected for chromophore absorption at 280 nm by subtracting 0.2× absorption at 679 nm for AF or subtracting 0.2× absorption at 494 nm for fluorescein. In various preparations, there were 1.3–1.5 AF groups per antibody molecule and 1.7–3.38 F groups per antibody molecule. The 125I-labeled antibodies were initially about 200,000 cpm/μg protein.
Exogenous Peptide Binding to Cells.
Typically, 200,000 cells in 180 μL RPMI 1640-based medium (10% heat-inactivated FCS) were mixed with 20 μL peptide at various concentrations in PBS or with PBS alone (control) in a total volume of 200 μL/well in round-bottomed 96-well plates. After incubating the plates at 37 °C (5% CO2) for various times, they were centrifuged, and the cells were washed one time (with cold PBS) and stained on ice by first adding an Fc blocker and then after 10 min, F- or AF-labeled antibody. After 45 min on ice, the cells were washed and analyzed using a flow cytometer (FACSCaliber; BD Sciences) for F antibody-stained cells or a Licor plate reader for AF antibody-stained cells (Licor).
Measurement of Number of Cognate pMHC per Cell (MP).
Standardized beads, with specified numbers of fluorescein equivalents per bead (Bang Laboratory), were run through the flow cytometer immediately before (and/or after) the stained cells. Calibration curves based on the beads’ fluorescence allowed conversion of fluorescence intensity of stained cells to moles of fluorescein per cell and hence (given the number of chromophore groups per antibody molecule), moles of antibody bound per cell. The latter value, multiplied by two to correct for bivalent antibody binding, yielded the number of antibody-bound MP per cell. The validity of this approach was indicated from the agreement found for MP values when T2–Kb cells were stained with fluorescein-labeled Y3 antibody or 125I-labeled Y3 (340,000 peptide–Kb complexes per cell vs. 320,000 complexes per cell).
For cells stained with F-25D1.16, background staining of nonpulsed cells was negligible (<10%). For cells stained with F-Y3, however, it was necessary to subtract from their fluorescence values found for cells that had been treated identically but not peptide-pulsed. To test the validity of this procedure, RMA-S cells were loaded with OVA, and the resulting OVA–Kb complexes were measured with both antibodies; with F-25D1.16, there were 194,864 complexes per cell, and with F-Y3, there were 176,016 complexes per cell.
For experiments in which the cells were stained with AF antibody or antibody doubly labeled with 125I and AF, cell fluorescence was read in the infrared (700 nm) using the Odysses program on a Licor plate reader. Cells stained with 125I-labeled antibody were read in a γ-detector (Packard), with the known specific radioactivity allowing determination of the number of antibody molecules bound per cell.
Peptides.
These were synthesized and purified by HPLC at the Koch Institute Biopolymer Facility.
Dissociation of Peptides from MP on Cells.
Cells were incubated with cytochalasin D (final concentration = 1–10 μg/mL) before and while they were being loaded with exogenous peptide. After the cytochalasin D-treated cells were peptide-pulsed, they were washed to remove unbound peptide, and the loss of cell surface MP was followed beginning about 0.5–1 h after the washed cells were resuspended in peptide-free medium. When pMHC dissociation was measured with Y3 antibody, Brefeldin A was added to a final concentration of 1 μg/mL to prevent the background cell surface level of Kb from increasing during the period of observation.
Supplementary Material
Acknowledgments
We thank Arup K. Chakraborty for valuable comments, especially for suggesting the connection between promiscuous peptide binding and immunodominance of some peptides, and Charles Whittaker for helpful discussions about bioinformatics. We thank Andrej Kosmrlj for reviewing the manuscript and Yuri Sykulev for discussions at the initiation of this study. Massachusetts Institute of Technology's Undergraduate Research Opportunities Program (UROP) supported X.H.H., K.W., V.K.T., C. Smith, K.K., and L.N. This work was supported in part by Licor, Inc. and a Cancer Center core grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201586109/-/DCSupplemental.
References
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falk K, Rötzschke O, Stevanović S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 4.Udaka K, Wiesmüller KH, Kienle S, Jung G, Walden P. Self-MHC-restricted peptides recognized by an alloreactive T lymphocyte clone. J Immunol. 1996;157:670–678. [PubMed] [Google Scholar]
- 5.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 6.Yewdell JW. Confronting complexity: Real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Heath WR, Carbone FR. Cross-presentation in viral immunity and self-tolerance. Nat Rev Immunol. 2001;1:126–134. doi: 10.1038/35100512. [DOI] [PubMed] [Google Scholar]
- 8.Clement CC, Rotzschke O, Santambrogio L. The lymph as a pool of self-antigens. Trends Immunol. 2011;32:6–11. doi: 10.1016/j.it.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan JF, et al. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat Immunol. 2010;11:350–354. doi: 10.1038/ni.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 11.Wearsch PA, Cresswell P. The quality control of MHC class I peptide loading. Curr Opin Cell Biol. 2008;20:624–631. doi: 10.1016/j.ceb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su RC, Miller RG. Stability of surface H-2K(b), H-2D(b), and peptide-receptive H-2K(b) on splenocytes. J Immunol. 2001;167:4869–4877. doi: 10.4049/jimmunol.167.9.4869. [DOI] [PubMed] [Google Scholar]
- 13.Binz AK, Rodriguez RC, Biddison WE, Baker BM. Thermodynamic and kinetic analysis of a peptide-class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry. 2003;42:4954–4961. doi: 10.1021/bi034077m. [DOI] [PubMed] [Google Scholar]
- 14.Gakamsky DM, Bjorkman PJ, Pecht I. Peptide interaction with a class I major histocompatibility complex-encoded molecule: Allosteric control of the ternary complex stability. Biochemistry. 1996;35:14841–14848. doi: 10.1021/bi961707u. [DOI] [PubMed] [Google Scholar]
- 15.Gakamsky DM, et al. An allosteric mechanism controls antigen presentation by the H-2K(b) complex. Biochemistry. 1999;38:12165–12173. doi: 10.1021/bi9905821. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher TN, et al. Direct binding of peptide to empty MHC class I molecules on intact cells and in vitro. Cell. 1990;62:563–567. doi: 10.1016/0092-8674(90)90020-f. [DOI] [PubMed] [Google Scholar]
- 17.Fahnestock ML, Tamir I, Narhi L, Bjorkman PJ. Thermal stability comparison of purified empty and peptide-filled forms of a class I MHC molecule. Science. 1992;258:1658–1662. doi: 10.1126/science.1360705. [DOI] [PubMed] [Google Scholar]
- 18.Sha WC, et al. Selective expression of an antigen receptor on CD8-bearing T lymphocytes in transgenic mice. Nature. 1988;335:271–274. doi: 10.1038/335271a0. [DOI] [PubMed] [Google Scholar]
- 19.Day PM, Yewdell JW, Porgador A, Germain RN, Bennink JR. Direct delivery of exogenous MHC class I molecule-binding oligopeptides to the endoplasmic reticulum of viable cells. Proc Natl Acad Sci USA. 1997;94:8064–8069. doi: 10.1073/pnas.94.15.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brophy SE, Jones LL, Holler PD, Kranz DM. Cellular uptake followed by class I MHC presentation of some exogenous peptides contributes to T cell stimulatory capacity. Mol Immunol. 2007;44:2184–2194. doi: 10.1016/j.molimm.2006.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura M, Saito Y, Jackson MR, Song ES, Peterson PA. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules isolated from transfected Drosophila melanogaster cells. J Biol Chem. 1992;267:23589–23595. [PubMed] [Google Scholar]
- 22.Chen W, Khilko S, Fecondo J, Margulies DH, McCluskey J. Determinant selection of major histocompatibility complex class I-restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J Exp Med. 1994;180:1471–1483. doi: 10.1084/jem.180.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hämmerling GJ, Rüsch E, Tada N, Kimura S, Hämmerling U. Localization of allodeterminants on H-2Kb antigens determined with monoclonal antibodies and H-2 mutant mice. Proc Natl Acad Sci USA. 1982;79:4737–4741. doi: 10.1073/pnas.79.15.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang LW, Bäumer W, Monteiro-Riviere NA. Cellular uptake mechanisms and toxicity of quantum dots in dendritic cells. Nanomedicine (Lond) 2011;6:777–791. doi: 10.2217/nnm.11.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosmrlj A, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Istrail S, et al. Comparative immunopeptidomics of humans and their pathogens. Proc Natl Acad Sci USA. 2004;101:13268–13272. doi: 10.1073/pnas.0404740101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutiérrez RA, Green PJ, Keegstra K, Ohlrogge JB. Phylogenetic profiling of the Arabidopsis thaliana proteome: What proteins distinguish plants from other organisms? Genome Biol. 2004;5:R53. doi: 10.1186/gb-2004-5-8-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sykulev Y, et al. Kinetics and affinity of reactions between an antigen-specific T cell receptor and peptide-MHC complexes. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 30.Dutz JP, Tsomides TJ, Kageyama S, Rasmussen MH, Eisen HN. A cytotoxic T lymphocyte clone can recognize the same naturally occurring self peptide in association with a self and nonself class I MHC protein. Mol Immunol. 1994;31:967–975. doi: 10.1016/0161-5890(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 31.Sykulev Y, et al. High-affinity reactions between antigen-specific T-cell receptors and peptides associated with allogeneic and syngeneic major histocompatibility complex class I proteins. Proc Natl Acad Sci USA. 1994;91:11487–11491. doi: 10.1073/pnas.91.24.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grotenbreg GM, et al. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Springer S, Doring K, Skipper JC, Townsend AR, Cerundolo V. Fast association rates suggest a conformational change in the MHC class I molecule H-2Db upon peptide binding. Biochemistry. 1998;37:3001–3012. doi: 10.1021/bi9717441. [DOI] [PubMed] [Google Scholar]
- 34.Jacob J, Baker B, Bryant RG, Cafiso DS. Distance estimates from paramagnetic enhancements of nuclear relaxation in linear and flexible model peptides. Biophys J. 1999;77:1086–1092. doi: 10.1016/S0006-3495(99)76958-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Insaidoo FK, et al. Loss of T cell antigen recognition arising from changes in peptide and major histocompatibility complex protein flexibility: Implications for vaccine design. J Biol Chem. 2011;286:40163–40173. doi: 10.1074/jbc.M111.283564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, et al. Systematic identification of immunodominant CD8+ T-cell responses to influenza A virus in HLA-A2 individuals. Proc Natl Acad Sci USA. 2011;108:9178–9183. doi: 10.1073/pnas.1105624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tscharke DC, et al. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moutaftsi M, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 39.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.