Abstract
APOBEC-3 proteins induce C-to-U hypermutations in the viral genome of various viruses and have broad antiviral activity. Generally only a small proportion of viral genomes (≤10−2) are hypermutated by APOBEC-3s but often many cytidines (up to 40%) are converted to uridine. The mechanism of this unique selective hypermutation remains unknown. We found that rat APOBEC-1 over-expression had a hypermutation pattern similar to APOBEC-3s on its substrate apoB mRNA. Transient plasmid transfection of rat APOBEC-1 resulted in 0.4% and 1.8% hypermutations with apoB mRNA in HepG2 and McA7777 cells, respectively. The low frequency of hypermutated apoB mRNA targets was enriched by 3D-PCR at 72–76°C with hypermutation levels increasing up to 67%. Up to 69.6% of cytidines in HepG2 and 75.5% in McA7777 cells were converted to uridines in the hypermutated apoB mRNA. When rat APOBEC-1 was over-expressed by adenovirus, the hypermutation frequency of apoB mRNA increased from 0.4% to ~20% and was readily detected by regular PCR. However, this higher expression efficiency only increased the frequency of hypermutation, not the number of affected cytidines in the hypermutated targets. Rat APOBEC-1 hypermutation was modulated by cofactors and was eliminated by an E181Q mutation, indicating the role of cofactors in the hypermutation. The finding of an APOBEC-3 hypermutation pattern with rat APOBEC-1 suggests that cofactors could also be involved in APOBEC-3 hypermutation. Utilizing HBV hypermutation, we found that KSRP increased APOBEC-3C and -3B hypermutation. These data show that like rat APOBEC-1 hypermutation, cellular factors may play a regulatory role on APOBEC-3 hypermutation.
Keywords: APOBEC-1, APOBEC-3G, hypermutation, editing, HBV, apoB
INTRODUCTION
Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) proteins are members of a protein family sharing the common characteristic of cytidine deaminase activity, consisting of APOBEC-1, APOBEC-2, APOBEC-3 (A to H), and AID (activation induced deaminase).1 Among them, APOBEC-3 family members have been of special interest in the past decade because of their potential role in antiviral innate immunity. APOBEC-3G (A3G) was first identified in the APOBEC-3 family as an anti-HIV-1 cellular factor based on the significant differential infectivity of Vif-defective HIV-1 virus in cultured cells having different levels of A3G expression.2 A3G deaminates cytidine to uridine in nascent minus-strand viral DNA during reverse transcription, resulting in G-to-A hypermutation in the plus-strand DNA and inhibiting the replication of HIV-1.3 HIV-1 Vif protein counteracts the antiviral activity of A3G by targeting it for ubiquitin-dependent degradation, enabling HIV-1 to replicate in target cells.1,4 Since these discoveries, A3G and other family members have been shown to have broad antiviral activity which include not only retroviruses such as other lentiviruses, murine leukemia virus (MLV), and human T-cell leukemia virus type 1 (HTLV-1) but also other viruses such as hepatitis B virus (HBV) and adeno-associated virus.1,5
APOBEC-3 proteins can hypermutate various viral genomes. However, the hypermutation efficiency varies significantly depending on the virus. It has been proposed that in the absence of a fully functional Vif protein, APOBEC-3 proteins can hypermutate HIV-1 genomes to such an extent that they cannot produce infectious progeny virus,4,6 although the effect on HIV-1 replication appears to be partially independent from deamination activity.4 The hypermutation levels of the HIV-1 genome were high enough to be detected by regular PCR amplification followed by cloning and sequencing.3 In contrast, the hypermutation of the HTLV-1 genome was very low despite the virus replicating in the same cell type as HIV-1.7 The viral genomic hypermutation of HTLV-1, HBV, and MLV was only detected by a selective amplification of the hypermutated targets in a process called differential DNA denaturation (3D)-PCR. 3D-PCR is based on the AT-richness in hypermutated DNA targets as a result of C-to-U mutations that lead to melting at a slightly lower temperature than the parent DNA. PCR at a lower denaturing temperature generates a selective amplification of the hypermutated genomes.7–10 By analyzing 3D-PCR selective mutant amplification, it has been found that there are extensive cytidine deaminations in the evaluated regions of HTLV-1, HBV and MLV viral genomes. However, although cytidine targets are extensively mutated, the frequency of mutation in the total population is estimated to be 10−2–10−4.5,7,10–12 For example, only ~10−2 HBV genomes in the total population were hypermutated by APOBEC-3A, but on average 20.5% and 40.1% of cytidines on the minus and plus strands in the evaluated region were mutated respectively in the HBV mutant amplification obtained by 3D-PCR at 88.4°C.5 In other words, although many cytidines at different sites in the proportion selectively amplified by 3D-PCR are hypermutated by APOBEC-3, the overall frequency of HBV hypermutation in the total population is very low. APOBEC-3A hypermutates extensively a small proportion of HBV virus. Recently it was reported that the overall hypermutation rate of HBV (<2–35%) by deep clone sequencing using regular PCR in human cirrhotic samples was much higher than the analyses of virions (<10−2) from cultured cells or serum estimated by 3D-PCR above.13 While the deep sequencing allowed many HBV with 1–3 deaminated cytidines to be detected, the data showed that among the clonal sequences with detected mutations, there were only about 30% sequences containing ≥8 G-to-A mutations. However, up to 86% (50/58) edited cytidines were found among the hypermutated portion, indicating that hypermutation occurred extensively on a relative small portion of HBV virus and the pattern is still identical with that observed by 3D-PCR. A similar pattern is also observed with APOBEC-3 hypermutation on HIV-1 although the hypermutation level is much higher and can be detected by regular PCR sequencing.3,14–17 Massive parallel sequencing analyses of HIV-1 env DNA17 showed that G-to-A hypermutations varied <1–85% depending on the site. 75.2 ± 9.1% was observed at the hotspot 7424 among selected sequences with one or more GG-to-AG mutations. Considering the total sequences analyzed, 43.1 ± 5.2% of HIV-1 env DNA are hypermutated by endogenous APOBEC-3 proteins. Up to 50% of all guanines in the evaluated region are mutated among the hypermutated sequences, indicating that like that in HBV, APOBEC-3 proteins hypermutate a proportion of HIV-1 extensively.
APOBEC-3G hypermutation in cells as described above significantly contrasts with the hypermutation activity of purified APOBEC-3G on minus single strand DNA in a cell-free reaction. In this cell free system, hypermutation induced by A3G is proportional to the APOBEC-3G concentration in dose dependent manner including both the number of mutated bases as well as the mutation frequency at a single base.18 Based on the A3G activity in a cell-free reaction, the extent of hypermutation observed in cells should require a high concentration of A3G that in turn should result in a high mutation frequency of virus in the population. However, what has been observed is that there is a divergence in hypermutation activity. As it is for HBV virions mentioned above, the hypermutation frequency is low (10−2–10−4) but the hypermutation is extensive (up to 40% of cytidines are mutated) in the small proportion selectively amplified by 3D-PCR. This difference between the in vivo and in vitro A3G hypermutation activities suggests the potential presence of cofactor regulation in vivo although A3G alone is functional in vitro. Studying why APOBEC-3 proteins extensively hypermutate only a small proportion of virus may help understand the APOBEC-3 hypermutation mechanism and develop potential antiviral applications.
APOBEC-1 is the founding member of the APOBEC family.19 It is an RNA editing enzyme whose primary cellular target is apoB (apolipoprotein B) mRNA. In conjunction with its cofactors, APOBEC-1s from different species are invariably highly specific and normally deaminate only a single cytidine residue at C6666 on the more than 14,000 nucleotide long apoB mRNA to create a premature translational stop codon.19–20 However, APOBEC-1 editing fidelity was severely compromised when the protein was over-expressed.21 Hypermutation induced by human APOBEC-1 over-expression can be further increased significantly by the cofactor ACF but decreased to a background level by the single amino acid replacements, P29F or E181Q within APOBEC-1. This decreased activity correlated with a markedly reduced protein interaction between APOBEC-1 and ACF.22
Rodent APOBEC-1 is different from human APOBEC-1 despite their amino acids having 69% identity and 79% similarity.23 It has been shown that rat APOBEC-1, but not human APOBEC1, curtailed HIV-1 replication in vitro.24 Additionally murine APOBEC-1 can extensively edit both RNA and DNA in vivo.10 These data indicate that rat APOBEC-1 may have hypermutation activity similar to APOBEC-3. Because of its known natural target, cofactors, and editing mechanism, rat APOBEC-1 is more characterized than APOBEC-3 and may more readily help reveal information about APOBEC-3. It has been known that over-expression of rat APOBEC-1 results in hypermutation, but its hypermutation pattern has not yet been elucidated. In this report, we evaluated rat APOBEC-1 induced hypermutation to identify its pattern, explored the role of cofactors in hypermutation, and investigated the effect of potential cofactors on APOBEC-3 hypermutation on the HBV viral genome. We found that rat APOBEC-1 over-expression hypermutated apoB mRNA at a low frequency but with extensive cytidine to uridine conversion, mimicking APOBEC-3 hypermutation on viral genomes. The induced hypermutation was modulated by cofactors, but was eliminated by the single amino acid mutation E181Q within APOBEC-1. These data suggest a role of APOBEC-1’s unique structure and temporal and spatial interaction with cofactors in determining its hypermutation activity. This pattern similarity led us to find that the APOBEC-1 cofactor KSRP increased APOBEC-3C and -3B hypermutation on the HBV viral genome. These data suggest that like rat APOBEC-1, APOBEC-3 may have cofactor regulated hypermutation.
RESULTS
Rat APOBEC-1 over-expression hypermutates apoB mRNA extensively
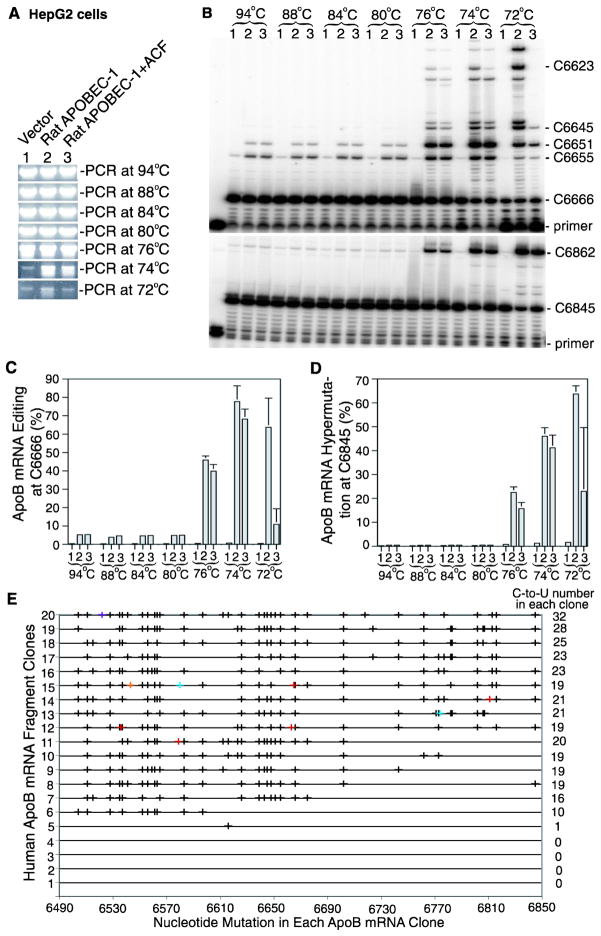
To investigate the rat APOBEC-1 hypermutation pattern, we first expressed the gene using plasmid transient transfection in the human liver cell line HepG2 which has no native APOBEC-1 expression. Two days after transfection, the apoB mRNA region flanking the physiological editing site 6666 was amplified by RT-PCR. Aliquots of the apoB mRNA RT-PCR were subjected to apoB hypermutation analyses by 3D-PCR followed by primer extension and sequencing analyses. As shown in Fig. 1A, 3D-PCR showed no significant differences between vector control and rat APOBEC-1 or rat APOBEC-1 + ACF from 94°C to 76°C. A difference became apparent when the denaturing temperature decreased to 74°C. ApoB was selectively amplified by 3D-PCR at 74°C with rat APOBEC-1 and rat APOBEC-1 + ACF, but not the vector control, suggesting the presence of hypermutated apoB mRNA. The selective apoB mutant amplification with rat APOBEC-1 was still observable at 72°C. A trace amount of apoB was observed with vector control even by 3D-PCR at 72°C and 74°C. This is probably due to the unusual AT richness of apoB mRNA in the evaluated sequence regions (G+C 29.6%) which results in non-specific amplification by 3D-PCR.
Figure 1. Rat APOBEC-1 over-expression hypermutates apoB mRNA in HepG2 cells.
Rat APOBEC-1 was expressed in HepG2 cells with or without rat ACF by transient plasmid transfection. Total RNAs were extracted after a 2 day transfection. The apoB mRNA (6471–6886 nt) was amplified by RT-PCR. The PCR reactions were diluted 1:10 and subjected to 3D-PCR amplification at different denaturing temperatures. The hypermutated apoB mRNA enriched by the 3D-PCR were further analyzed by primer extension and sequencing. A, Detection of 3D-PCR amplifications at different denaturing temperatures by 1% agarose gel electrophoresis with ethidium bromide staining. B, Detection of the hypermutated apoB mRNA in the 3D-PCR amplifications by primer extension analyses. The 3D-PCR amplifications as in figure 1A were subjected to primer extension analyses with primers for editing at the physiological site 6666 (top panel) or for hypermutation at site 6845 (bottom panel). The resultant primer extension products were separated by 8% polyacrylamide urea gel electrophoresis and quantified using a PhosphoImager. C, and D, Graphical presentation of the data obtained from three repeats as in figure 1B for editing levels at site 6666 (C) and hypermutation levels at site 6845 (D). Each bar represents the mean and standard deviation with n=3. E, Sequencing analyses of the 3D-PCR amplifications at 74°C. The 3D-PCR amplifications at 74°C were cloned and 20 clones were randomly selected for DNA sequencing to determine apoB mRNA hypermutation. The C-to-U mutations detected in each clone are graphically presented in black by each individual clone number vs sites of mutation. Other minor nucleotide conversions are marked as follows, A/G, red; T/C, bright blue; A/C, orange; and A/G, purple. The graph right side numbers indicate how many C-to-U conversions are detected in the single clone.
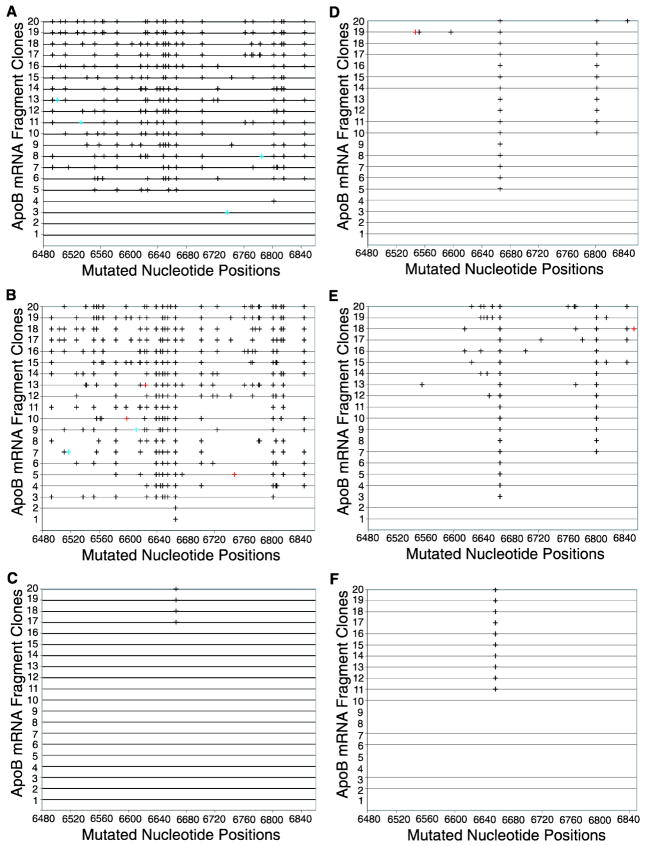
Figure 3. Hypermutation of apoB mRNA in HepG2 cells induced by rat APOBEC-1 over-expression using adenovirus.
Rat or human APOBEC-1 encoded in adenoviruses was expressed in HepG2 cells with or without co-expression of rat or human ACF, respectively. Total RNAs were extracted after a 3 day viral exposure. The apoB mRNA (6471–6886 nt) was amplified by RT-PCR. The PCR amplifications were cloned and 20 clones were randomly selected for DNA sequencing. The C-to-U mutations detected in each clone are graphically presented in black by each individual clone number vs sites of mutation. Other minor mutations are marked as follows, A/G, red; and T/C, bright blue. A, B, and C. ApoB mRNA hypermutation induced by rat APOBEC-1 alone (A), rat APOBEC-1 + rat ACF (B), and rat APOBEC-1-E181Q + rat ACF (C). D, E, and F. ApoB mRNA hypermutation induced by human APOBEC-1 alone (D), human APOBEC-1 + human ACF (E), and human APOBEC-1-E181Q + human ACF (F).
Hypermutated apoB in the regular apoB RT-PCR amplification was further enriched by selective 3D-PCR amplification at 74°C. To further verify hypermutation, the 3D-PCR amplifications at different denaturing temperatures were analyzed by primer extension. Primer extension is a general quantitative analysis method for apoB mRNA editing. It is a modified sequencing analysis for cytidine presence in a sequence at a specific site by using a specific local primer to anneal with apoB template followed by the primer extension. Primer extension analysis provides a reliable quantitative measurement of C-to-U editing frequency in a population of apoB mRNAs.22 Two representative primer extension analyses are presented in figure 1 to reflect editing at the physiological site 6666 and hypermutation at site 6845. As shown in Fig. 1B, 1C, 1D, transient plasmid transfection of rat APOBEC-1 resulted in a 5.2% C-to-U mutation rate at site 6666 by PCR at 94°C while the hypermutation at site 6845 was 0.4%. No significant changes were observed with 3D-PCR at 88°C, 84°C, and 80°C. However, the rat APOBEC-1 induced mutation rate at site 6666 increased from 5.2% to 45.9% with 3D-PCR at 76°C. The hypermutation rate at site 6845 also increased from 0.4% to 22.5%. As the hypermutated apoB was further selectively enriched by 3D-PCR at the lower denaturing temperatures, 74°C and 72°C, the mutation rate at site 6666 further increased to 77.7% and 63.7%, respectively (Fig. 1B, 1C). The hypermutation rate at site 6845 also further increased to 46.0% and 63.7% (Fig. 1B, 1D). The primer extension at site 6666 also showed multiple ladders above site 6666, indicating simultaneous C-to-U mutations of multiple cytidines following site 6666. The results of rat APOBEC-1 + ACF paralleled the rat APOBEC-1 alone results and further confirmed the hypermutation effect of rat APOBEC-1. These data demonstrate that the hypermutated apoB mRNA is present in HepG2 cells over-expressing rat APOBEC-1. However, the hypermutated apoB proportion is low because the hypermutation rate is only 0.4% with PCR at 94°C and becomes as high as 77.7% after being enriched by 3D-PCR at 74°C. Background levels of editing at site 6666 and hypermutation at site 6845 were observed for 3D-PCR vector control amplification at 74°C and 72°C (Fig. 1B). The trace amount of 3D-PCR amplification at 74°C and 72°C was found to contain only wild-type apoB mRNA which is amplified due to high regional AT content.
To further investigate the extensiveness of apoB mRNA hypermutation, the 3D-PCR amplifications at 74°C were cloned and sequenced. As shown in Fig. 1E, 15 out of 20 random clones had 10–32 simultaneous C-to-U conversions in the evaluated region containing 46 cytidines, indicating that apoB mRNA was extensively hypermutated, affecting up to 69.6% of cytidines in the region. Together with the low proportion nature by the 3D-PCR amplification at 74°C, these data demonstrate that rat APOBEC-1 over-expression extensively hypermutates a small proportion of apoB mRNA, mimicking the hypermutation pattern of APOBEC-3 proteins.
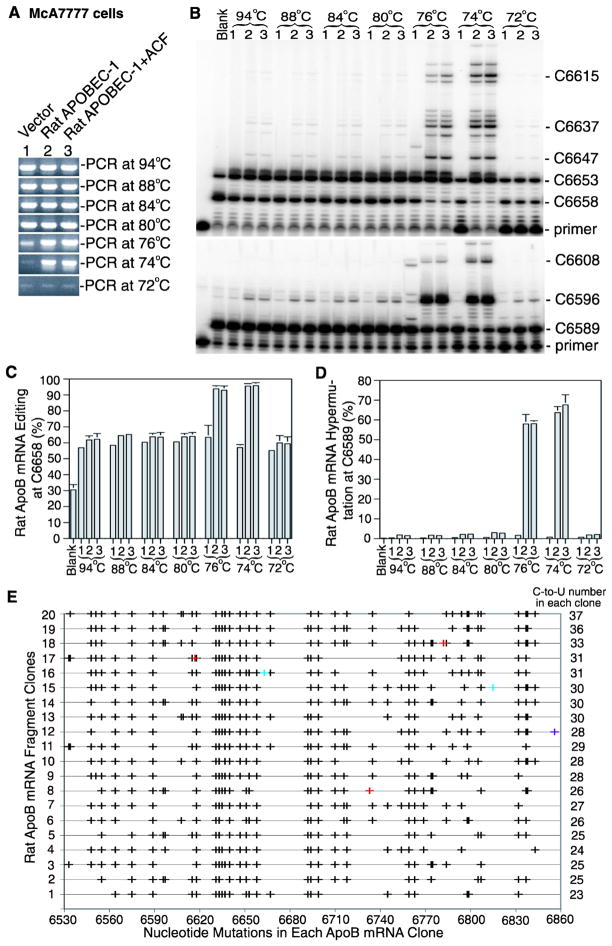
In contrast to human liver that has no APOBEC-1 expression, APOBEC-1 is normally expressed in rat liver. To determine if the hypermutation results induced by rat APOBEC-1 in human cells are also observed in rat cells, rat APOBEC-1 was also over-expressed in the rat liver cell line McA7777 by plasmid transient transfection. As shown in Fig. 2A, hypermutated apoB mRNA in McA7777 cells was selectively amplified by 3D-PCR at 76°C and 74°C with either rat APOBEC-1 alone or rat APOBEC-1 plus ACF while the amplification from the vector control diminished as the denaturing temperature was lowered. Primer extension analyses showed that hypermutated rat apoB could only be detected in the 3D-PCR amplifications at 76°C and 74°C (Fig. 2B, 2D). The blank control representing cells receiving no treatment had 30.5% editing at the physiological specific site 6658 (equivalent to human site 6666) reflecting native rat APOBEC-1 expression. With just vector alone treatment, the editing at site 6658 increased from 30.5% to 57%, indicating that the native editing was sensitive to vector exposure. With either rat APOBEC-1 alone or APOBEC-1 plus ACF treatment, the native site editing increased further to about 62% (Fig. 2B, 2C). However, the hypermutation rates as determined at site 6589 were <0.4% for both blank and vector controls (Fig. 2B). The treatment of rat APOBEC-1 alone or APOBEC-1 plus ACF increased the hypermutation from <0.4% to 1.8% or 1.5%, respectively. The editing levels remained comparable with all 3D-PCR amplifications at denaturing temperatures higher than 76°C while the hypermutation ranged 2%–3%. The hypermutation rates increased from 1.8% to 58.0% and 63.7% for 3D-PCR at 76°C and 74°C, respectively, with rat APOBEC-1 over-expression while the editing at the native site 6658 increased from 64% to 93.9% and 95.5%, respectively (Fig. 2C, 2D). Parallelling rat APOBEC-1 alone, rat APOBEC-1 plus ACF further confirmed rat APOBEC-1 hypermutation. Like human apoB, rat apoB was also amplified with the vector control by 3D-PCR having low efficiency even at 72°C and 74°C. However, there was no hypermutation observed with these temperatures by primer extension at site 6596 and only normal levels of apoB mRNA editing at the physiological site 6658 (Fig. 2B, 2D), indicating that 3D-PCR amplifications with vector control at 72°C and 74°C are a result of presence of high AT content in the evaluated sequence regions of rat apoB (G+C 33.1%).
Figure 2. Rat APOBEC-1 over-expression hypermutates apoB mRNA in McA7777 cells.
Rat APOBEC-1 was expressed in McA7777 cells with or without rat ACF by transient plasmid transfection as in Fig. 1. Total RNAs were extracted after a 2 day transfection. The rat apoB mRNA (6507–6878 nt) was amplified by RT-PCR and was followed by apoB mRNA hypermutation analyses with 3D-PCR, primer extension, and sequencing. A, Detection of apoB mRNA 3D-PCR amplifications at different denaturing temperatures by 1% agarose gel electrophoresis. B, Primer extension analyses of the hypermutated apoB mRNA in the 3D-PCR amplifications. The 3D-PCR amplifications as in figure 2A were subjected to primer extension analyses with primers for editing at the physiological site 6658 (top panel) or for hypermutation at site 6589 (bottom panel) followed by gel separation and PhosphoImager quantification. The blank control represents untreated McA7777 cells. C, and D, Graphical presentation of the data obtained from three repeats as in figure 2B for editing levels at site 6658 (C) and hypermutation levels at site 6589 (D). Each bar represents the mean and standard deviation with n=3. E, Sequencing analyses of the 3D-PCR amplifications at 74°C. The 3D-PCR amplifications at 74°C were cloned and 20 clones were randomly selected for DNA sequencing. The C-to-U mutations detected in each clone are graphically presented in black by each individual clone number vs sites of nucleotide mutation. Other minor mutations are marked as follows, A/G, red; and T/C, bright blue. The graph right side numbers indicate how many C-to-U conversions are detected in the single clone.
Further cloning and sequencing of 3D-PCR amplification at 74°C showed that there were 23–37 simultaneous C-to-U conversions in the evaluated region containing 49 cytidines, indicating that up to 75.5% of the cytidines in the region were hypermutated (Fig. 2E). Consistent with that in HepG2 cells, the data from McA7777 cells suggest that rat APOBEC-1 over-expression hypermutates extensively a small proportion of rat apoB mRNA.
Rat APOBEC-1 hypermutation can be directly detected by regular PCR with adenoviral expression
ApoB mRNA hypermutation is correlated to the expression levels of APOBEC-1. To explore rat APOBEC-1 hypermutation when more highly expressed, rat APOBEC-1 was over-expressed by adenovirus in HepG2 cells. Consistent with previous reports,22 apoB mRNA hypermutation was readily detected by regular RT-PCR amplification followed by direct cloning and sequencing. As shown in Fig. 3A, 16 out of 20 random clones were found to have 6–23 simultaneous C-to-U conversions in the evaluated region affecting a total of 46 cytidines, indicating that 80% of apoB mRNA was hypermutated based on the percentage of positive clones and up to 50% of cytidines were mutated in the region. These data indicate that adenoviral expression of rat APOBEC-1 increased the proportion of hypermutated apoB mRNA from 0.4% (the mutation rate at site 6845 as determined by primer extension analyses) to 80% (the proportion determined by clonal analyses) compared to the plasmid transfection as in figure 1, but the quantities of mutated cytidines were comparable, 50% vs 69.6%. Co-expression of rat APOBEC-1 and ACF resulted in 18 out of 20 clones with 11–26 simultaneous C-to-U conversions (Fig. 3B), indicating that 90% of apoB mRNA was hypermutated. Up to 57% of cytidines were converted to uridines in the region, similar to results using rat APOBEC-1 alone.
In contrast, human APOBEC-1 over-expression in HepG2 cells resulted in only 1–3 C-to-U conversions in 16 out of 20 randomly selected clones and these C-to-U conversions were predominantly found at the physiologically specific site 6666 and site 6802 even in the presence of cofactor ACF (Fig. 3D, 3E). These findings likely reflect the stronger protein interaction of human APOBEC-1 associating with a mooring sequence motif located 4–6 nucleotides downstream from site 6666 and a mooring sequence like motif for site 6802. On the other hand, the E181Q mutation eliminated hypermutation induced by rat APOBEC-1 and left only editing at the physiological site 6666 as was found for human APOBEC-1 (Fig. 3C, 3F), potentially reflecting a decreased interaction between rat APOBEC-1 and cofactors like ACF. Taken together, these data suggest that the unique structure of rat APOBEC-1 and its interaction with its auxiliary cofactors may play important roles in hypermutation induced by over-expression. The expression levels of rat APOBEC-1 affect the frequency of apoB mRNA hypermutation, not the number of mutated cytidines within apoB mRNA.
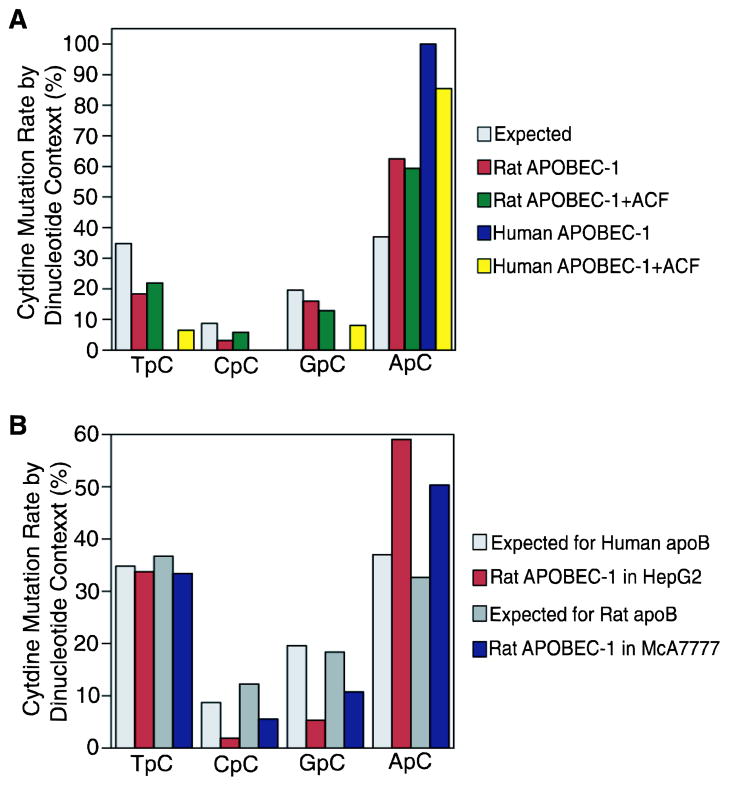
It has been shown that different APOBEC-3 proteins have different immediate 5′ nucleotide preference for cytidine deamination.5–8,13 To evaluate APOBEC-1’s hypermutation preference, rat and human APOBEC-1 induced apoB mRNA hypermutation (Fig. 3) was analyzed and compared to those obtained by 3D-PCR in Fig. 1E and 2E. Within the human apoB evaluated region (GenBank ID: X04714, 6493–6861, 369 bp), there are 46 cytidines which can be divided into 16 TpC, 4 CpC, 9 GpC and 17 ApC according to their immediate 5′ nucleotides, T, C, G, or A. As shown in Fig. 4A, human APOBEC-1 hypermutated ApC exclusively. Co-expression of ACF reduced human APOBEC-1 specificity on ApC from 100% to 85%. Compared to human APOBEC-1, rat APOBEC-1 was less specific and hypermutated 62.5% ApC along with 18% TpC, 3% CpC, and 16% GpC. Co-expression of rat ACF minimally decreased rat APOBEC-1 specificity for ApC from 62.5% to 59.4%.
Figure 4. Cytidine deamination dinucleotide context analyses of APOBEC-1 hypermutation.
APOBEC-1 hypermutation sequences obtained in Fig. 1E, 2E, and 3 were analyzed according to the 5′ immediate nucleotides of each hypermutated cytidine. The data were summarized and presented in graphs to show their dinucleotide preferences. A, Dinucleotide context analyses of rat and human APOBEC-1 hypermutation on human apoB mRNA by adenoviral expression as in HepG2 cells, Fig. 3. B, Dinucleotide context analyses of rat APOBEC-1 hypermutation on human or rat apoB mRNA by plasmid transfections as in HepG2 cells, Fig. 1E or McA7777 cells, Fig. 2E, respectively.
Rat APOBEC-1 hypermutates both human and rat apoB mRNA as C-to-U mutations and there were no G-to-A mutation after considering PCR error (see Supplemental Figure 1 which presents the top 10 clone sequences of Fig. 1E and 2E). As shown in Fig. 4B, rat APOBEC-1 hypermutation on human apoB mRNA by 3D-PCR (Fig. 1E) showed a 5′ nucleotide preference (59.1% for ApC) comparable to that obtained by regular PCR (Fig. 4A, 62.5% for ApC). Rat APOBEC-1 hypermutated rat apoB mRNA (3D-PCR in Fig. 2E) by 33.4% TpC, 5.5% CpC, 10.7% GpC and 50.3% ApC, thus being less specific compared to that on human apoB mRNA (Fig. 4B). The decreased preference for ApC from 59.1% in HepG2 cells to 50.3% in McA7777 cells indicates that cofactors in rat cells may further contribute to rat APOBEC-1 hypermutation.
APOBEC-1 cofactors variably affect rat APOBEC-1 hypermutation
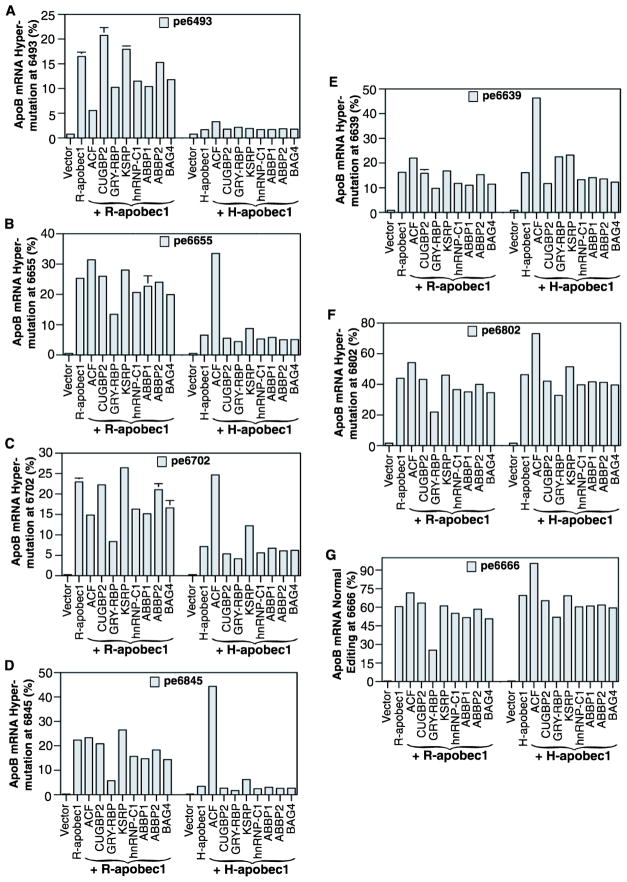
APOBEC-1 alone is not functional, requiring cofactors for physiological apoB mRNA editing at site 6666. Based on their ability to interact with APOBEC-1 and/or apoB mRNA, a group of proteins has been identified as APOBEC-1 cofactors including ACF, CUGBP2, GRY-RBP, KSRP, hnRNP-C1, ABBP1, ABBP2, and BAG4.25 Among them, ACF forms the minimal functional unit with APOBEC-1 for apoB mRNA editing in vitro. To further investigate the role of APOBEC-1 cofactors on rat APOBEC-1 hypermutation, we co-expressed rat APOBEC-1 with these rat or human cofactors in HepG2 cells using adenoviral expression which increases the proportion of hypermutated apoB mRNA to levels readily detectable by regular apoB mRNA RT-PCR (Fig. 3). Hypermutation levels were then quantitatively determined by primer extension analyses at multiple sites with selection based on sequencing results (Fig. 3). Representative data from sites 6493, 6655, 6702, 6845, 6639, 6802, and 6666 are graphically presented in Fig. 5.
Fig. 5. Rat APOBEC-1 hypermutation is regulated by its auxiliary cofactors.
Rat or human APOBEC-1 and cofactors encoded in adenoviruses were co-expressed in HepG2 cells and total RNAs were extracted after a 2 day viral exposure. The apoB mRNA (6471–6886 nt) was amplified by RT-PCR and the apoB amplification were subjected to primer extension analyses with primers specific for sites 6493, 6655, 6702, 6845, 6639, 6802, and 6666. The resultant primer extension products were separated by 8% polyacrylamide urea gel electrophoresis and quantified using a PhosphoImager. The data obtained are summarized and graphically presented. The pe6493, etc. stands for the primer extension analyses at site 6394, etc. Each bar represents the mean and standard deviation with n=3.
As shown in Fig. 5, hypermutation levels of rat APOBEC-1 varied at different sites and were variably affected by different cofactors. Rat APOBEC-1 induced about a 20% hypermutation rate at sites 6493, 6639, and 6845. The levels increased to about 25% at sites 6655 and 6702. The levels further increased to about 40% at site 6802 and 60% at the physiological site 6666. These data indicate a 2 to 3 fold increase of hypermutation activity at site 6666 having the mooring sequence motif and at site 6802 having the mooring like motif, likely due to the preferential binding of cofactors at these locations. ACF moderately increased rat APOBEC-1 activity at sites 6639, 6655, 6666, and 6802 while decreasing the activities by 66% and 35% at sites 6493 and 6702, respectively. GRY-RBP decreased rat APOBEC-1 hypermutation by 40%–75% at all sites. KSRP slightly increased rat APOBEC-1 hypermutation. All these cofactors except CUGBP2 are endogenously expressed in HepG2 cells.26 The adenoviral over-expressions increased their expression levels by 2–3 fold in general (data not shown). The observed hypermutation changes indicate that rat APOBEC-1 hypermutation can be modulated by these cofactors. Human APOBEC-1 induced hypermutation rates were comparable to rat APOBEC-1 at sites 6666 and 6802 due to the presence of the mooring or mooring like motifs in apoB mRNA. Hypermutation at site 6639 was also comparable between rat and human APOBEC-1. However, the hypermutation rate induced by human APOBEC-1 was significantly lower than that of rat APOBEC-1 at all other sites, especially at distal sites 6493 and 6845 where rat APOBEC-1 hypermutation was 6–10 fold higher. On the other hand, human APOBEC-1 hypermutation was more sensitive to ACF. Taken together, these data indicate that like human APOBEC-1, cofactors also play an important regulatory role in rat APOBEC-1 hypermutation.
It is noteworthy that plasmid (Fig. 1) and adenoviral (Fig. 5) induced expression caused different ACF effects on rat APOBEC-1 hypermutation. Rat ACF decreased hypermutation in plasmid transfection while an increase was observed in adenoviral expression where ACF expression was increased 2–3 fold. The rat ACF expression levels by plasmid transfection are much lower and the impact could be limited especially considering existing endogenous human ACF expression.
APOBEC-1 cofactor KSRP affects APOBEC-3C, -3B hypermutation on HBV viral genome
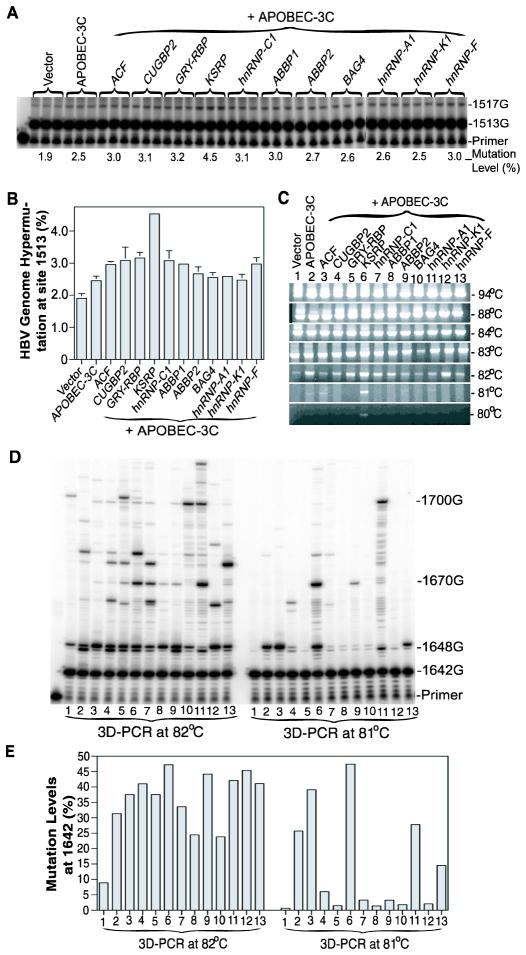
The contrast between in vivo and in vitro APOBEC-3G hypermutation activities suggests the potential presence of cofactor regulation. The hypermutation pattern similarity between APOBEC-3 and rat APOBEC-1 together with the cofactor requirement in rat APOBEC-1 hypermutation further suggests that there could potentially be common cofactors. To search for potential cellular factors involved in APOBEC-3 hypermutation, we utilized APOBEC-3C hypermutation on the HBV viral genome in HepG2 cells to explore the effect of APOBEC-1 cofactors. HBV DNA and A3C in plasmids were co-transfected in HepG2 cells with the APOBEC-1 human cofactors including ACF, CUGBP2, GRY-RBP, KSRP, hnRNP-C1, ABBP1, ABBP2, and BAG4. hnRNP-A1, K, and F were also included because of the reported regulatory role of hnRNP-K in HBV replication.27 After co-expression for 3 days, HBV viral genomes were extracted and the hypermutation of HBV was analyzed by PCR amplification of the HBV X-gene region followed by 3D-PCR at different denaturing temperatures, amplicon primer extension, and sequencing analyses. The representative data are shown in Fig. 6.
Fig. 6. The effect of APOBEC-1 cofactors on APOBEC-3C hypermutation.
APOBEC-1 cofactors encoded in pcDNA3.2 plasmids were co-transfected with HBV and APOBEC-3C into HepG2 cells and the HBV viral genome was extracted after 72 h. The HBV x-gene was amplified by regular PCR and the resultant amplifications were analyzed by direct primer extension, 3D-PCR at different denaturing temperatures, and follow-up primer extension analyses of 3D-PCR amplifications. A and B, Direct primer extension analyses of HBV x-gene hypermutation at site 1513 in regular PCR amplifications followed by 8% polyacrylamide urea gel separation (A) and graphical presentation of the hypermutation quantification using a PhosphoImager (B). Each bar represents the mean and standard deviation with n=3. C, HBV x-gene hypermutation analyses by 3D-PCR at different denaturing temperatures followed by 1% agarose gel electrophoresis and ethidium bromide staining. D and E, Primer extension analyses of HBV x-gene hypermutation levels in 3D-PCR amplifications: 3D-PCR amplifications at 82°C and 81°C from figure 6C (lanes 1–13) were primer extended with a primer specific for site 1642. The primer extension products were separated by 8% polyacrylamide urea gel electrophoresis (D) and quantified using a PhosphoImager. The data are summarized and presented graphically (E).
The HBV X-gene was readily detected by regular PCR at a denaturing temperature of 94°C. As shown in Fig. 6A, the hypermutation of HBV in the X gene was detectable even with the regular PCR amplified at 94°C by the primer extension analyses at site 1513 although the rate was low. APOBEC-3C alone had 2.5% hypermutation activity that was increased up to 4.5% by the APOBEC-1 cofactor KSRP (Fig. 6B). The 3D-PCR amplifications at different denaturing temperatures showed that the A3C hypermutated HBV was selectively amplified at 82°C and the co-expresssion of KSRP and A3C resulted in higher HBV amplification than A3C alone while the vector control amplification that reflected endogenous APOBEC-3 expression was diminishing (Fig. 6C). When the denaturing temperature was lowered to 81°C, only KSPR + A3C resulted in a readily detectable band, indicating that there were more extensive mutations with KSRP + A3C than A3C alone.
As shown in Fig. 6D, 6E, primer extension analyses at site 1642 showed that the A3C hypermutation levels were variably affected by different cofactors. In 3D-PCR amplifications at 82°C, A3C alone had a 31% mutation rate and KSRP increased it up to 47%. In the 3D-PCR amplicons at 81°C, the vector control was decreased to a background level, 0.6%. A3C alone had a 26% hypermutation activity and KSRP increased it to 47%. There was detectable staining of the 81°C 3D-PCR amplifications for A3C alone and its co-expression with ACF or KSRP, indicating that there were enough DNA templates in the 3D-PCR amplifications for the primer extension detection (Fig. 6C). These data demonstrate that KSRP increases A3C hypermutation on HBV virus. The decreases observed with the other treatments could not be evaluated because there were insufficient DNA amplifications at the 81°C 3D-PCR for hypermutation determination.
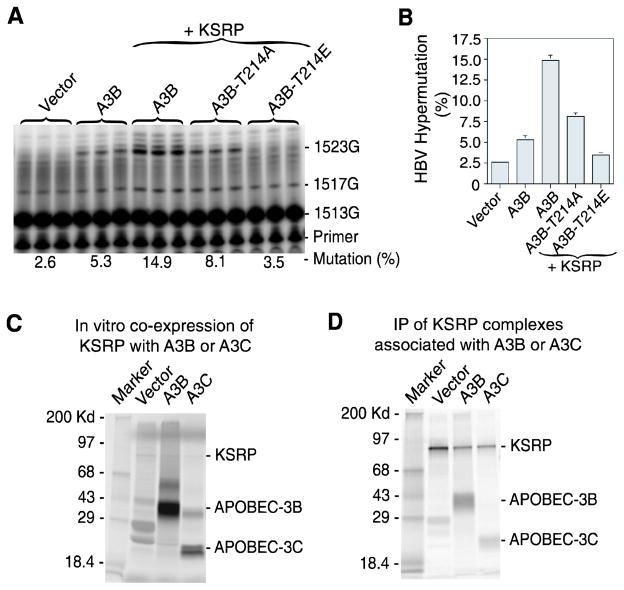
It has been reported that APOBEC-3B has a strong interaction with several hnRNP proteins and has a stronger HBV expression inhibition compared with other APOBEC-3 proteins.28 In addition, A3B has two deaminase domains while A3C only has one. It also has a conserved threonine phosphorylation site at 214 identical with APOBEC-3G at Thr-218.29 Phosphorylation of A3B Thr-214 is predicted to attenuate its intrinsic activity. To further investigate the effect of KSRP, A3B hypermutation on HBV was evaluated. As shown in Fig. 7A, A3B hypermutation on HBV was 5.3% by direct primer extension analyses of the regular PCR amplification at site 1513. Co-expression of KSRP increased hypermutation from 5.3% to 14.9%. The phosphorylation-null A3B-T214A mutant decreased the hypermutation of A3B from 14.9% to 8.1%. The phosphorylation mimicking A3B-T214E mutant decreased hypermutation to near background levels. These data further demonstrate that like that of A3C, KSRP significantly increases A3B hypermutation and the effect of KSRP is A3B specific because the phosphorylation mimicking mutant decreases its activity significantly.
Fig. 7. The effect of cellular factor KSRP on APOBEC-3B hypermutation.
A and B, APOBEC-3B hypermutation evaluation. KSRP was co-transfected with HBV and APOBEC-3B or its mutants into HepG2 cells and the HBV viral genome was extracted. The HBV x-gene was amplified by regular PCR and the resultant amplifications were directly analyzed by primer extension at site 1513 followed by 8% polyacrylamide urea gel separation with quantification using a PhosphoImager (A). The results from (A) are graphically presented (B). Each bar represents the mean and standard deviation with n=3. C and D, Protein interaction analyses. KSRP was co-translated with A3B, A3C, or vector control by an in vitro coupled transcription/translation system in the presence of 35S-methionine. The protein complexes formed during the in vitro translation were immuno-precipitated by an antibody against KSRP. The precipitated protein complex was separated by a 12% SDS-PAGE denaturing gel and detected by a PhosphoImager. The results were presented as co-translation content analyses (C) and immuno-precipitated (IP) complex analyses (D).
HBV hypermutation occurs within a virion during maturation in the cell cytoplasm. KSRP and A3B or A3C should be within the virion to induce the observed hypermutation effect. To further investigate the KSRP effect on A3B and A3C, we evaluated the protein interaction between KSRP and A3B or A3C in vitro. KSRP was co-translated with A3B or A3C by an in vitro coupled transcription/translation system in the presence of 35S-methionine. The protein complexes formed during the in vitro translation were immunoprecipitated by an antibody against KSRP followed by SDS-PAGE denaturing gel electrophoresis. As shown in Fig. 7B, both A3B and A3C were precipitated with KSRP, demonstrating that there is a direct protein interaction between them.
Taken together, these data demonstrate that among the cellular factors tested, KSRP had apparent increased effect on A3C and A3B hypermutation of HBV genome. The increased hypermutation rate was even detectable by primer extension analyses with regular PCR amplifications (A3C from 2.5% to 4.5%, Fig. 6A; A3B from 5.3% to 14.9%, Fig. 7A). The increased effect for A3C was further confirmed by 3D-PCR selective amplifications at 82°C and 81°C and follow-up primer extension analyses. These data demonstrate that A3C and A3B hypermutation activity can be increased by the known APOBEC-1 cofactor KSRP, suggesting a potential role of cofactors modifying APOBEC-3 hypermutation activity in vivo.
DISCUSSION
How hosts prevent the APOBEC family proteins from generalized mutagenesis while maintaining their physiological functions has been a long-standing question. Cofactor regulation has been elucidated for APOBEC-1 and more recently for AID.30 APOBEC-3 proteins are the only members of the APOBEC family without reported regulatory cofactors. The cellular factor hnRNP-K has been reported to have a regulatory role in HBV viral replication27 and also interacts with APOBEC-3B.28 But the function of hnRNP-K in APOBEC-3B hypermutation is still unknown. Whether or not there are cofactors regulating APOBEC-3 activity remains to be elucidated. APOBEC-3 hypermutation is characterized by extensive cytidine deamination in the evaluated regions with a relative small proportion of virus.5,17,10–12,177 APOBEC-1 is the founding member of the APOBEC family and is the only member of the APOBEC family whose natural substrate, cofactors, and editing mechanism are well understood. In this study, we found that the over-expression of rat APOBEC-1, in contrast to human APOBEC-1, hypermutated its natural substrate apoB mRNA in a way similar to APOBEC-3 viral hypermutation. The hypermutation pattern similarity between APOBEC-3 and rat APOBEC-1 together with the cofactor requirement of rat APOBEC-1 hypermutation suggest the potential presence of cofactor regulation for APOBEC-3 hypermutation. Utilizing HBV hypermutation in HepG2 cells, we investigated the effect of APOBEC-1 cofactors on A3C and A3B hypermutation and found a stimulatory effect by KSRP. This is the first report demonstrating the regulation of APOBEC-3 hypermutation by a cellular factor.
Under physiological conditions, APOBEC-1 deaminates a single cytidine at site 6666 in apoB mRNA, leading to the formation of apoB-48. This editing normally occurs in the nucleus after apoB pre-mRNA splicing. Site specific deamination is achieved by the cooperation of APOBEC-1 with auxiliary cofactors that facilitate editing site recognition utilizing a mooring sequence and efficiency elements flanking the editing site.31; 32; 33; 34 No editing has been observed with APOBEC-1 alone in vitro. When APOBEC-1 is over-expressed, apoB mRNA editing specificity is lost and apoB mRNA is hypermutated in both the nucleus and cytoplasm.21; 35 In this paper, we further found that the over-expression of rat APOBEC-1 in HepG2 cells resulted in extensive hypermutation of cytidines in the evaluated region of apoB mRNA (up to 69.6% in HepG2 and 75.5% in McA7777 cells) while the proportion of hypermutated apoB varies depending on rat APOBEC-1 expression levels. This pattern is identical to APOBEC-3 induced hypermutation on viral genomes. The proportion of hypermutated apoB mRNA by transient plasmid transfection was 0.4% in HepG2 cells (Fig. 1). Upon increased rat APOBEC-1 expression using adenovirus, the proportion increased up to about 20% as determined by primer extension analyses at distal sites 6493 and 6845 and 60% at the major site 6666 (Fig. 5). However, the quantities of mutated cytidines in the hypermutated apoB mRNA were similar, up to 69.6% (plasmid expression) vs 57% (adenoviral expression) C-to-U mutations. It is interesting to note that the apoB mRNA hypermutation levels induced by rat APOBEC-1 by plasmid transient transfection vs adenoviral transduction are comparable to the APOBEC-3 hypermutation levels in HBV (<10-2) vs HIV-1(43.1±5.2%). The amount of rat APOBEC-1 hypermutation is 0.4% by plasmid transfection but is increased to about 20% by adenoviral transduction or up to 60% if counted at the physiological site 6666. Despite APOBEC-1 native expression levels being very low in tissues like liver or small intestine, up to >90% editing at the physiological site 6666 is achieved because of cofactor interactions. These data suggest that higher levels of apoB mRNA hypermutation could result from higher APOBEC-1 expression or increased local APOBEC-1 concentration or indirectly by cofactor interaction.
It appears that the structure of rat APOBEC-1 plays an important role in its hypermutation. Although sharing 79% amino acid sequence homology, rat APOBEC-1 hypermutated many cytidines in apoB mRNA while human APOBEC-1 had a strong preference for the cytidines at sites 6666 and 6802 which are associated with the mooring or mooring like sequences (Fig. 3). This is probably related to a stronger protein interaction between human APOBEC-1 and ACF that restricts the activity of human APOBEC-1 to the local cytidines through ACF binding to the local mooring motifs.22 The protein interaction between rat APOBEC-1 and ACF is much weaker than human APOBEC-1 (data not shown). The weakened interaction makes it possible for rat APOBEC-1 to deaminate multiple cytidines along apoB mRNA as shown in the observation of 60% editing at the mooring site 6666 vs the 22% mutation rate at distal site 6845 for rat APOBEC-1 compared to 69% vs 3.4% rates at the same sites for human APOBEC-1 (Fig. 5). In addition, the single amino acid mutation E181Q eliminated hypermutation for both rat and human APOBEC-1 (Fig. 3). For human APOBEC-1, the loss of hypermutation function in the E181Q mutant was associated with a reduced protein interaction between APOBEC-1 and ACF.22 The rat APOBEC-1 E181Q mutation had a similar decreased protein interaction with ACF although it is less than the human mutant (data not shown). With a decreased protein interaction, the rat APOBEC-1 mutant E181Q loses its hypermutation function but retains a sufficient interaction to still have specific editing activity, albeit at a lower level. These data suggest that cofactor(s) are required for rat APOBEC-1 hypermutation. Rat APOBEC-1 appears to have a different structure than human APOBEC-1 that allows it to induce a greater level of hypermutation on multiple cytidines along apoB mRNA. This is consistent with the finding that rat APOBEC-1, but not human APOBEC1, curtailed HIV-1 replication in vitro24 and murine APOBEC1 can extensively edit both RNA and DNA in vivo.10
It remains to be elucidated whether there are cofactors involved in APOBEC-3 hypermutation in vivo and how they function. The hypermutation pattern similarity between rat APOBEC-1 and APOBEC-3 suggests that cellular factors might be involved in the interaction between APOBEC-3 proteins with viral single-stranded DNA to achieve a low frequency but extensive hypermutation. APOBEC-1 cofactors were identified by their ability to interact with APOBEC-1 or apoB mRNA. ACF plays a critical role in APOBEC-1 editing on apoB mRNA because it interacts with both APOBEC-1 and apoB mRNA. It is possible that a cofactor functioning like ACF that interacts with APOBEC-3G and/or single-stranded DNA, could induce the higher level of hypermutation seen with HIV-1. Lack of an appropriate quantitative method to measure hypermutation levels has hindered previous studies of APOBEC-3 hypermutation. 3D-PCR based methods have been used but different studies are not readily comparable. Here we found that the primer extension method that has been used for APOBEC-1 studies can also be used for HBV hypermutation quantitation. Up to 15% HBV hypermutation could be observed by direct primer extension analyses of regular PCR HBV amplifications. With this method, we demonstrate that the APOBEC-1 auxiliary cofactor KSRP increases APOBEC-3C and APOBEC-3B hypermutation activity on the HBV viral genome. KSRP is a single-stranded nucleic acid binding protein that increases RNA decay at multiple levels.36 Further investigation on potential cofactors should be helpful to better understand the APOBEC-3 hypermutation mechanism and help develop potential APOBEC-3 antiviral applications.
MATERIALS AND METHODS
Cell culture, plasmid transfection, and adenoviral infection
Both HepG2 and McA7777 were purchased from ATCC. HepG2 cells were maintained in EMEM containing 10% fetal bovine serum. McA7777 cells were cultured in DMEM containing 5% fetal bovine serum and 20% horse serum. For transient plasmid transfection, HepG2 cells cultured on collagen coated plates and McA7777 cells were transfected with rat APOBEC-1 plasmid using FuGene HD (Roche). A total of 2 μg plasmid DNA and 6 μl FuGene HD were added to the cells on each plate for each test. The cells were harvested after 2 days transfection. Empty vector was used as a control. An additional blank control containing untreated cells was also added for McA7777 cells. For adenoviral over-expression of APOBEC-1 with or without cofactors, HepG2 cells were plated on collagen pre-coated plates as above. The next day, media were replaced with 1 ml fresh medium and different adenoviral preparations (~5 × 108 pfu/ml) were added. After culture overnight in the presence of adenovirus, the media were replaced with 2 ml fresh growth media. The cells were incubated for another 1–2 days and total cellular RNA was extracted using the Trizol reagent (Invitrogen). Three separate samples were prepared for each analysis.
Plasmid and adenoviral constructs
Plasmids (pcDNA3.2) encoding rat APOBEC-1, APOBEC-3C, and APOBEC-3B was constructed with a 9-amino acid HA tag preceded by a 3-alanine C-terminal spacer. A plasmid encoding ACF was constructed with a FLAG tag (DYKDDDDK) immediately after the N-terminal methionine. Briefly, the RT-PCR amplifications of full length cDNA for rat APOBEC-1 from rat small intestine, APOBEC-3C from the human acute monocytic leukemia cell line THP-1, and APOBEC-3B from human macrophages were cloned into pENTR1A vector (Invitrogen) using restriction enzyme sites. After the inserted genes were confirmed by plasmid DNA sequencing, they were transferred into the expression vector pcDNA3.2-DEST (Invitrogen) by a DNA recombination reaction. All plasmid constructs were verified by sequencing and in vitro protein translation. Their protein expression in HepG2 cells was confirmed by Western blotting (data not shown).
Adenoviruses encoding rat/human APOBEC-1 and cofactors were prepared as previously reported.22 The expression of these genes or APOBEC-1 mutants in HepG2 cells was confirmed by the detection of their resultant activity, apoB mRNA editing and mRNA levels determinated by quantitative RT-PCR (data not shown).
The plasmid encoding HBV viral genome was a gift from Dr. Josef Kock.11 The production of HBV virus was confirmed by PCR amplification of the x-gene as previously reported and viral genome hypermutation was evaluated using the PCR amplification of the x-gene.5; 8; 12
ApoB mRNA hypermutation analyses by 3D-PCR, primer extension, and sequencing
To determine apoB mRNA hypermutation, rat APOBEC-1 was over-expressed in HepG2 or McA7777 cells by transient plasmid transfection or adenoviral infection and total RNAs were extracted after 2–3 days of over-expression. After pre-treatment with DNase, the human apoB mRNA fragment from 6471 to 6885 or rat apoB from 6507 to 6878 26 was amplified from the total RNA by RT-PCR as follows: 2 min at 94°C; 35 cycles of 30 sec at 94°C, 1 min at 55°C, 2 min at 72°C; and 7 min at 72°C. For 3D-PCR analyses, the first PCR amplifications were diluted 1:10 times and 5 μl aliquot each were used for further 3D-PCR amplification with the same primers at different denaturing temperature by 5 min at selected denaturing temperatures (94°C– 72°C) followed by 30 cycles of 1 min at the denaturing temperature; 1 min at 56°C; 2 min at 72°C and a final 7 min extension at 72°C. The 3D-PCR amplifications were detected by 1% agarose gel electrophoresis with ethidium bromide staining. For sequencing analyses, the PCR or 3D-PCR amplifications were cloned into pCR4-TOPO vector by the TA-cloning method (Invitrogen). After transformation, 20 bacterial clones containing a single molecular copy of the amplified apoB mRNA fragment were randomly selected for sequencing analyses.
For primer extension analyses, either sense or anti-sense primers located 4–7 nucleotides away from a chosen site were synthesized and purified by polyacrylamide gel electrophoresis.22 After being 5′-end labeled by [gamma-32P]-ATP with T4 nucleotide kinase at 37°C for 40 min, the primers were utilized for the determination of apoB mRNA mutation levels at the chosen sites by thermo cycle primer extension as described previously.37 Briefly, RT-PCR or 3D-PCR amplifications above were purified to remove dNTP using a QIAEX II Gel Extraction kit (Qiagen). An aliquot of the purified apoB DNA was taken for thermo cycle primer extension in the presence of ddGTP for antisense primers or ddCTP for sense primers. The resultant primer extension products were resolved by electrophoresis in an 8% polyacrylamide-urea sequencing gel and the ratio of edited to unedited apoB mRNA at a given site was determined by a PhosphoImager. The primers used for primer extension were: pe6493-AS, ttgaatgaattcagataatc; pe6639-S, tgccaaaatcaactttaatg; pe6655-AS, tatactgatcaaattgtatcat; pe6666-AS, tatctttaatatactgatc; pe6702-S, agtatattaaagatagttatg; pe6802-S, tcatatccgtgtaaatttag; pe6845-S, acatttgtttattgaaaatattg; and rat pe6589S, ctatagaattacagataatgat.
HBV viral genome hypermutation analyses
HepG2 cells cultured on collagen coated plates in EMEM containing 10% fetal bovine serum were co-transfected with equal amount of plasmids containing the HBV viral genome, A3C or A3B, and cofactors through FuGene HD (Roche) according to the manufacturer’s instructions. A total 2 μg plasmid DNA and 6 μl FuGene HD were added to HepG2 cells on each 35-mm 6-well plate for each test. The HBV viruses were harvested 72 h post-transfection by lysing the cells in a buffer containing 50 mM Tris-HCl, pH 8.0, 1.5 mM MgCl2, and 0.5% NP-40. The viral extractions were pre-treated with 30 U micrococcal nuclease (New England Lab) in the presence of 2 mM CaCl2 for 1 h at 37°C and then digested with 20 μl of 20 mg/ml proteinase K (Invitrogen) for 30 min at 55°C in the presence of 5 mM EDTA. Finally the HBV viral genomic DNAs were collected and purified by the QIAEX II DNA purification kit (Qiagen) according to the manufacturer’s instructions.
To determine A3C or A3B induced HBV viral genome hypermutation, the HBV x-gene was amplified from the HBV DNA extraction by regular PCR denaturing at 94°C and using primers of X-F: ttctcgccaacttacaaggcctttc and X-R: aaaaagttgcatggtgctggtg. The PCR reactions were diluted 1:10 and a 4 μl aliquot was used for further 3D-PCR amplification using primers of X-FII: cgcaaatatacatcgtatccat and X-R. The samples were denatured at various temperatures (94°C–80°C) for 5 min followed by 35 cycles of 1 min at the denaturing temperature; 1 min at 55°C; 2 min at 72°C and a final 7 min extension at 72°C. The PCR amplifications were detected by 1% agarose gel electrophoresis with ethidium bromide staining (Fig. 6).
For hypermutation level quantitation by primer extension, the PCR amplifications were purified to remove dNTP by a QIAEX II Gel Extraction kit (Qiagen). An aliquot of the purified HBV x-gene DNA was subjected to a thermo-cycle primer extension reaction in the presence of ddGTP with 5′-32P-end labeled primers, pe1513, tccccttctccgtctgccgtt or pe1642, ccaccgtgaacgcccaccaaa as described previously.37 The primer extension products were resolved by electrophoresis in an 8% polyacrylamide-urea sequencing gel and hypermutation levels were determined by a PhosphoImager.
Protein interaction analyses as determined by an in vitro translation system
To determine protein interactions, cofactor KSRP and A3B or A3C were co-translated by TNT quick coupled transcription/translation systems (Promega) at 30°C for 2 h in the presence of L-35S-Methionine (1175 Ci/mmol, PerkinElmer). The protein complexes formed during the in vitro translation were immunoprecipitated by an antibody against KSRP (Novus) incubating at room temperature for 30 min followed by precipitation with Protein G Dynabeads (Invitrogen) according to the manufacturer’s instructions. The immunoprecipitated protein complexes were then separated on a 12% SDS-PAGE denaturing gel and the target proteins were quantified by a PhosphoImager. The empty vector was used as blank control for a comparison to A3B or A3C translation. As a verification of protein translation, an aliquot before immunoprecipitation was denatured directly in SDS-sample buffer and separated on a 12% SDS-PAGE gel.
Supplementary Material
Rat APOBEC-1 was expressed in HepG2 or McA7777 cells by transient plasmid transfection. The apoB mRNA was amplified by RT-PCR after a 2 day transfection. The hypermutated apoB mRNA was selectively amplified by the 3D-PCR at 74°C and were cloned for DNA sequencing analyses as in Fig. 1E and 2E. The DNA sequences of ten clones are aligned with apoB mRNA hypermutations in human HepG2 (A) and rat McA7777 (B) cells. Individual mutations in each clone are presented as nucleotides at their corresponding locations and the dots represent nucleotides identical to the wild type reference apoB mRNA.
Abbreviations
- apo
apolipoprotein
- APOBEC-3G
apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G
- HIV-1
human immunodeficiency virus type 1
- HBV
hepatitis B virus
- MLV
murine leukemia virus
- HTLV-1
human T-cell leukemia virus type 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 3.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 4.Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol Mol Biol Rev. 2009;73:211–232. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry M, Guetard D, Suspene R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 7.Mahieux R, Suspene R, Delebecque F, Henry M, Schwartz O, Wain-Hobson S, Vartanian JP. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J Gen Virol. 2005;86:2489–2494. doi: 10.1099/vir.0.80973-0. [DOI] [PubMed] [Google Scholar]
- 8.Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suspene R, Henry M, Guillot S, Wain-Hobson S, Vartanian JP. Recovery of APOBEC3-edited human immunodeficiency virus G->A hypermutants by differential DNA denaturation PCR. J Gen Virol. 2005;86:125–129. doi: 10.1099/vir.0.80426-0. [DOI] [PubMed] [Google Scholar]
- 10.Petit V, Guetard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, Vartanian JP. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol. 2009;385:65–78. doi: 10.1016/j.jmb.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 11.Kock J, Blum HE. Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J Gen Virol. 2008;89:1184–1191. doi: 10.1099/vir.0.83507-0. [DOI] [PubMed] [Google Scholar]
- 12.Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Fujimoto Y, Ochi H, Abe H, Maekawa T, Yatsuji H, Shirakawa K, Takaori-Kondo A, Chayama K. Dual effect of APOBEC3G on Hepatitis B virus. J Gen Virol. 2007;88:432–440. doi: 10.1099/vir.0.82319-0. [DOI] [PubMed] [Google Scholar]
- 13.Vartanian JP, Henry M, Marchio A, Suspene R, Aynaud MM, Guetard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P, Dejean A, Wain-Hobson S. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 2010;6:e1000928. doi: 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 16.Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoepfel SA, Di Giallonardo F, Daumer M, Thielen A, Metzner KJ. In-depth analysis of G-to-A hypermutation rate in HIV-1 env DNA induced by endogenous APOBEC3 proteins using massively parallel sequencing. J Virol Methods. 171:329–338. doi: 10.1016/j.jviromet.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 20.Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M, Gotto AM, Li WH, Chan L. Apolipoprotein B-48 is the product of a messenger RNA with an organ- specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 21.Sowden M, Hamm JK, Smith HC. Overexpression of APOBEC-1 results in mooring sequence-dependent promiscuous RNA editing. J Biol Chem. 1996;271:3011–3017. doi: 10.1074/jbc.271.6.3011. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Eggerman TL, Bocharov AV, Baranova IN, Vishnyakova TG, Csako G, Patterson AP. Hypermutation induced by APOBEC-1 overexpression can be eliminated. RNA. 2010;16:1040–1052. doi: 10.1261/rna.1863010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadjiagapiou C, Giannoni F, Funahashi T, Skarosi SF, Davidson NO. Molecular cloning of a human small intestinal apolipoprotein B mRNA editing protein. Nucleic Acids Res. 1994;22:1874–1879. doi: 10.1093/nar/22.10.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 25.Anant S, Davidson NO. Identification and regulation of protein components of the apolipoprotein B mRNA editing enzyme. A complex event. Trends Cardiovasc Med. 2002;12:311–317. doi: 10.1016/s1050-1738(02)00180-9. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Eggerman TL, Patterson AP. ApoB mRNA editing is mediated by a coordinated modulation of multiple apoB mRNA editing enzyme components. Am J Physiol Gastrointest Liver Physiol. 2007;292:G53–G65. doi: 10.1152/ajpgi.00118.2006. [DOI] [PubMed] [Google Scholar]
- 27.Ng LF, Chan M, Chan SH, Cheng PC, Leung EH, Chen WN, Ren EC. Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS Med. 2005;2:e163. doi: 10.1371/journal.pmed.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Zhang X, Tian C, Wang T, Sarkis PT, Fang Y, Zheng S, Yu XF, Xu R. Cytidine deaminase APOBEC3B interacts with heterogeneous nuclear ribonucleoprotein K and suppresses hepatitis B virus expression. Cell Microbiol. 2008;10:112–121. doi: 10.1111/j.1462-5822.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 29.Demorest ZL, Li M, Harris RS. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G protein. J Biol Chem. 2011;286:26568–26575. doi: 10.1074/jbc.M111.235721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storck S, Aoufouchi S, Weill JC, Reynaud CA. AID and partners: for better and (not) for worse. Curr Opin Immunol. 2011;23:337–344. doi: 10.1016/j.coi.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Backus JW, Smith HC. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992;20:6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Driscoll DM, Lakhe-Reddy S, Oleksa LM, Martinez D. Induction of RNA editing at heterologous sites by sequences in apolipoprotein B mRNA. Mol Cell Biol. 1993;13:7288–7294. doi: 10.1128/mcb.13.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah RR, Knott TJ, Legros JE, Navaratnam N, Greeve JC, Scott J. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem. 1991;266:16301–16304. [PubMed] [Google Scholar]
- 34.Hersberger M, Innerarity TL. Two efficiency elements flanking the editing site of cytidine 6666 in the apolipoprotein B mRNA support mooring-dependent editing. J Biol Chem. 1998;273:9435–9442. doi: 10.1074/jbc.273.16.9435. [DOI] [PubMed] [Google Scholar]
- 35.Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briata P, Chen CY, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. KSRP, many functions for a single protein. Front Biosci. 2011;16:1787–1796. doi: 10.2741/3821. [DOI] [PubMed] [Google Scholar]
- 37.Patterson AP, Chen Z, Rubin DC, Moucadel V, Iovanna JL, Brewer HB, Jr, Eggerman TL. Developmental regulation of apolipoprotein B mRNA editing is an autonomous function of small intestine involving homeobox gene Cdx1. J Biol Chem. 2003;278:7600–7606. doi: 10.1074/jbc.M201601200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rat APOBEC-1 was expressed in HepG2 or McA7777 cells by transient plasmid transfection. The apoB mRNA was amplified by RT-PCR after a 2 day transfection. The hypermutated apoB mRNA was selectively amplified by the 3D-PCR at 74°C and were cloned for DNA sequencing analyses as in Fig. 1E and 2E. The DNA sequences of ten clones are aligned with apoB mRNA hypermutations in human HepG2 (A) and rat McA7777 (B) cells. Individual mutations in each clone are presented as nucleotides at their corresponding locations and the dots represent nucleotides identical to the wild type reference apoB mRNA.