David Wilson, Anthony Keefe, and Jack Szostak have made a wonderful contribution to the changing world of peptide and protein in vitro evolution and selection. In a previous issue of PNAS (1), they demonstrated that the expected (and observed) weak affinities of peptides (from, for example, bead or phage display) can be enhanced by using “mRNA display.” The present work is a large step forward.
The work builds on the earlier work of Roberts and Szostak (2). These authors, after inspecting the state of phage display for peptide evolution and selection, realized that the limiting feature of phage display was the “bacterial transformation requirement”; phage display libraries are thus limited, effectively, to <109 different peptides in the starting library. Synthetic peptide display and selection on beads is not better; screening and selection of beads with one peptide sequence per bead cannot be done easily with 109 beads. The solution to this limitation was to move toward ribosome display (3) or “mRNA display.”
In either embodiment, the solution is technically complex but conceptually simple. One prepares libraries of randomized mRNAs (containing on the order of 1013 to 1014 different messages), with each randomized message being flanked by fixed sequences to allow amplification after rounds of selection. [These mRNA libraries would be perfect libraries for in vitro evolution and selection of RNA aptamers, but the goal of these experiments is peptides and proteins (encoded by the mRNA libraries), not aptamers (4)]. In the case of ribosome display, the mRNA is stalled after translation of the randomized codons to allow selection of the appropriate peptides protruding from the ribosome that had just read each mRNA, with specific mRNAs still bound to each ribosome. Ribosome display is similar to phage display, with the exception that the libraries can be larger because amplification requires enzymology rather than bacterial transformation (specifically in the first round of phage display). In both cases the “displayed” peptide (or protein) is a rather small object dangling from a very large phage coat or from a very large ribosome. I have always thought that selection of peptides as binding partners (for target proteins, as is common) would be complicated by the size and surface of the displaying object (phage or ribosome); some high affinity peptides might be lost through unpredictable interactions between the displayed peptide and the enormous protein complex to which it was attached. After all, a ribosome has a mass of more than 2,000,000 Da, and typical peptide or protein libraries contain selectable molecules with masses of less than 10,000 Da.
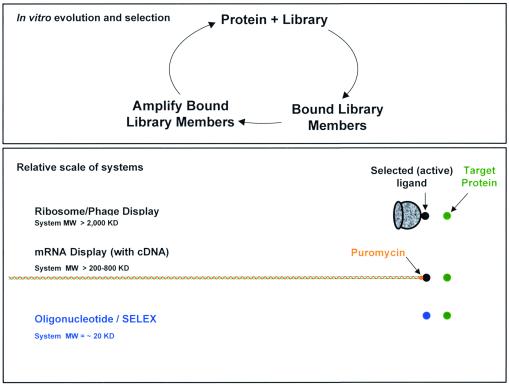
mRNA display solves the “large display object problem” in an elegant fashion, while maintaining the major advantage of ribosome display, which is large diversity not constrained by bacterial transformation. After translation of random peptides, as above, the encoding mRNAs are covalently pinned to the newly synthesized peptides through the puromycin reaction (the puromycin having been previously placed on the 3′ end of each mRNA). Purification of the mRNA-peptide fusions, free of ribosomes, allows selection of appropriate peptides with only the encoding mRNA immediately adjacent to the peptide. In mRNA display, the mRNAs are more than ten times the size of the displayed peptide (three nucleotides per codon, plus extra nucleotides for amplification and the puromycin reaction—but this appendage is smaller than a phage or ribosome. In Fig. 1 are displayed the systems (and the sizes of those systems) that allow selection of peptides or proteins (and aptamers—see below). The authors double the size of the mRNA by doing reverse transcription before selection, for reasons discussed below. Peptide libraries about 88 aa in length were used in this publication.
I have always thought that selection of peptides as binding partners would be complicated by the size and surface of the displaying object… mRNA display solves the “large display object problem” in an elegant fashion.
Figure 1.
Systems for in vitro selection.
Gratifyingly, some of the peptides selected as binding partners to the target protein streptavidin had low dissociation constants. A few had KDs of between 5 and 10 nM, substantially lower than the KDs of peptides also aimed at streptavidin found through phage display. The authors conclude that the low KDs flow largely from the larger starting library diversity (“> 104-fold more sequences sampled”), the length of the peptides sampled (adding another 50-fold to the sample size for a 38-mer, corresponding to the different 38-mers within a peptide of 88 aa), and the slight bias in favor of HPQ sequences in their libraries (given that HPQ was a known consensus motif for streptavidin binding from previous work). I would add to their list the avoidance of presumptive interferences by phages or ribosomes (or beads, in other work) as another advantage of mRNA display. The appendage in this paper is a double-stranded and monotonous helical polyanion, rather than a textured protein-rich phage or ribosome; I think the double helix is a step in the right direction.
The authors conclude that “it should be possible to increase the affinities of peptides for their targets by constraining them as loops in known protein scaffolds”; in fact, the entire technology platform of Phylos (see their web site for more details, www.phylos.com) is aimed at exactly that approach. Interestingly, in the present work, all of the selected and sequenced clones shifted the frame (through deletion of two nucleotides or addition of one nucleotide), suggesting among other things that 1013–1014 sequences provide ample opportunity for mutational events that thwart the intentions of the investigator. That is, the investigators tried to “inform” their peptide libraries toward good binding structural motifs, and the selection pressure was more than sufficient to ignore that design. Nevertheless, mRNA display is going to be developed enthusiastically because (as the authors note) “complete expression and activity analysis of the proteome will require the use of hundreds of thousands of protein-binding and inhibiting reagents.” I couldn't agree more.
I read this paper from the perspective of both an interested scientist and a person with a great love for nucleic acid aptamers (5–7). As has been published repeatedly (including by Szostak, in an earlier phase of his productive life), nucleic acid aptamers, aimed at protein targets, can show KDs well below 1 nM (in fact, low pM is common; see the SomaLogic web site, www.somalogic.com, and ref. 8). Because the selection of the binding peptides included a randomized mRNA region (in the mRNA-peptide fusions) of more than 250 nucleotides, why didn't the authors select aptamers in competition with the selected peptides? In fact, the authors show (see figure 2B of ref. 1) that RNA binding activity is not responsible for the binding of the seventh round pool of their mRNA-peptide fusions. The authors (wisely) have eliminated head-to-head competition between peptides and RNA aptamers through two steps in their protocol. The binding buffers for selection include tRNA at 1 μg/ml, and the mRNA is prevented from assuming any globular (aptamer) shapes through reverse transcription [so that the mRNA-peptide fusions during the selection are actually mRNA/cDNA-peptide fusions (see figure 1 of ref. 1)]. Interestingly, the elution of the bound peptides is by biotin exchange at the active site of streptavidin, and it is not at all clear where the dominant epitope on streptavidin would lie for a classic SELEX experiment aimed at finding an aptamer. Over the years to come, the KDs for lots of peptides and proteins identified by mRNA display will be compared with the KDs for lots of aptamers identified by the SELEX process. Eventually we will all know whether what Bruce Eaton called “side chain envy” is based on a fundamental advantage of amino acids over nucleotides that more than overcomes the structural stability of short oligonucleotide motifs (9).
Does mRNA display advance the field of peptide and protein selection? At the simplest level, the answer is a resounding “yes.” Since the 1979 publication of Perelson and Oster (10), and even before, one knew that library diversity must help as one searches through sequence space for interesting molecules (probably interesting, most often, because the selected molecules have shape—that is, sequence diversity must be far larger than shape diversity). At the very least, mRNA display brings the diversity up to the concentration limits of mRNAs and ribosomes (the real limit, I imagine) and has already shown an improvement in the affinities of peptides for a classic protein target. However, these experiments are not easy; I have incubated thousands of in vitro translation experiments, minus the further steps of puromycin chemistry and library amplification, and the work was demanding and tedious. Hopefully, the rather daunting experimental protocols will be placed onto robots so that average experimentalists of ordinary skills routinely will be able to use mRNA display.
Footnotes
See companion article on page 3750 in issue 7 of volume 98.
References
- 1.Wilson D S, Keefe A D, Szostak J W. Proc Natl Acad Sci USA. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. . (First Published March 13, 2001; 10.1073/pnas.061028198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts R W, Szostak J W. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanes J, Jermutus L, Weber-Bornhauser S, Bosshard H R, Plückthun A. Proc Natl Acad Sci USA. 1998;95:14130–14135. doi: 10.1073/pnas.95.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gold L. J Biol Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 5.Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Brody E N, Gold L. J Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 8.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 9.Eaton B E, Gold L, Zichi D A. Chem Biol. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 10.Perelson A S, Oster G F. J Theor Biol. 1979;81:645–670. doi: 10.1016/0022-5193(79)90275-3. [DOI] [PubMed] [Google Scholar]