Abstract
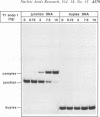
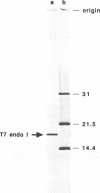
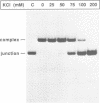
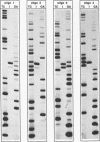
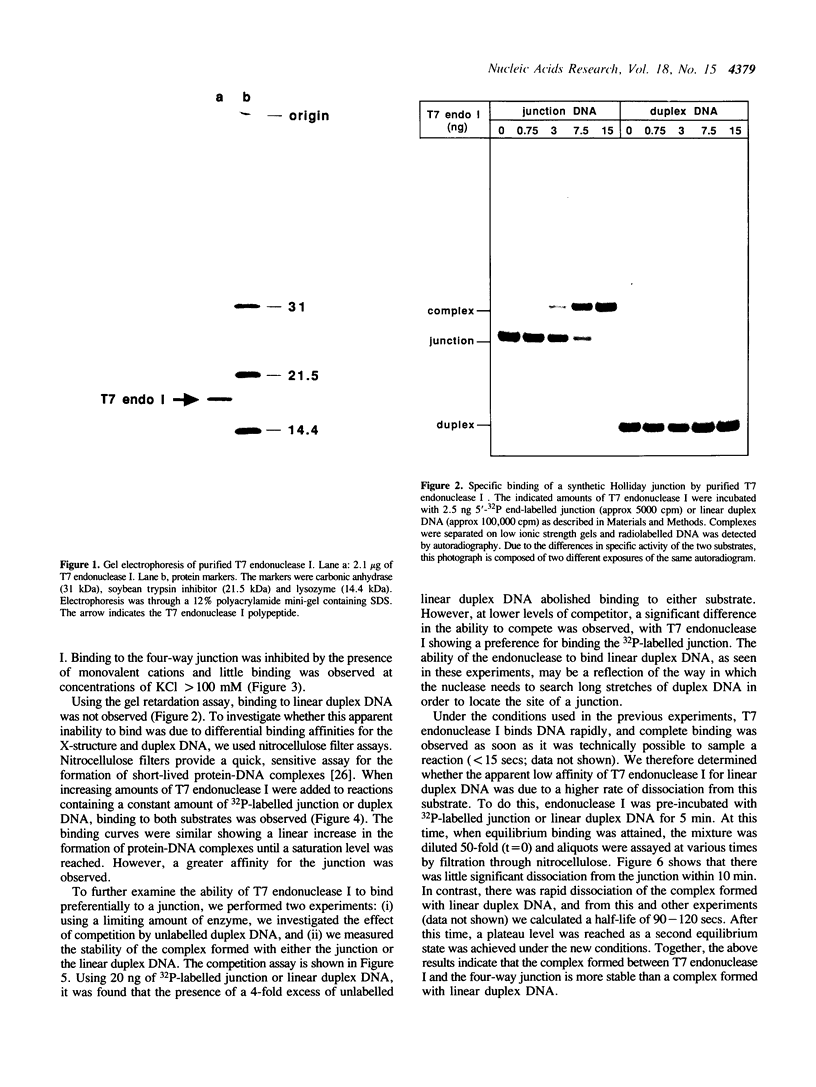
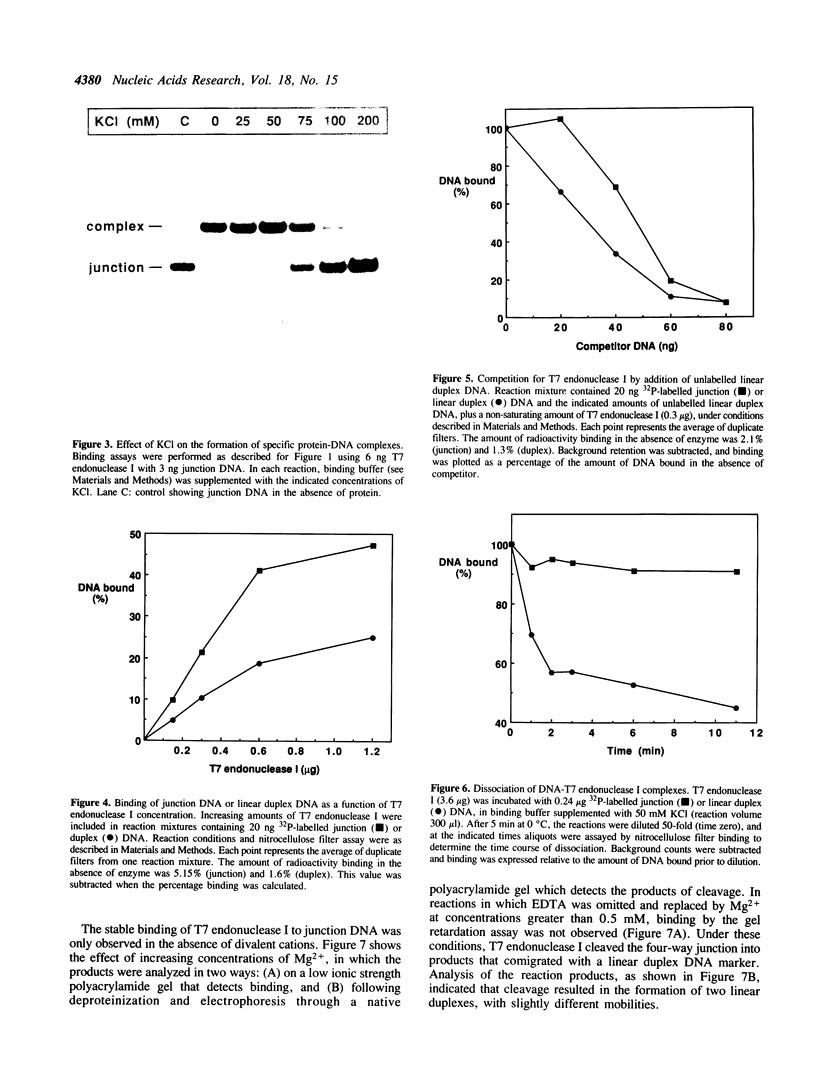
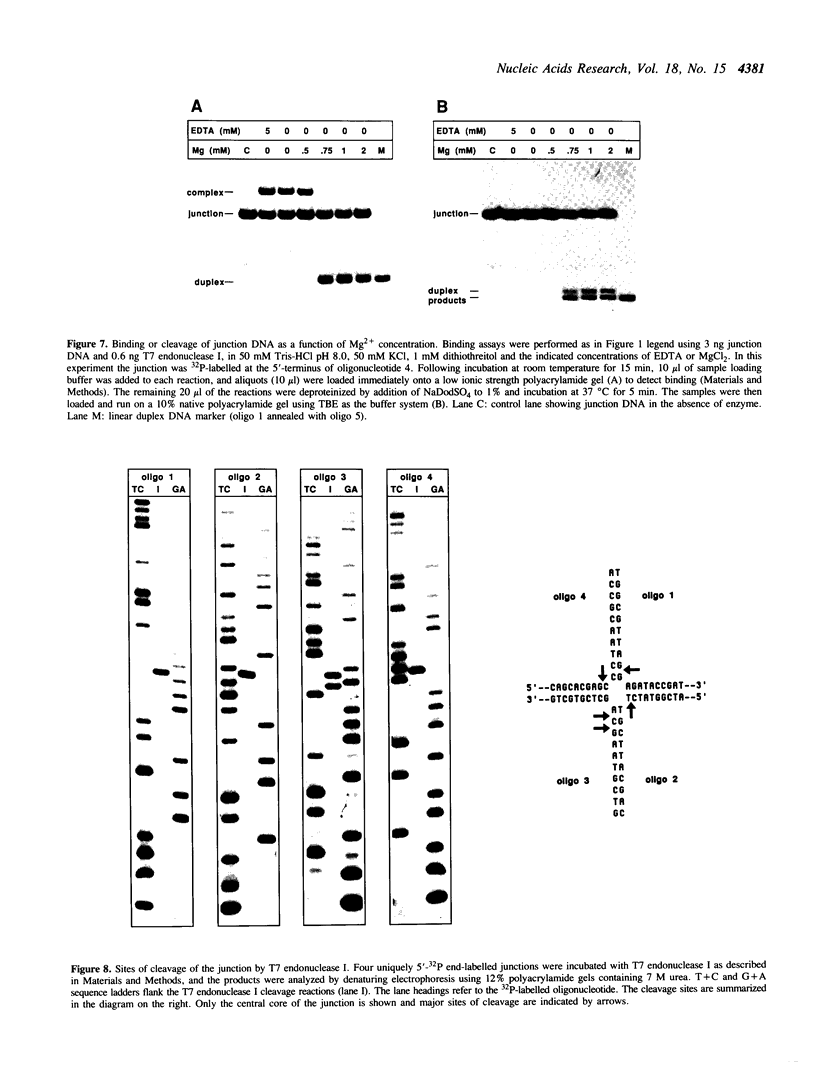
T7 endonuclease I binds specifically to four-way junctions in duplex DNA and promotes their resolution into linear duplexes. Under conditions in which the nuclease activity is blocked by the absence of divalent cations, the enzyme forms a distinct protein-DNA complex with the junction, as detected by gel retardation and filter binding assays. The formation of this complex is structure-specific and contrasts with the short-lived binding complexes formed on linear duplex DNA. The binding complex between T7 endonuclease I and a synthetic Holliday junction analog has been probed with hydroxyl radicals. The results indicate that the nuclease binds all four strands about the junction point.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth K. A., Powell D., Trupin M., Mosig G. Regulation of two nested proteins from gene 49 (recombination endonuclease VII) and of a lambda RexA-like protein of bacteriophage T4. Genetics. 1988 Oct;120(2):329–343. doi: 10.1093/genetics/120.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center M. S., Studier F. W., Richardson C. C. The structural gene for a T7 endonuclease essential for phage DNA synthesis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):242–248. doi: 10.1073/pnas.65.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Suzuki M. 'SPKK' motifs prefer to bind to DNA at A/T-rich sites. EMBO J. 1989 Dec 20;8(13):4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie P., McFadden G., Morgan A. R. The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem. 1987 Oct 25;262(30):14826–14836. [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Evans D. H., Kolodner R. Construction of a synthetic Holliday junction analog and characterization of its interaction with a Saccharomyces cerevisiae endonuclease that cleaves Holliday junctions. J Biol Chem. 1987 Jul 5;262(19):9160–9165. [PubMed] [Google Scholar]
- Jensch F., Kosak H., Seeman N. C., Kemper B. Cruciform cutting endonucleases from Saccharomyces cerevisiae and phage T4 show conserved reactions with branched DNAs. EMBO J. 1989 Dec 20;8(13):4325–4334. doi: 10.1002/j.1460-2075.1989.tb08619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Garabett M. Studies on T4-head maturation. 1. Purification and characterization of gene-49-controlled endonuclease. Eur J Biochem. 1981 Mar 16;115(1):123–131. [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- Kerr C., Sadowski P. D. The involvement of genes 3,4,5 and 6 in genetic recombination in bacteriophage T7. Virology. 1975 May;65(1):281–285. doi: 10.1016/0042-6822(75)90031-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee D., Sadowski P. D. Genetic recombination of bacteriophage T7 in vivo studied by use of a simple physical assay. J Virol. 1981 Dec;40(3):839–847. doi: 10.1128/jvi.40.3.839-847.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lu M., Guo Q., Seeman N. C., Kallenbach N. R. DNase I cleavage of branched DNA molecules. J Biol Chem. 1989 Dec 15;264(35):20851–20854. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Carter W. A., Portugal J., Lilley D. M. The tertiary structure of the four-way DNA junction affords protection against DNase I cleavage. Nucleic Acids Res. 1990 May 11;18(9):2599–2606. doi: 10.1093/nar/18.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie A. I., Clegg R. M., von Kitzing E., Duckett D. R., Diekmann S., Lilley D. M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989 Oct 26;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- Müller B., Jones C., Kemper B., West S. C. Enzymatic formation and resolution of Holliday junctions in vitro. Cell. 1990 Jan 26;60(2):329–336. doi: 10.1016/0092-8674(90)90747-3. [DOI] [PubMed] [Google Scholar]
- Paetkau V., Langman L., Bradley R., Scraba D., Miller R. C., Jr Folded, concatenated genomes as replication intermediates of bacteriophage T7 DNA. J Virol. 1977 Apr;22(1):130–141. doi: 10.1128/jvi.22.1.130-141.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Fontaine A. An endonuclease specific for single-stranded DNA selectively damages the genomic DNA and induces the SOS response. J Biol Chem. 1985 Mar 10;260(5):3173–3177. [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., Kemper B., West S. C. Interaction of a four-way junction in DNA with T4 endonuclease VII. J Biol Chem. 1990 Jun 5;265(16):9285–9289. [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Resolution of model Holliday junctions by yeast endonuclease is dependent upon homologous DNA sequences. Cell. 1988 Feb 26;52(4):621–629. doi: 10.1016/0092-8674(88)90474-6. [DOI] [PubMed] [Google Scholar]
- Pham T. T., Coleman J. E. Cloning, expression, and purification of gene 3 endonuclease from bacteriophage T7. Biochemistry. 1985 Sep 24;24(20):5672–5677. doi: 10.1021/bi00341a058. [DOI] [PubMed] [Google Scholar]
- Picksley S. M., Parsons C. A., Kemper B., West S. C. Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol. 1990 Apr 20;212(4):723–735. doi: 10.1016/0022-2836(90)90233-C. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Sadowski P. D. Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B. J Biol Chem. 1971 Jan 10;246(1):209–216. [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y., Ogawa H. Intermediates in genetic recombination of bacteriophage T7 DNA. Biological activity and the roles of gene 3 and gene 5. J Mol Biol. 1978 Nov 5;125(3):255–273. doi: 10.1016/0022-2836(78)90402-3. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]
- von Kitzing E., Lilley D. M., Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990 May 11;18(9):2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]