Two recent reports provide atomic resolution information detailing the interaction of the class II release factor, RF3, with the bacterial ribosome. Differences in the composition of the two crystal forms allow us to learn a considerable amount about how translational GTPases engage the ribosome to facilitate and define conformational rearrangements involved in protein synthesis.
Keywords: translation, termination, ribosome, RF3, GTPase, switch loop
Abstract
Two recent reports provide atomic resolution information detailing the interaction of the class II release factor, RF3, with the bacterial ribosome. Differences in the composition of the two crystal forms allow us to learn a considerable amount about how translational GTPases engage the ribosome to facilitate and define conformational rearrangements involved in protein synthesis.
Many older molecular biologists (and even a younger set) fondly remember a 1971 movie about protein synthesis, in which Stanford graduate students personified the various molecular components of the process (http://www.youtube.com/watch?v=u9dhO0iCLww). Particularly memorable in the movie is the burst of smoke that appears each time a GTPase engages the ribosome and hydrolyzes GTP; initiation factor 2 (IF2) escorts initiator tRNA to the P site, T factor (EFTu) escorts elongator aa-tRNAs to the A site, and G factor (EFG) promotes the indexing step (translocation). To those of us in the translation field, the movie is astoundingly accurate in its overall description of the translation cycle, decades before pre-steady state kinetics and high-resolution structural approaches defined the molecular specifics. Indeed, the only GTPase not depicted (and moreover its function not yet defined) in this early movie is the class II release factor, RF3. This perspective focuses on two recent structures that have elucidated the molecular details of this factor and its interactions with the ribosome, and as a result, rationalize some of its biochemical activities.
Termination of protein synthesis takes place when a stop codon at the end of an open reading frame enters the decoding center of the small ribosomal subunit. Class I RFs have codon recognition motifs that discriminate stop from sense codons with extraordinary fidelity—a sense codon is misread in cells with a frequency of only one in 105 events (Jorgensen et al. 1993). Following stop-codon recognition, class I RFs engage the peptidyl transferase catalytic center of the large ribosomal subunit, catalyzing a hydrolytic reaction that releases the growing polypeptide chain. At its simplest level, this is the end of termination. On a more complex level, an mRNA and a deacylated tRNA still remain on the intact ribosome at the completion of peptide release, a complex that must be dissociated in order for the ribosome to move on to its next round of translation. This is where RF3 enters the scene.
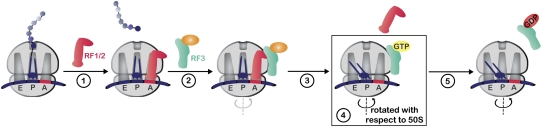
RF3 was biochemically identified in 1969 (Milman et al. 1969) as a factor (S for stimulatory) that increased the rate of peptide release catalyzed by the class I release factors (RF1 and RF2). Soon thereafter, the gene prfC (for RF3) was identified by a genetic screen in which absence of this gene allowed more read-through of a nonsense codon inserted in β-galactosidase (Grentzmann et al. 1994; Mikuni et al. 1994). While apparently nonessential to the bacterial cell, a biochemical role for the GTPase in termination was identified by Ehrenberg and colleagues, who showed that RF3 greatly increased the amount of release when substoichiometric amounts of class I RF were present in the reaction; RF3 appears to increase the rate of dissociation of the class I RFs after the completion of catalysis (Fig. 1; Freistroffer et al. 1997). Interestingly, RF3 has no impact on the actual rate of peptide release catalysis (krel) on stop codons under normal circumstances (Freistroffer et al. 2000).
FIGURE 1.
Model of RF3 function on the ribosome, with boxed step indicating the structure defined in Zhou et al. (2011) and Jin et al. (2011). Steps are outlined as follows: (1) binding of class I release factor (RF1/2) to stop codon programmed ribosomes and catalysis of peptide release; (2) RF3 binding to the ribosome—it is unclear at this stage whether RF3 binds in its GTP- or GDP-bound state; additionally, the ribosome may be in a nonratcheted or ratcheted state (rotation lightly indicated); (3) RF1/2 dissociating from the ribosome; (4) RF3:GDPNP:ribosome complex characterized in Zhou et al. (2011) and Jin et al. (2011); ratcheted state of ribosome is indicated; (5) departure of RF3:GDP from the ribosome following GTP hydrolysis.
CryoEM structures solved over the past several years revealed that RF3:GDPNP-bound ribosomes are stabilized in a rotated state (i.e., the ribosomal subunits are rotated with respect to one another), with the deacylated P-site tRNA arranged in a “hybrid” configuration (Klaholz et al. 2004; Gao et al. 2007). The hybrid state of binding, long ago hypothesized by Bretscher as a mechanism to facilitate translocation (Bretscher 1968), was first visualized in chemical footprinting analysis by Moazed and Noller (1989). Interestingly, ribosomes engaging EFG (with nonhydrolyzable GTP) (Frank and Agrawal 2000; Valle et al. 2003b) and ribosome recycling factor (RRF) (Gao et al. 2005; Dunkle et al. 2011) also are found in a rotated configuration, suggesting common mechanisms that promote these events. So far, EFTu is the only GTPase that does not appear to interact with the rotated form of the ribosome (Valle et al. 2003a; Schmeing et al. 2009). What is interesting to consider is whether the rotation is identical in each of these ratcheted structures and how the ribosomal interface is stabilized in the various states.
Atomic resolution structural information has emerged more slowly for RF3, and for all of the GTPases for that matter, because of an unfortunate crystal lattice packing determinant. Ribosomal protein L9 from the neighboring ribosome contacts the “factor binding domain” of another ribosome in all of the known crystal forms, thus precluding the binding of the translational GTPases. This problem was recently solved by the Ramakrishnan group, who deleted L9 from the bacterial strain, and the floodgates were opened to solve structures of the ribosome bound first by EFTu ternary complex (Schmeing et al. 2009) and EFG (Gao et al. 2009), and now by RF3 (Jin et al. 2011; Zhou et al. 2011).
The Ramakrishnan (Jin et al. 2011) structure, refined to 3.8Å resolution, contains E. coli RF3:GDPCP bound to T. thermophilus ribosomes with a deacylated tRNA bound in the P/E state (Jin et al. 2011), as previously reported by cryoEM. The investigators report a 9.3° counterclockwise rotation of the 30S subunit relative to the 50S subunit and an additional 3° swivel of the head of the 30S subunit (toward the E site). The higher resolution Noller (Zhou et al. 2011) structure (3.3Å) of E. coli RF3:GDPNP bound to E. coli ribosomes does not contain deacylated tRNA (though it was in the crystallization buffer), but the ribosome is also ratcheted: 7° counterclockwise with a more significant 14° swivel of the head of the 30S subunit (Zhou et al. 2011). This structure benefited from the addition of the antibiotic viomycin, similar to earlier observations with paromomycin contributing to high-quality RRF:ribosome structures (Dunkle et al. 2011) and isolated 30S subunit structures with bound near-cognate tRNAs (Ogle et al. 2001). Crystals grown by the same group without viomycin had seemingly identical structures, but were overall lower resolution. Why the Zhou et al. (2011) structure did not retain the deacylated tRNA is not clear, though the investigators do show that overall low [Mg2+] in the drop weakens the stability of the interaction with the tRNA. Zhou et al. (2011) nevertheless point out that their observed rotation of the head opens the constriction between the P and the E site (Schuwirth et al. 2005), which would allow the tRNA to move to occupy the now aligned E and P sites on the 50S and 30S subunits, respectively. The absence of the tRNA in the Zhou et al. (2011) structure may account for differences in the amount of head rotation observed (3° vs. 14°). Consistent with this idea, the ratcheted RRF structure, with a P/E hybrid bound tRNA, is more modestly rotated like the Jin et al. (2011) structure (9° of counterclockwise rotation and only a 4° head swivel).
Now, with as many as five high-resolution ribosome structures in a ratcheted or hybrid state (Zhang et al. 2009; Ben-Shem et al. 2010; Dunkle et al. 2011; Jin et al. 2011; Zhou et al. 2011), we can begin to systematically understand how previously documented intersubunit bridges (Frank et al. 1995; Yusupov et al. 2001) are impacted by the ratcheting motions. As seen in earlier studies, the axis of rotation is centered about bridge B3—a central bridge formed by a cross-strand adenosine-stacking motif (otherwise known as A minor interactions) involving residues A1418 and A1483 of the 16S rRNA (in h44) with H71 of the 23S rRNA. In ratcheted and unratcheted structures, including here for RF3, this bridge and several other centrally located bridges (B2a and B4), are maintained through concerted movements of the RNA and protein elements on the two subunits. Other bridges, including B5, B7a, and B8, appear to be fully disrupted. Zhou et al. (2011); specifically note that these disrupted bridges are often replaced with new ones, such as those they designate R5, R7a, and R8. Somewhat different between the Zhou et al. (2011) and Jin et al. (2011) structures is the disruption of B1a/B1b in the Zhou et al. (2011) structure; this difference may stem from the greater head swivel reported therein.
Despite some minor differences, a comprehensive look at the high-resolution ratcheted structures reveals common themes. A ratcheted state forms when the 30S subunit rotates counterclockwise relative to the 50S subunit, with the axis of rotation centering near bridge B3; as well, head swiveling accompanies subunit ratcheting. It will be interesting to determine whether the ratcheted forms of the ribosomal subunits are stabilized by some of the “near” A-minor motifs previously identified in the 16S rRNA (Noller 2005).
Of mechanistic interest is the fact that the ratcheted ribosomal state induced by RF3 binding provides a rationale for how this GTPase triggers the dissociation of class I release factors post termination. Crystal structures with RF1 or RF2 bound to pre- or post-termination ribosomes are unambiguously in a classical, nonratcheted state (Korostelev et al. 2008; Laurberg et al. 2008; Weixlbaumer et al. 2008; Jin et al. 2010). As such, RF3 may initially bind the ribosome in the classical state, or perhaps post-termination ribosome complexes sample a ratcheted state. In the RF3 structures, we can see that the ratcheted state of the ribosome stabilized by RF3-GDPNP would sterically clash with bound class I factors. In particular, clashes would be anticipated between domain IV of RF1/2 and the head of the 30S subunit, as well as between domain I and the L11 stalk. Thus, in stabilizing the ratcheted form of the ribosome, RF3-GTP effectively promotes dissociation of the class I RF. After GTP hydrolysis by RF3, the now GDP-bound form likely has decreased affinity, presumably due to the introduction of steric clashes between domain III of RF3-GDP and the 30S subunit. While these ideas are consistent with the ribosome itself functioning as the guanine nucleotide exchange factor (GEF) for RF3 as previously proposed (Zavialov et al. 2001), it is also possible that RF3 binds to the ribosome in the GTP bound form, as has been broadly seen for the other translational GTPases. Notably, RF3 leaves the ribosome in a ratcheted state that could function as an ideal substrate for RRF binding to promote subsequent ribosome recycling.
What do we learn about GTPase positioning from these new RF3 structures? RF3 binds the ribosome at the subunit interface, in a position that overlaps with other GTPases, including IF2 (Allen et al. 2005), EFG, and EFTu. RF3 itself undergoes several structural rearrangements from its free GDP-bound form to its GDPNP-bound ribosome state. Both groups observed a ∼55° rotation of RF3′s domain III relative to its G domain, placing domain III in contact with the 30S shoulder. This rearrangement results from the ordering, most evident from the higher resolution Zhou et al. (2011) structure, of switch loops I and II relative to their disordered form in the isolated RF3 structures.
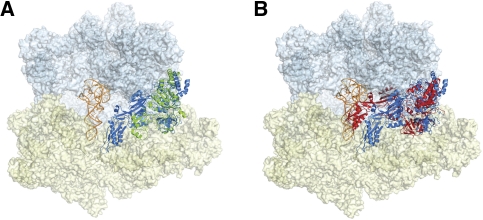
Like other GTPases, RF3′s G domain (domain I) interacts directly with the sarcin–ricin loop (SRL), though the orientation of RF3 is distinct from that observed for EFG and EFTu. RF3′s G domain is rotated by 45° relative to those factors, allowing RF3 to make contacts with L6 as opposed to the L11 stalk. This orientation is of particular interest, because if the highly related EFG could interact in this mode during the early stages of translocation, it would reconcile previously reported hypothetical steric clashes between domain IV of EFG and the A-site-bound tRNA in the pre-translocation state (Fig. 2). Indeed, there were hints of such a binding configuration for EFG in an EFG-cryoEM structure trapped with thiostrepton (Stark et al. 2000). In addition, EFG similarly contains switch loops that become ordered upon GTP binding (Gao et al. 2009), suggesting that the same rearrangement of its G domain may be possible.
FIGURE 2.
(A) Crystal structure of RF3-GDPNP (green) on the ribosome (30S subunit in pale yellow and 50S subunit in pale blue). EFG (blue) aligned by G domain to the RF3 position in Zhou et al. (2011) allows domain IV of EFG to avoid steric clashes with the A site tRNA (orange). (B) EFG as seen in crystal structures with GDP and fusidic acid (red) (Gao et al. 2009) shows many steric clashes with A site tRNA as opposed to the EFG as aligned with RF3 G domain (again in blue).
Although both groups agree that the SRL interacts with the G domain of RF3, there is some disagreement as to which key nucleotide and amino acid interactions might be relevant to GTP hydrolysis. With its higher resolution information, the Zhou et al. (2011) structure reveals the “enclosure” that surrounds GDPNP, and thus, the detailed atomic interactions. In broad terms, the nucleotide is positioned by extensive contacts with backbone and side-chain groups of the conserved P loop found in RF3. The base moiety is exposed to the backbone of the sarcin–ricin loop of 23S rRNA, where a direct interaction is seen between the phosphate of U2656 and the 2-amino group of the guanine. These structural details are difficult to reconcile with the model based on EFTu proposed by Voorhees et al. (2010) in which nucleotide A2662 of the SRL coordinates a conserved histidine to function as a general base in the GTPase reaction. In the Zhou et al. (2011) structure, histidine 92 seems unlikely to participate in this manner as it is positioned 8Å away from the γ phosphate, and A2662 does not contact GDPNP or RF3. Instead, Zhou et al. (2011) propose that structural ordering of RF3 upon binding to the ribosome is sufficient to position a critical Mg2+ ion at the β-γ phosphate linkage, stabilizing the developing negative charge, and thus, simply promoting catalysis. It is also possible, as the investigators note, that the state observed here is an intermediate in the pathway and that subsequent conformational rearrangements may affect orientation for catalysis.
GTPases function at each stage of translation throughout biology. IF2 in bacteria (or eIF5B in eukaryotes) functions during initiation to facilitate subunit joining (with perhaps some more minor contributions to loading initiator tRNA into the P site). EFTu and EFG in bacteria (or eEF1A and eEF2 in eukaryotes) function during each round of elongation to load the appropriate aminoacyl-tRNA into the A site and translocate the complex, respectively. Each of these bacterial factor GTPases has a clear homolog in eukaryotes that functions in an equivalent manner to promote specific events in a GTP-dependent fashion. RF3 and its seeming equivalent in eukaryotes, eRF3, don't fit this pattern. RF3, more closely related to EFG than to EFTu, is nonessential in E. coli, and is not even found in many bacterial species. eRF3, more closely related to EFTu than to EFG, is essential in yeast and is found throughout the eukaryotic kingdom (Atkinson et al. 2008). The nonessential nature of RF3 may suggest that its role in termination is secondary. Indeed, a recent report argued that RF3 plays a primary role in post peptidyl transfer quality control rather than in termination per se (Zaher and Green 2011). In this role, RF3 appears to stimulate the catalytic rate of a premature peptide release (krel) reaction following a misincorporation event on the ribosome. Atomic resolution views of RF3 bound in the presence of a class I RF, perhaps trapped prior to catalysis with a catalytically inactive variant, would provide insight into how this factor might increase krel in these situations.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032011.111.
REFERENCES
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V 2008. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol 8: 290 doi: 10.1186/1471-2148-8-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M 2010. Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Bretscher MS 1968. Translocation in protein synthesis: a hybrid structure model. Nature 218: 675–677 [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Agrawal RK 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322 [DOI] [PubMed] [Google Scholar]
- Frank J, Verschoor A, Li Y, Zhu J, Lata RK, Radermacher M, Penczek P, Grassucci R, Agrawal RK, Srivastava S 1995. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem Cell Biol 73: 757–765 [DOI] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M 1997. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J 16: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M 2000. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci 97: 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J 2005. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell 18: 663–674 [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. 2007. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129: 929–941 [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham RH 1994. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci 91: 5848–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci 107: 8593–8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Ramakrishnan V 2011. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci 108: 15798–15803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F, Adamski FM, Tate WP, Kurland CG 1993. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol 230: 41–50 [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, Van Heel M 2004. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427: 862–865 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y 1994. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci 91: 5798–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman G, Goldstein J, Scolnick E, Caskey T 1969. Peptide chain termination. 3. Stimulation of in vitro termination. Proc Natl Acad Sci 63: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148 [DOI] [PubMed] [Google Scholar]
- Noller HF 2005. RNA structure: reading the ribosome. Science 309: 1508–1514 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 A resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W 2000. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100: 301–309 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J 2003a. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J 2003b. Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R 2011. A primary role for release factor 3 in quality control during translation elongation in Escherichia coli. Cell 147: 396–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124 [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH 2009. Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Trakhanov S, Noller HF 2011. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA 18: 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]