Eukaryotic ribonuclease (RNase) P and RNase MRP are closely related ribonucleoprotein complexes involved in the metabolism of various RNA molecules including tRNA, rRNA, and some mRNAs. While evolutionarily related to bacterial RNase P, eukaryotic enzymes of the RNase P/MRP family are much more complex. Saccharomyces cerevisiae RNase P consists of a catalytic RNA component and nine essential proteins; yeast RNase MRP has an RNA component resembling that in RNase P and 10 essential proteins, most of which are shared with RNase P. The structural organizations of eukaryotic RNases P/MRP are not clear. Here the authors present the results of RNA–protein UV crosslinking studies performed on RNase P and RNase MRP holoenzymes isolated from yeast. The results indicate locations of specific protein-binding sites in the RNA components of RNase P and RNase MRP and shed light on the structural organizations of these large ribonucleoprotein complexes.
Keywords: RNA–protein interactions, ribonuclease MRP, ribonuclease P, Saccharomyces cerevisiae
Abstract
Eukaryotic ribonuclease (RNase) P and RNase MRP are closely related ribonucleoprotein complexes involved in the metabolism of various RNA molecules including tRNA, rRNA, and some mRNAs. While evolutionarily related to bacterial RNase P, eukaryotic enzymes of the RNase P/MRP family are much more complex. Saccharomyces cerevisiae RNase P consists of a catalytic RNA component and nine essential proteins; yeast RNase MRP has an RNA component resembling that in RNase P and 10 essential proteins, most of which are shared with RNase P. The structural organizations of eukaryotic RNases P/MRP are not clear. Here we present the results of RNA–protein UV crosslinking studies performed on RNase P and RNase MRP holoenzymes isolated from yeast. The results indicate locations of specific protein-binding sites in the RNA components of RNase P and RNase MRP and shed light on the structural organizations of these large ribonucleoprotein complexes.
INTRODUCTION
RNA-based RNase P is a ribonucleoprotein complex found in practically all organisms (Altman 2010). RNase P is universally responsible for the maturation of the 5′ end of tRNA and is involved in the metabolism of a variety of other RNA molecules (Coughlin et al. 2008; Altman 2010; Marvin et al. 2011a); in addition, human RNase P was suggested to play a role in transcription (Reiner et al. 2006, 2008). The well-conserved RNA component of RNase P (Chen and Pace 1997) is the catalytic subunit of the enzyme in all domains of life (Guerrier-Takada et al. 1983; Pannucci et al. 1999; Thomas et al. 2000; Kikovska et al. 2007; Li et al. 2009). The protein composition of RNase P varies: While bacterial enzymes have a single small protein (Stark et al. 1978), archaeal RNases P have four to five proteins (Hall and Brown 2002; Cho et al. 2010), and eukaryotic RNases P contain a large multicomponent protein part (nine essential proteins in Saccharomyces cerevisiae; for review, see Marvin and Engelke 2009; Esakova and Krasilnikov 2010).
RNase MRP (Chang and Clayton 1987a,b; Karwan et al. 1991) is a site-specific endoribonuclease closely related to eukaryotic RNase P, but with a distinct substrate specificity (Esakova et al. 2011). RNase MRP is an essential ribonucleoprotein complex found exclusively and universally in eukaryotes (Piccinelli et al. 2005). While RNase MRP was originally identified as a mitochondrial enzyme involved in the processing of RNA primers in mtDNA replication, later studies have demonstrated that the vast majority of RNase MRP is localized to outside the mitochondria (Chang and Clayton 1987a; Karwan et al. 1991; Gill et al. 2006 and references therein); moreover, the protein composition of mitochondrial RNase MRP is distinct (Lu et al. 2010).
Outside the mitochondria, RNase MRP was shown to participate in the maturation of the 5′ end of 5.8S rRNA (Schmitt and Clayton 1993; Chu et al. 1994; Lygerou et al. 1996a); recent data indicate that RNase MRP may be involved in other steps of rRNA maturation (Lindahl et al. 2009; Schneider et al. 2010). In addition, RNase MRP was shown to be involved in the regulation of the cell cycle in yeast by cleaving the 5′ UTR of Cyclin B2 mRNA (Cai et al. 2002; Gill et al. 2004, 2006).
The RNA component of RNase MRP (Lopez et al. 2009) contains a part closely resembling the catalytic domain of RNase P. This strongly suggests that, similar to RNase P, RNase MRP relies on its RNA moiety for catalysis (for review, see Esakova and Krasilnikov 2010). The protein composition of RNase MRP is also very similar to that of RNase P. S. cerevisiae RNase P has nine essential protein components: Pop1, Pop3, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1, and Rpr2 (for review, see Esakova and Krasilnikov 2010). All of these proteins, except for Rpr2, are also found in RNase MRP (Chamberlain et al. 1998). Yeast RNase MRP has 10 known protein subunits: in addition to proteins found in RNase P, RNase MRP has two essential proteins: Snm1 (Schmitt and Clayton 1994) and Rmp1 (Salinas et al. 2005).
The structural organizations of eukaryotic RNase P and RNase MRP and the roles of their individual parts are not clear. High-resolution structural information is available only for the P3 RNA subdomain of RNases MRP/P in a complex with protein components Pop6 and Pop7 (Perederina et al. 2010a,b). The eukaryotic P3 RNA subdomain was suggested to act as a protein-binding hub that played an important role in the evolutionary transition to the protein-rich eukaryotic RNases P/MRP (Ziehler et al. 2001; Perederina and Krasilnikov 2010).
Structures of all archaeal RNase P proteins (without any RNA) have been determined (for review, see Esakova and Krasilnikov 2010). The proteins found in archaeal RNase P (aPop4 [Rpp29], aRpr2 [Rpp21], aRpp1 [Rpp30], aPop5, and aPop3) have homologs in eukaryotic RNase P and RNase MRP (Jarrous and Gopalan 2010), making their structures relevant to the eukaryotic enzymes, although eukaryotic proteins tend to be larger. Aside from the structure of the P3 RNA subdomain in a complex with Pop6 and Pop7 (Perederina et al. 2010b), no relevant high-resolution information on RNA–protein interactions in eukaryotic RNases P/MRP is currently available.
RNA–protein interactions in eukaryotic RNases P/MRP have been studied using pull-down, three-hybrid, UV-crosslinking, and footprinting assays, mostly in the context of partially assembled complexes (Pluk et al. 1999; Jiang et al. 2001; Houser-Scott et al. 2002; Welting et al. 2004, 2007; Aspinall et al. 2007; Perederina et al. 2007, 2011; Esakova et al. 2008; Reiner et al. 2011); however, the structural organizations of eukaryotic enzymes of the RNase P/MRP family—in particular, the locations of the binding sites of individual proteins on RNA—remain unclear.
To clarify the locations of the individual protein components of S. cerevisiae RNase P and RNase MRP on the RNA components, we performed a UV-crosslinking study of RNA–protein interactions in the context of fully assembled, active RNase P/MRP holoenzymes isolated from yeast.
RESULTS
To determine the locations of the UV-crosslinking sites on the RNA components of RNase P and RNase MRP holoenzymes, the holoenzymes were isolated from 11 yeast strains, each containing a histidine tag fused to one of the 11 RNase P/MRP protein components (shared RNase P/MRP proteins Pop1, Pop3, Pop4, Pop5, Pop6, Pop7, Pop8, and Rpp1; RNase P protein Rpr2; RNase MRP proteins Snm1 and Rmp1). The TAP tag (Rigaut et al. 1999), which was fused to the protein component Pop4 found in both RNase P and RNase MRP, was used as the purification handle for the holoenzymes, allowing for a simultaneous isolation of RNase P and RNase MRP as an ∼1:1 mixture (Esakova et al. 2008).
Following the isolation of holoenzymes, the RNase P/MRP mixture was subjected to UV irradiation, the holoenzymes were disassembled under denaturing conditions, His-tagged proteins of interest with covalently attached crosslinked RNA were pulled down using a metal affinity resin, and the RNA that was crosslinked to the protein of interest and copurified with it was treated with Proteinase K, extracted with phenol, and analyzed for the locations of the crosslinks to the protein of interest using primer extension. The primers used in the primer extension were specific to either RNase P or RNase MRP RNAs; for details, see Materials and Methods. To help distinguish between RNA–protein and RNA–RNA crosslinks, RNA was phenol-extracted from isolated samples of RNase P and RNase MRP holoenzymes, subjected to UV irradiation, and used as a negative control. Additionally, the locations of the RNA–RNA crosslinks did not depend on the specific protein used to pull down crosslinked RNA, allowing for easy and reliable differentiation between RNA–RNA and RNA–protein crosslinks.
Typical experimental results of the primer extension analysis of the RNA–protein UV-crosslinking are shown in Figure 1; the results are summarized in Figure 2, A and B. While all 11 RNase P/MRP protein components were analyzed, no RNA–protein crosslinks have been identified for proteins Pop3, Pop6, Pop8, Rpp1, Rpr2, Rmp1, or Snm1. It should be noted that the presence of a close contact between RNA and a protein by itself does not guarantee that a detectable crosslink (that typically requires specific amino acids) (Urlaub et al. 2008) can be formed as it has been observed previously (e.g., in the spliceosome) (Urlaub et al. 2008).
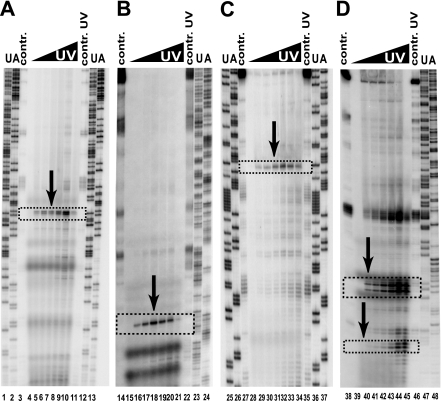
FIGURE 1.
Typical results of the primer extension analysis of RNase P/MRP RNA crosslinked to specific proteins. (A) RNase MRP RNA crosslinked to Pop1; (B) RNase P RNA crosslinked to Pop7; (C) RNase MRP RNA crosslinked to Pop7; (D) RNase P RNA crosslinked to Pop4. (Lanes 1,2,12,13,23–26,37,47,48) Sequencing markers; (lanes 3,14,27,38) primer extension reactions for deproteinated RNAs (controls); (lanes 11,22,35,46) primer extension reactions for UV-treated (1280 mJ/cm2) deproteinated RNAs (controls); (lanes 4–10,15–21,28–34,39–45) primer extension reactions for UV-crosslinked RNAs, the UV dose increasing from 0 to 640 mJ/cm2. The arrows and boxed regions indicate the locations of the RNA–protein crosslinks.
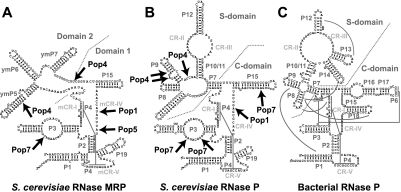
FIGURE 2.
Secondary structure diagrams of the RNA components of (A) yeast RNase MRP; (B) yeast RNase P; and (C) bacterial (Thermotoga maritima) RNase P. (Arrows) The identified sites of RNA–protein crosslinking in eukaryotic RNases P/MRP. (Gray lines) Tertiary interactions in the bacterial RNase P. The diagrams are based on Esakova and Krasilnikov (2010).
DISCUSSION
Interactions between proteins and RNA in yeast RNases P/MRP
While RNA-based RNases P from all domains of life have a well-conserved RNA core (Chen and Pace 1997), the evolution of RNase P was accompanied by a dramatic, and not yet understood, change in the size and composition of the protein part: instead of the single small protein found in bacterial RNase P, eukaryotic RNase P has multiple proteins that constitute the bulk of the enzyme (Marvin and Engelke 2009; Esakova and Krasilnikov 2010). To better understand the roles of the proteins in eukaryotic RNase P and the closely related RNase MRP, we used UV-crosslinking to analyze RNA–protein interactions in the holoenzymes isolated from S. cerevisiae; the results are presented in Figure 2, A and B.
Pop1 is the largest (100 kDa) protein component found in both RNase P and RNase MRP (Lygerou et al. 1994, 1996b). In humans, certain mutations in Pop1, similar to mutations in the RNA component of RNase MRP (Ridanpää et al. 2001), result in multisystemic disorders (Glazov et al. 2011). Pop1 appears to play multiple roles in the assembly and function of RNases P and MRP (Ziehler et al. 2001; Xiao et al. 2006).
Our results indicate that Pop1 interacts directly with the RNA component of both RNase P and RNase MRP, with the identified binding site located in the immediate vicinity of the active site (Fig. 2A,B). At the same time, it is likely that Pop1, being a large protein, may interact with other parts of the RNA component of RNases P/MRP as well.
Protein components Pop6 and Pop7 (as well as their human homologs) were shown to form a heterodimer that binds directly to the in vitro–transcribed P3 RNA subdomain of both RNase P and RNase MRP (Pluk et al. 1999; Welting et al. 2004, 2007; Perederina et al. 2007; Hands-Taylor et al. 2010), and the crystal structure of a complex of the Pop6/Pop7 heterodimer with the P3 RNA subdomain has been reported (Perederina et al. 2010b). Our results obtained for the active holoenzymes (Fig. 2A,B) confirm the relevance of the results produced by partial reconstitution with in vitro–transcribed RNA components; the locations of UV-induced crosslinks between Pop7 and the P3 RNA subdomain are consistent with the recently published crystal structure of the reconstituted Pop6/Pop7-P3 RNA subdomain complex (Perederina et al. 2010b).
However, we did not identify crosslinks between Pop6, the other member of the Pop6/Pop7 heterodimer, and RNA, even though interactions between Pop6 and the P3 RNA (albeit less extensive than those for Pop7) were observed in the crystal structure (Perederina et al. 2010b) and were consistent with in vitro reconstitution experiments (Hands-Taylor et al. 2010). Apparently, this discrepancy reflects the fact that the presence of an RNA–protein contact does not guarantee that a UV-induced crosslink will form. Indeed, one of the two cross-links between Pop7 and the P3 RNA subdomain of RNase P (Fig. 2B) is not observed in RNase MRP (Fig. 2A) even though the P3 RNA subdomains of the two enzymes are sufficiently similar to be interchangeable (Lindahl et al. 2000; Perederina et al. 2010b).
Pop7 also forms a crosslink to the P15 stem of the RNA component of RNase P (Fig. 2B). Footprinting studies of yeast RNase P holoenzyme (Esakova et al. 2008) show that while most of the P15 stem of RNase P is not affected by the binding of proteins, a 2-nt-long area is protected from modifying agents in the presence of RNase P proteins. The location of the protected area coincides with the identified crosslinking site of Pop7. Considering the relatively small size of the protected area (Esakova et al. 2008), it is unlikely that any proteins other than Pop7 are involved in interactions with the distal part of the P15 stem in RNase P.
While the putative catalytic domain of RNase MRP (Domain 1) (Fig. 1A) is expected to be structurally similar to the catalytic (C-) domain of RNase P (Fig. 2B; Esakova et al. 2008; Esakova and Krasilnikov 2010), the P15 stem of RNase MRP is shorter than its counterpart in RNase P and does not reach the position of the crosslink with Pop7 in the latter. Accordingly, no P15–Pop7 crosslinks are observed in RNase MRP (Fig. 2A).
Protein component Pop5 formed a crosslink with the RNA component of RNase MRP in the immediate vicinity of the catalytic site, consistent with the results of the footprinting studies performed for a partially reconstituted RNase MRP (Perederina et al. 2011). We did not observe a corresponding crosslink in RNase P (Fig. 2B). Considering the similarity between the two enzymes, it is unlikely that the absence of the Pop5 crosslink in RNase P reflects a significant difference between the C-domain of RNase P and Domain 1 of RNase MRP, but rather is the result of a relatively small conformational variation that prevented the crosslink from forming in RNase P.
A human homolog of Pop4 (Rpp29) appeared to interact directly with the RNA component of human RNase P (Jiang et al. 2001; Reiner et al. 2011). Yeast three-hybrid studies and pull-down experiments suggested interactions between Pop4 and S. cerevisiae RNase P as well (Chu et al. 1997; Houser-Scott et al. 2002). Our results show that Pop4 interacts with the central part of the specificity (S-) domain of the RNA component of the RNase P holoenzyme (Fig. 2B). It should be noted that while we observed Pop4 crosslinks in two distinct parts of the RNA (a group of two crosslinks at the bulge of the P9 stem and the isolated third crosslink near the base of the P9 stem) (Fig. 2B), all three crosslinks are likely to be grouped together in three-dimensional (3D) space (because the isolated crosslink is located on the opposite RNA strand and about half of a helical turn from the other two crosslinks).
Footprinting results obtained for archaeal RNase P (Xu et al. 2009) indicate that a binary complex formed by the archaeal homolog of Pop4 (Rpp29) and the archaeal homolog of Rpr2 (Rpp21) interacts with the S-domain of RNase P RNA. The RNA region protected in the presence of Rpp29/Rpp21 (aPop4/aRpr2) (Xu et al. 2009) includes the region roughly corresponding to the location of the Pop4 crosslink in yeast RNase P RNA. Thus the location of the Pop4 crosslink is consistent with at least some degree of similarity in the overall location of Pop4 in yeast RNase P and that of its homolog in archaeal RNase P.
The results of three-hybrid studies and pull-down experiments (Chu et al. 1997; Houser-Scott et al. 2002; Welting et al. 2004; Aspinall et al. 2007) suggest that Pop4 is involved in interactions with the RNA component of RNase MRP. Our crosslinking data indicate that Pop4 binds to the ymP5 stem of the RNA components of RNase MRP (Fig. 2A), a structural element that does not have a clear parallel in RNase P, although the presence of a small bulge in the immediate vicinity of the Pop4 binding sites of both RNase P and RNase MRP (Fig. 2A,B) could potentially indicate a certain commonality in the mode of interactions between Pop4 and RNA in the two holoenzymes.
Truncations of the ymP5 stem that remove the nucleotide forming the crosslink to Pop4 are lethal (Shadel et al. 2000; Li et al. 2004), whereas truncations leaving the crosslinking site intact result in either a conditional phenotype (when the ymP5 stem is truncated in the immediate vicinity of the crosslinking site) or no noticeable growth defects (when only three terminal ymP5 base pairs are truncated) (Li et al. 2004). The results of footprinting studies (Esakova et al. 2008) are also consistent with Pop4 interacting with the central part of ymP5.
The second crosslinking site of Pop4 is located at the border of the RNA Domain 1 and Domain 2 (Fig. 2A), suggesting that Pop4 could potentially be involved in the structural stabilization of the mutual orientations of the two RNA domains in RNase MRP. It is interesting to note that Pop4 overexpression or a F207S mutation partially suppresses the effects of simultaneous truncation of multiple RNase MRP stems, perhaps by helping to stabilize RNase MRP structure (Li et al. 2004).
A G122A mutation in the RNA component of yeast RNase MRP affects the function of the enzyme, but the resulting yeast phenotype can be suppressed by the overexpression of protein component Snm1 (Schmitt and Clayton 1994) or mutations in protein component Pop4 (Chu et al. 1997). The Pop4–ymP5 stem crosslink in the RNase MRP holoenzyme is located in the immediate vicinity of G122, indicating that Pop4 binds in this region. However, even though we did not observe Snm1–RNA crosslinks, simultaneous Snm1 binding to this part of the RNA cannot be excluded.
3D mapping of RNA–protein interactions in yeast RNase P
In spite of significant differences in the peripheral structural elements, eukaryotic and bacterial RNases P share the core elements (Chen and Pace 1997): conserved elements CR-I, CR-IV, and CR-V (Fig. 2B), which form the catalytic center of the enzymes; and conserved elements CR-II and CR-III, which are responsible for the recognition of the T-loop and D-loop in the tRNA (Krasilnikov et al. 2004; Torres-Larios et al. 2006; Reiter et al. 2010).
Considering the similarity of the major substrates of the bacterial and eukaryotic RNases P (tRNA), one can expect that while the peripheral elements differ, the overall mutual orientation of the well-conserved catalytic core (comprising CR-I, CR-IV, and CR-V) and the T-/D-loop recognizing element (CR-II/ CR-III) is very similar in the bacterial and eukaryotic RNases P because this expected similarity is dictated by the similarities in the juxtapositions of the T-/D-loops and RNase P cleavage sites in corresponding tRNAs. Accordingly, in the absence of an available high-resolution structure of eukaryotic RNase P, the locations and structures of the conserved core elements in bacterial RNase P can serve as a first approximation for the locations of the corresponding elements in eukaryotic enzymes. The resulting outline of the core elements of the RNA component is similar to that in the recently proposed model of the RNA component of archaeal RNase P (Zwieb et al. 2011).
To better understand the positions of the identified crosslinks in three dimensions, we have mapped them onto the structure of the corresponding elements in bacterial RNase P (Reiter et al. 2010), keeping in the RNA model only the major core elements that are conserved throughout the three domains of life; the result is shown in Figure 3.
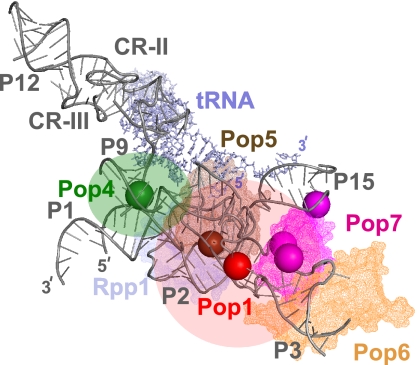
FIGURE 3.
UV-induced RNA–protein crosslinks mapped onto the outline of the RNA component of RNase P. The outline is based on the crystal structure of bacterial RNase P (Reiter et al. 2010). (Gray) RNase P RNA; (light gray) tRNA product; only the RNA elements that are universally conserved from bacteria to eukaryotes are shown. (Solid spheres) The locations of the identified crosslinking sites for individual proteins. (Red) The location of the Pop1 crosslink; (green) the location of the Pop4 crosslink; (brown) the location of the Pop5 crosslink (carried over from RNase MRP); (magenta) the locations of the Pop7 crosslinks. Corresponding approximate locations of proteins are shown as semitransparent in matching colors. The structures of Pop6 and Pop7 are modeled according to Perederina et al. (2010b); Pop5 and Rpp1 are modeled according to Perederina et al. (2011). RNA structural elements are marked according to the nomenclature used in Figure 2B.
Most of the identified protein-binding sites are positioned near the central part of the complex, consistent with the results of footprinting studies (Tranguch et al. 1994; Esakova et al. 2008) showing that auxiliary elements in the RNA component are largely unaffected by interactions with RNase P proteins.
The 3D structure of the RNA component of bacterial RNase P is stabilized by a network of tertiary RNA–RNA interactions involving mostly auxiliary parts of the RNA (Kazantsev et al. 2005; Torres-Larios et al. 2005; Reiter et al. 2010). However, most of the RNA elements involved in the stabilization of the structure are not found in eukaryotic RNase P, suggesting that one of the roles of the dramatically enlarged protein part of eukaryotic RNase P is to replace the missing RNA elements and stabilize the structure of the catalytic RNA (for review, see Esakova and Krasilnikov 2010). The results of mapping RNA–protein interactions (Fig. 3) support this idea because proteins Pop6/Pop7, Pop1, and Pop4 are positioned to be able to mediate tertiary interactions.
The protein components Pop6 and Pop7, acting as a heterodimer, bind to the P3 loop region of the P3 RNA subdomain of yeast RNase P (Perederina et al. 2007, 2010b). In addition to the P3 subdomain, Pop7 interacts with the P15 stem of RNase P RNA (Fig. 2B). The elements involved in the tertiary RNA–RNA interactions that stabilize the position of the P15 stem in bacterial RNase P are absent in yeast RNase P. As crosslinking data demonstrate (Fig. 3), it is the protein component Pop7 that mediates the tertiary interaction between the core and the P15 stem in yeast RNase P.
The protein component Pop1 crosslinks to RNase P RNA in the immediate vicinity of the catalytic site (Figs. 2A,B, 3). Its possible participation in the folding of the RNA-based catalytic site and/or the coordination of metal ions essential for activity warrants further investigation.
The results of yeast three-hybrid studies (Ziehler et al. 2001; Houser-Scott et al. 2002) suggested interactions between Pop1 and RNase P RNA in S. cerevisiae, although it was not clear if this interaction was direct or mediated by other proteins. Mutations in the P3 subdomain of RNase P RNA affected Pop1 binding (Ziehler et al. 2001). At the same time, crosslinking, pull-down, and immunoprecipitation studies involving in vitro transcribed RNase MRP RNA in HeLa cell extract (Pluk et al. 1999) suggested that Pop1 interacted with the closely related RNase MRP RNA indirectly, through interactions with other proteins, possibly homologs of Pop6/Pop7.
Our results indicate that Pop1 forms a crosslink in the immediate vicinity of the P3 RNA subdomain and the Pop6/Pop7 heterodimer (Fig. 3). An interaction between Pop1 and Pop6/Pop7 can explain how mutations in the phylogenetically conserved part of the P3 RNA subdomain loop can affect Pop1 binding (Ziehler et al. 2001), even while the structural data indicate that the mutated nucleotides are involved in interactions with Pop7 (Perederina et al. 2010b). The existence of direct Pop1–P3 RNA interactions also cannot be excluded. In any case, the proximity of the identified Pop1-binding site to the P3 RNA subdomain supports the suggested role of the P3 RNA subdomain in the assembly and structural stabilization of eukaryotic RNases P/MRP (Ziehler et al. 2001; Perederina and Krasilnikov 2010; Perederina et al. 2010b).
Based on our crosslinking results, Pop4 occupies a central position in yeast RNase P (Fig. 3). It was previously suggested (Chen et al. 2011) that in archaeal RNase P, the protein component Rpp29 (a homolog of yeast Pop4) acting in a pair with the protein component Rpp21 (a homolog of yeast Rpr2) (Tsai et al. 2006; Xu et al. 2009; Sinapah et al. 2011) serves to enable long-distance interactions between the catalytic (C-) and the specificity (S-) domains of the RNA component of RNase P. Mapping of the crosslinking sites of Pop4 into the expected outline of the RNA component of yeast RNase P (Fig. 3) demonstrates that the localization of Pop4 is consistent with the protein's role in the stabilization of the mutual orientation of the C-domain and S-domain of yeast RNase P RNA.
Indeed, our crosslinking results position Pop4 in the immediate vicinity of the P1 stem of the catalytic domain of the RNase P RNA (Fig. 3). Although we did not observe crosslinks between Pop4 and the P1 stem (it should be noted that a large part of the P1 stem was not accessible to the analysis that involved primer extension), the proximity of the identified Pop4-binding site and the P1 stem supports the existence of Pop4–P1 stem interactions. The footprinting results obtained for the in vitro–transcribed human RNase P RNA in a complex with the human Pop4 homolog (Reiner et al. 2011) also position Pop4 so that it can bridge the two RNA domains, although the location of the protected area differs from that of the crosslinks observed in the yeast holoenzyme.
In addition to serving structural roles, the large protein part of eukaryotic RNase P was suggested to serve to widen the spectrum of recognized substrates (Coughlin et al. 2008; Marvin and Engelke 2009; Marvin et al. 2011a). Based on our data (Fig. 3) several RNase P proteins are positioned to interact with substrates: Pop1, Pop4, Pop5, and, potentially, Rpp1, consistent with recent results of enzyme–substrate crosslinking studies (Marvin et al. 2011b).
MATERIALS AND METHODS
S. cerevisiae strains
Yeast strains EK-Pop1NHis, EK-Pop3, EK-Pop5, EK-Pop6, EK-Pop7, EK-Pop8, EK-Rpp1, EK-Rpr2, EK-Rmp1, and EK-Snm1 were based on yeast strain YSW1 (Salinas et al. 2005), a generous gift from Mark Schmitt, which contained a TAP tag (Rigaut et al. 1999) attached to the carboxyl terminus of the RNase P/MRP protein Pop4. In EK-Pop3, EK-Pop5, EK-Pop6, EK-Pop7, EK-Pop8, EK-Rpp1, EK-Rpr2, EK-Rmp1, and EK-Snm1, His6 tags were fused to the carboxyl termini of corresponding proteins using standard PCR-based methods and kanMX4 as the selectable marker. In EK-Pop1NHis, the promoter region of the genomic Pop1 was replaced with the ADH promoter followed by a fused amino-terminal His6 tag, because carboxy-terminal Pop1 tags were not tolerated (Lygerou et al. 1994); kanMX4 was used as the selectable marker. Yeast strain EK-Pop4 was based on yeast strain LSY389-34A (Salinas et al. 2005), a generous gift from Mark Schmitt; a His8 tag followed by the standard TAP tag (Rigaut et al. 1999) was fused to the carboxyl terminus of the genomic Pop4 using PCR-based methods and TRP1 as the selectable marker. Sequences of modified regions in all yeast strains were verified by sequencing. The insertion of the tags did not have any noticeable effect on yeast viability.
Isolation of RNase P/RNase MRP holoenzymes
Active RNase P and RNase MRP were isolated from yeast strains EK-Pop1NHis, EK-Pop3, EK-Pop4, EK-Pop5, EK-Pop6, EK-Pop7, EK-Pop8, EK-Rpp1, EK-Rpr2, EK-Rmp1, and EK-Snm1 as an ∼1:1 RNase P/ RNase MRP mixture using the TAP tag attached to the carboxyl terminus of RNase P/MRP protein component Pop4 as a purification handle and following a previously described procedure (Salinas et al. 2005; Esakova et al. 2008). The addition of the fused His6/His8 tags did not require any modifications of the isolation procedure.
UV crosslinking analysis of RNA–protein interactions in RNase P and RNase MRP
To analyze the binding of each of the 11 individual proteins found in RNases P and MRP (Pop1, Pop3, Pop4, Pop5, Pop6, Pop7, Pop8, Rpp1, Rpr2, Snm1, and Rmp1), UV crosslinking and the subsequent analysis were performed for each of the 11 generated yeast strains carrying His-tags fused to the individual proteins of interest (EK-Pop1NHis, EK-Pop3, EK-Pop4, EK-Pop5, EK-Pop6, EK-Pop7, EK-Pop8, EK-Rpp1, EK-Rpr2, EK- EK-Snm1, and EK-Rmp1, respectively). The crosslinking analysis procedure was similar to the one previously described in Perederina et al. (2011).
RNase P/RNase MRP holoenzymes were isolated from yeast as an ∼1:1 mixture (above), diluted to ∼0.5 μM of each in a buffer containing 20 mM Tris-HCl (pH 7.8), 150 mM NaCl, 2.5 mM MgCl2, 0.05 mM EDTA, 5% (v/v) glycerol, and subjected to UV (254 nm) irradiation in a UV SpectroLinker XL-1500 crosslinker (Spectronics). The irradiation dose varied from 40 to 640 mJ/cm2; the samples (15-μL drops on glass plates) were kept on ice during irradiation. After UV irradiation, 20 μL of a denaturing buffer containing 4% SDS, 100 mM Tris-HCl (pH 7.8), 20% (v/v) glycerol, 50 mM DTT were added, and the samples were incubated for 10 min at 60°C. To facilitate the complete disassembly of RNA–protein complexes, after the initial round of incubation, additional 30 mM DTT was added to the samples, and incubation was continued for another 10 min; then the cycle of 30 mM DTT addition and 10 min of incubation at 60°C was repeated three more times. The samples containing disassembled RNases P/MRP were transferred in 1 mL of a binding buffer containing 8 M urea, 15 mM Tris-HCl (pH 7.8), 500 mM NaCl, 10 mM Na-imidazole (pH 7.4), 1% (v/v) Tween 20, and incubated with 50 μL of Ni-IDA resin (Macherey-Nagel) for 2 h at room temperature with intensive agitation. After incubation, the resin was washed five times with 1 mL of the binding buffer (above). Bound proteins were eluted three times with 100 μL of a buffer containing 4 M urea, 15 mM Tris-HCl (pH 7.8), 250 mM NaCl, 300 mM Na-imidazole (pH 7.4), and 0.5% (v/v) Tween 20; an aliquot was taken to analyze the quality of protein isolation using silver-stained SDS-polyacrylamide gels (Fig. 4); the rest of the sample was treated with 300 μg of Proteinase K for 20 min at 37°C. Crosslinked RNA that was pulled down by the specific His-tagged protein was extracted with phenol, precipitated with ethanol, and dissolved in a buffer containing 5 mM Tris-HCl (pH 8.0), 10 mM KCl, and 0.1 mM EDTA.
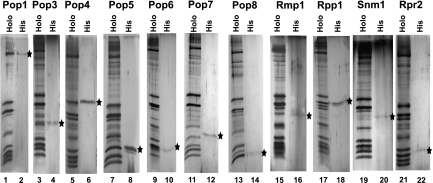
FIGURE 4.
Isolation of His-tagged proteins from RNase P/MRP holoenzymes. (Lanes 1,2) Protein Pop1 (yeast strain EK-Pop1NHis); (lanes 3,4) protein Pop3 (yeast strain EK-Pop3); (lanes 5,6) protein Pop4 (yeast strain EK-Pop4); (lanes 7,8) protein Pop5 (yeast strain EK-Pop5); (lanes 9,10) protein Pop6 (yeast strain EK-Pop6); (lanes 11,12) protein Pop7 (yeast strain EK-Pop7); (lanes 13,14) protein Pop8 (yeast strain EK-Pop8); (lanes 15,16) protein Rmp1 (yeast strain EK-Rmp1); (lanes 17,18) protein Rpp1 (yeast strain EK-Rpp1); (lanes 19,20) protein Snm1 (yeast strain EK-Snm1); (lanes 21,22) protein Rpr2 (yeast strain EK-Rpr2). Odd lanes are isolated mixtures of RNase P/MRP holoenzymes; even lanes are pull-downs of tagged proteins (marked by black stars). Silver-stained SDS-polyacrylamide gels.
Locations of the crosslinks on the RNase P and RNase MRP RNAs were identified using primer extension with SuperScript II (Invitrogen) reverse transcriptase following a protocol described in Esakova et al. (2008). To ensure a reliable and redundant coverage of RNA, the following primers were used for analysis: For RNase P: PRTP1B (complementary to nucleotides 343–369); PRTP11 (329–366); PRTP15 (284–315); PRTP2A (260–290); PRTP23 (227–268); PRTP25A (200–230); PRTP27 (167–208); PRTP3 (112–138); PRTP4 (95–130). For RNase MRP: RTP1AL (complementary to nucleotides 324–340); RTP15A (243–276); RTP2A (213–248); RTP21 (203–242); RTP25 (168–201); RTP3B (133–174); RTP4 (91–126). This set of primers allowed for reliable coverage of the entire length of RNase P and MRP RNAs except for the last 30/25 nucleotides at the 3′ ends of RNase P/MRP, respectively.
RNA that was phenol-extracted from isolated holoenzymes and subjected to UV irradiation at a dose twice the maximum dose given to the test samples (640 mJ/cm2) was used as a control for RNA–RNA crosslinks; the same phenol-extracted RNA, but not subjected to UV irradiation, was used as a control for the reverse transcriptase stops not related to crosslinking.
DNA sequencing reactions obtained using plasmids pYRP (for RNase P) and p31/51 (for RNase MRP) (Perederina et al. 2007) and corresponding primers were used as ladders. 32P-labeled products of primer extension were resolved on 5% denaturing polyacrylamide gels and analyzed using a PhosphorImager (Molecular Dynamics).
ACKNOWLEDGMENTS
We thank P. Bevilacqua, D. Engelke, J. Reese, and M. Schmitt for valuable suggestions; M. Schmitt for generous gifts of yeast strains and plasmids; C. Quan for technical assistance; and L. Krasilnikova for help with the manuscript preparation. This work was supported by NIH Grant GM085149 to A.S.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030874.111.
REFERENCES
- Altman S. 2010. History of RNase P and overview of its catalytic activity. In Ribonuclease P (ed. F Liu and S Altman), Protein Reviews 10, pp. 1–15. Springer, New York. [Google Scholar]
- Aspinall TV, Gordon JMB, Bennett HJ, Karahalios P, Bukowski J-P, Walker SC, Engelke DR, Avis JM 2007. Interactions between subunits of Saccharomyces cerevisiae RNase MRP support a conserved eukaryotic RNase P/MRP architecture. Nucleic Acids Res 35: 6439–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Aulds J, Gill T, Cerio M, Schmitt ME 2002. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics 161: 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA 1987a. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J 6: 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA 1987b. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science 235: 1178–1184 [DOI] [PubMed] [Google Scholar]
- Chen J-L, Pace NR 1997. Identification of the universally conserved core of ribonuclease P RNA. RNA 3: 557–560 [PMC free article] [PubMed] [Google Scholar]
- Chen W-Y, Xu Y, Cho I-M, Oruganti SV, Foster MP, Gopalan V 2011. Cooperative RNP assembly: Complementarity rescue of structural defects by protein and RNA subunits of archaeal RNase P. J Mol Biol 411: 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V 2010. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci 107: 14573–14578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci 91: 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Zengel JM, Lindahl L 1997. A novel protein shared by RNase MRP and RNase P. RNA 3: 382–391 [PMC free article] [PubMed] [Google Scholar]
- Coughlin DJ, Pleiss JA, Walker SC, Whitworth GB, Engelke DR 2008. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc Natl Acad Sci 105: 12218–12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Krasilnikov AS 2010. Of proteins and RNA: The RNase P/MRP family. RNA 16: 1725–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Perederina A, Quan C, Schmitt ME, Krasilnikov AS 2008. Footprinting analysis demonstrates extensive similarity between eukaryotic RNase P and RNase MRP holoenzymes. RNA 14: 1558–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakova O, Perederina A, Quan C, Berezin I, Krasilnikov AS 2011. Substrate recognition by ribonucleoprotein ribonuclease MRP. RNA 17: 356–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME 2004. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol Cell Biol 24: 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Aulds J, Schmitt ME 2006. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J Cell Biol 173: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Zankl A, Donskoi M, Kenna TJ, Thomas GP, Clark GR, Duncan EL, Brown MA 2011. Whole-exome re-sequencing in a family quartet identifies POP1 mutations as the cause of a novel skeletal dysplasia. PLoS Genet 7: e1002027 doi: 10.1371/journal.pgen.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857 [DOI] [PubMed] [Google Scholar]
- Hall TA, Brown JW 2002. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA 8: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hands-Taylor KLD, Martino L, Tata R, Babon JJ, Bui TT, Drake AF, Beavil RL, Pruijn GJM, Brown PR, Conte MR 2010. Heterodimerization of the human RNase P/MRP subunits Rpp20 and Rpp25 is a prerequisite for interaction with the P3 arm of RNase MRP RNA. Nucleic Acids Res 38: 4052–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR 2002. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc Natl Acad Sci 99: 2684–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Gopalan V 2010. Archaeal/eukaryal RNase P: Subunits, functions and diversification. Nucleic Acids Res 39: 7885–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Guerrier-Takada C, Altman S 2001. Protein–RNA interactions in the subunits of human nuclear RNase P. RNA 7: 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwan R, Bennett JL, Clayton DA 1991. Nuclear RNase MRP processes RNA at multiple discrete sites: interaction with an upstream G box is required for subsequent downstream cleavages. Genes Dev 5: 1264–1276 [DOI] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci 102: 13392–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikovska E, Svard SG, Kirsebom LA 2007. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci 104: 2062–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107 [DOI] [PubMed] [Google Scholar]
- Li X, Zaman S, Langdon Y, Zengel JM, Lindahl L 2004. Identification of a functional core in the RNA component of RNase MRP of budding yeast. Nucleic Acids Res 32: 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Willkomm DK, Hartmann RK 2009. Minor changes largely restore catalytic activity of archaeal RNase P RNA from Methanothermobacter thermoautotrophicus. Nucleic Acids Res 37: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Fretz S, Epps N, Zengel JM 2000. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA 6: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Bommankanti A, Li X, Hayden L, Jones A, Khan M, Oni T, Zengel JM 2009. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA 15: 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MD, Rosenblad MA, Samuelsson T 2009. Conserved and variable domains of RNase MRP RNA. RNA Biol 6: 208–220 [DOI] [PubMed] [Google Scholar]
- Lu Q, Wierzbicki S, Krasilnikov AS, Schmitt ME 2010. Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components. RNA 16: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev 8: 1423–1433 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B 1996a. Accurate processing of a eukaryotic precursor ribosomal RNA by Ribonuclease MRP in vitro. Science 272: 268–270 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Seraphin B 1996b. hPop1: An autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J 15: 5936–5948 [PMC free article] [PubMed] [Google Scholar]
- Marvin MC, Engelke DR 2009. Broadening the mission of an RNA enzyme. J Cell Biochem 108: 1244–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin MC, Clauder-Munster S, Walker SC, Sarkeshik A, Yates JR III, Steinmetz LM, Engelke DR 2011a. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA 17: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin MC, Walker SC, Fierke CA, Engelke DR 2011b. Binding and cleavage of unstructured RNA by nuclear RNase P. RNA 17: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW 1999. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci 96: 7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Krasilnikov AS 2010. The P3 domain of eukaryotic RNases P/MRP: Making a protein-rich RNA-based enzyme. RNA Biol 7: 534–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS 2007. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA 13: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS 2010a. Crystallization and preliminary X-ray diffraction analysis of the P3 RNA domain of yeast ribonuclease MRP in a complex with RNase P/MRP protein components Pop6 and Pop7. Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS 2010b. Eukaryotic ribonucleases P/MRP: The crystal structure of the P3 domain. EMBO J 29: 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Khanova E, Quan C, Berezin I, Esakova O, Krasilnikov AS 2011. Interactions of a Pop5/Rpp1 heterodimer with the catalytic domain of RNase MRP. RNA 17: 1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinelli P, Rosenblad MA, Samuelsson T 2005. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res 33: 4485–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJM, van Venrooij WJ 1999. RNA–protein interactions in the human RNase MRP ribonucleoprotein complex. RNA 5: 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N 2006. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev 20: 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N 2008. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS ONE 3: e4072 doi: 10.1371/journal.pone.0004072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Alfiya-Mor N, Berrebi-Demma M, Wesolowski D, Altman S, Jarrous N 2011. RNA binding properties of conserved protein subunits of human RNase P. Nucleic Acids Res 39: 5704–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridanpää M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. 2001. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104: 195–203 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Salinas K, Wierzbicki S, Zhou L, Schmitt ME 2005. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J Biol Chem 280: 11352–11360 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13: 7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA 1994. Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes Dev 8: 2617–2628 [DOI] [PubMed] [Google Scholar]
- Schneider MD, Bains AK, Rajendra TK, Dominski Z, Matera AG, Simmonds AJ 2010. Functional characterization of Drosophila MRP (mitochondrial RNA processing) RNA gene. RNA 16: 2120–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Buckenmeyer GA, Clayton DA, Schmitt ME 2000. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene 245: 175–184 [DOI] [PubMed] [Google Scholar]
- Sinapah S, Wu S, Chen Y, Pettersson BM, Gopalan V, Kirsebom LA 2011. Cleavage of model substrates by archaeal RNase P: Role of protein cofactors in cleavage-site selection. Nucleic Acids Res 39: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark BC, Kole R, Bowman EJ, Altman S 1978. Ribonuclease P: An enzyme with an essential RNA component. Proc Natl Acad Sci 75: 3717–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Chamberlain J, Engelke DR, Gegenheimer P 2000. Evidence for an RNA-based catalytic mechanism in eukaryotic nuclear ribonuclease P. RNA 6: 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A 2005. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437: 584–587 [DOI] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Pan T, Mondragon A 2006. Structure of ribonuclease P—a universal ribozyme. Curr Opin Struct Biol 16: 327–335 [DOI] [PubMed] [Google Scholar]
- Tranguch AJ, Kindelberger DW, Rohlman CE, Lee JY, Engelke DR 1994. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry 33: 1778–1787 [DOI] [PubMed] [Google Scholar]
- Tsai H-Y, Pulukkunat DK, Woznick WK, Gopalan V 2006. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc Natl Acad Sci 103: 16147–16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H, Hartmuth K, Lührmann R 2008. Protein–RNA crosslinking in native ribonucleoprotein particles. In Handbook of RNA biochemistry (ed. RK Hartmann et al.). Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- Welting TJM, van Venrooij WJ, Pruijn GJM 2004. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res 32: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welting TJM, Peters FMA, Hensen SMM, van Doorn NL, Kikkert BJ, Raats JMH, van Venrooij WJ, Pruijn GJM 2007. Heterodimerization regulates RNase MRP/RNase P association, localization, and expression of Rpp20 and Rpp25. RNA 13: 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Hsien J, Nugent RL, Coughlin DJ, Fierke CA, Engelke DR 2006. Functional characterization of the concerved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP. RNA 12: 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP 2009. Solution structure of an archaeal RNase P binary protein complex: Formation of the 30-kDa complex between Pyrococcus furiosus RPP21 and RPP29 is accompanied by coupled protein folding and highlights critical features for protein–protein and protein–RNA interactions. J Mol Biol 393: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR 2001. An essential protein-binding domain of nuclear RNase P RNA. RNA 7: 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Nakao Y, Nakashima T, Takagi H, Goda S, Andersen ES, Kakuta Y, Kimura M 2011. Structural modeling of RNase P RNA of the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem Biophys Res Commun 414: 517–522 [DOI] [PubMed] [Google Scholar]