The CRISPR-Cas system encodes an RNA-based adaptive immune response in bacteria and archaea. The authors applied a differential deep-sequencing approach to define the expression and processing patterns of the RNAs encoded by the multiple CRISPR loci in the genome of Thermus thermophilus HB8. The 3′ ends of the CRISPR RNAs are generated by two alternative mechanisms that depend on the CRISPR repeat-sequence. These observations are consistent with the operation of multiple crRNA processing systems within a single bacterial strain.
Keywords: CRISPR, biogenesis, crRNA, deep-sequencing, processing mechanism
Abstract
The CRISPR-Cas system represents an RNA-based adaptive immune response system in prokaryotes and archaea. CRISPRs (clustered regularly interspaced short palindromic repeats) consist of arrays of short repeat-sequences interspaced by nonrepetitive short spacers, some of which show sequence similarity to foreign phage genetic elements. Their cistronic transcripts are processed to produce the mature CRISPR RNAs (crRNAs), the elements that confer immunity by base-pairing with exogenous nucleic acids. We characterized the expression and processing patterns of Thermus thermophilus HB8 CRISPRs by using differential deep-sequencing, which differentiates between 5′ monophosphate and 5′ non-monophosphate-containing RNAs and/or between 3′ hydroxyl and 3′ non-hydroxyl-containing RNAs. The genome of T. thermophilus HB8 encodes 11 CRISPRs, classified into three distinct repeat-sequence types, all of which were constitutively expressed without deliberately infecting the bacteria with phage. Analysis of the differential deep sequencing data suggested that crRNAs are generated by endonucleolytic cleavage, leaving fragments with 5′ hydroxyl and 3′ phosphate or 2′,3′-cyclic phosphate termini. The 5′ ends of all crRNAs are generated by site-specific cleavage 8 nucleotides upstream of the spacer first position; however, the 3′ ends are generated by two alternative, repeat-sequence-type–dependent mechanisms. These observations are consistent with the operation of multiple crRNA processing systems within a bacterial strain.
INTRODUCTION
Bacteria and archaea have developed various defense mechanisms against foreign mobile genetic elements (phages and plasmids), including a new mechanism utilizing regulatory RNAs originating from CRISPRs (clustered regularly interspaced short palindromic repeats) (Sturino and Klaenhammer 2004; Horvath and Barrangou 2010). Currently more than 500 bacteria are predicted to contain CRISPRs. A CRISPR locus comprises a 5′ leader sequence followed by short repeats, which are separated by spacer sequences (for reviews, see Sorek et al. 2008; Deveau et al. 2010; Horvath and Barrangou 2010; Karginov and Hannon 2010; Marraffini and Sontheimer 2010; Jore et al. 2011a). The average length of the repeats and spacers is 31 and 36 nucleotides (nt), respectively (based on the CRISPRdb database, October 2010 download) (Grissa et al. 2007). The number of repeats in CRISPR arrays included in this database range from two to 588, with an average of 49 and median of 13 repeats per CRISPR array. The repeat-sequences are typically invariant within a given CRISPR array. Paralogous and orthologous relationships of repeats were also observed (Kunin et al. 2007). Spacer sequences, on the other hand, are highly variable even within a CRISPR array but tend to exhibit similar lengths. Clusters of genes encoding CRISPR-associated (Cas) proteins are often located in close proximity to the CRISPR locus, defining CRISPR-Cas modules. The CRISPR-Cas modules vary in their Cas protein composition and order and in the CRISPR repeat-sequence and RNA structure, features that are used to classify and characterize these modules into three major types and 10 major subtypes (Haft et al. 2005; Makarova et al. 2011a,b). The various Cas proteins were shown to be involved in each of the three major stages of CRISPR-based immunity: adaptation, CRISPR transcript biogenesis, and interference (for review, see Marraffini and Sontheimer 2010). Interestingly, different CRISPR-Cas modules may differ in the mechanism employed in one or more of these stages (for review, see Makarova et al. 2011b).
CRISPR loci are transcribed to produce long precursor (pre-) CRISPR RNAs (crRNAs) that are subsequently cleaved at specific positions within the repeats, yielding mature crRNA fragments (for reviews, see Sorek et al. 2008; van der Oost et al. 2009; Deveau et al. 2010; Horvath and Barrangou 2010; Karginov and Hannon 2010; Marraffini and Sontheimer 2010; Al-Attar et al. 2011; Jore et al. 2011a; Terns and Terns 2011) The maturation of the crRNA 5′ end usually occurs by site-specific endonucleolytic cleavage by a Cas protein 8 nt upstream of the spacer within the preceding repeat region. An exception was demonstrated in Streptococcus pyogenes, where the initial cleavage involves a trans-encoded small guide RNA with complementarity to the repeat, recruiting RNase III rather than a Cas endonuclease for processing (Deltcheva et al. 2011). The mechanism of crRNA 3′ end maturation varies among species and may include retention of the original 3′ end as in Escherichia coli (Jore et al. 2011b), or further trimming as in Pyrococcus furiosus (Hale et al. 2008, 2009) and Pectobacterium atrosepticum (Przybilski et al. 2011).
Several functionally analogous Cas proteins are currently known to be involved in the endonucleolytic cleavage of pre-crRNAs. These include Cse3 of E. coli (Brouns et al. 2008) and of Thermus thermophilus HB8 (Sashital et al. 2011), Cas6 of P. furiosus (Carte et al. 2008; Hale et al. 2009), and Csy4 of Pseudomonas aeruginosa and P. atrosepticum (Haurwitz et al. 2010; Przybilski et al. 2011). All of these proteins were shown to cleave the pre-crRNA 8 nt upstream of the spacer, within the preceding repeat region. Biochemical characterization of the pre-crRNA cleavage products revealed that they exhibit 5′ hydroxyl (OH) and 2′,3′-cyclic phosphate (<2′,3′P) ends (Carte et al. 2008; Haurwitz et al. 2010; Jore et al. 2011b). The pre-crRNA repeats recognized by E. coli Cse3, T. thermophilus HB8 Cse3, and P. aeruginosa Csy4 are different in sequence but similar in secondary structures, directing the cleavage to the same position in respect to the secondary structure (Haurwitz et al. 2010; Jore et al. 2011b; Sashital et al. 2011). The repeat of the pre-crRNA associated with Cas6 of P. furiosus forms a different secondary structure. It seems therefore that there are combinations of CRISPR repeats and Cas protein subtypes that are associated with specific processing mechanisms (Haft et al. 2005; Kunin et al. 2007). Up to date, only one functional endogenous pre-crRNA processing mechanism was described for a single bacterial strain. However, the occurrence of multiple CRISPR-Cas subtypes in some bacterial genomes suggests that different processing mechanisms may operate in parallel.
Here we used a differential RNA sequencing approach to study the expression and processing of pre-crRNAs in T. thermophilus HB8 encoding multiple CRISPR operons on its chromosome and on one of its two plasmids, which were classified into three different repeat-sequence types (I, II, and III) (Fig. 1; Grissa et al. 2007; Agari et al. 2010). More than 30 Cas proteins are encoded in T. thermophilus HB8 and, according to a recent classification, are organized into four distinct subtypes: subtype III-A (previously Mtube or CASS6), subtype III-B (Polymerase-RAMP module), subtype I-E (Ecoli or CASS2), and a hybrid module that includes Cas6 (Fig. 1; Makarova et al. 2011a,b; K. Makarova, pers. comm.). The variability in the CRISPR repeat-sequences and the existence of multiple Cas subtypes, two of which were shown to be involved in distinct pre-crRNA biogenesis mechanisms, suggest that multiple processing mechanisms can operate in parallel in T. thermophilus HB8. We show that all predicted CRISPRs (Grissa et al. 2007) were constitutively transcribed and processed to crRNAs. Consistent with the biogenesis mechanisms revealed in other species, we found that the 5′ end of all crRNAs in T. thermophilus HB8 predominantly retained 8 nt of the repeat-sequences upstream of their variable spacer sequence. In contrast, the 3′ end processing exhibited dependence on the repeat-sequence type, where crRNAs of repeat-sequence type II retained their original 3′ end, while additional 3′ end processing was observed for types I and III. Our results suggest that at least two different pre-crRNA processing systems function in T. thermophilus HB8.
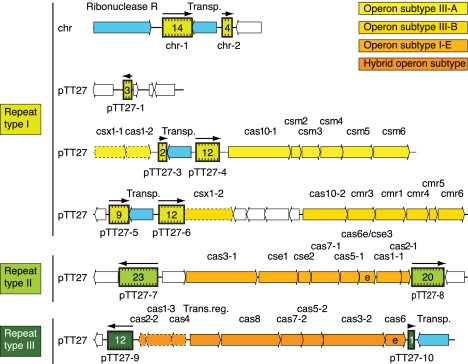
FIGURE 1.
Schematic representation of the genomic organization of T. thermophilus HB8 CRISPRs and CRISPR-associated (Cas) genes on the chromosome and plasmid pTT27. CRISPRs are indicated by colored boxes in different shades of green, one for each repeat-sequence type. The number of spacers for each CRISPR is given within each box. Transcriptional direction is indicated by black arrows above the boxes. Open reading frames are shown as boxes with arrowheads indicating their transcriptional direction. Genes unrelated to CRISPRs are white; CRISPR-associated genes are colored and named (on top) according to the classification and nomenclature of the proteins in the CRISPR-Cas systems (Makarova et al. 2011a,b; K. Makarova, pers. comm.). For clarity we added a serial number for paralogous genes (e.g., cas1-1, cas1-2, etc.). The four different colors correspond to four CRISPR-Cas system subtypes: light yellow indicates III-A (Mtube); yellow, III-B (RAMP); light orange, I-E (Ecoli); and orange, a hybrid of I-A (Aperm) and I-B (Tneap-Hmari). The gene Csx1-2 (dotted box) was colored according to its adjacent subtype, although it was not originally classified as part of this subtype. Two genes (cse3 and cas6) are marked by the letter “e,” indicating that these Cas proteins are orthologous to Cas proteins that are known to be involved in endonucleolytic cleavage of CRISPR transcripts. Transposons (Transp.) as well as a ribonuclease close to the CRISPRs are indicated by blue boxes with arrowheads indicating transcription direction. One transcriptional regulator associated with CRISPR-Cas systems is displayed as well (Trans.reg.).
RESULTS
Deep-sequencing approach to identify differentially phosphorylated RNAs
Different classes of RNA molecules in prokaryotes and eukaryotes have distinct chemical groups at their 5′ and/or 3′ end. Taking into consideration this variability, various approaches were developed to generate cDNA libraries enriched with RNA of specific combination of 5′ and/or 3′ end modifications, which are subsequently subjected to sequencing (Lau et al. 2001; Pak and Fire 2007; Hafner et al. 2008; Sharma et al. 2010). As described below, we generated several cDNA libraries to search for small noncoding RNAs (ncRNAs) in T. thermophilus HB8, including small interfering RNAs, as T. thermophilus HB8 also encodes an Argonaute protein (Wang et al. 2008, 2009).
Total RNA from T. thermophilus HB8 cells was isolated (Chomczynski and Sacchi 1987) and fractionated in three different size ranges: 19–35 nt (short), 35–50 nt (medium), and 50–100 nt (long). Each size fraction was divided into two aliquots. One was directly used for ligation of the 3′ and 5′ sequencing adapters, whereas the second was treated with alkaline phosphatase to remove 5′ and 3′ phosphoryl groups (mono-, di-, and triphosphate) from the RNAs, followed by phosphorylation of their 5′ termini and simultaneous removal of <2′,3′P by T4 polynucleotide kinase (Fig. 2; Supplemental Fig. S1). 3′ Aminoacylated tRNAs were not made accessible for adapter ligation by dephosphorylation and kinase treatment since the RNA was not deacylated prior to 3′ adapter ligation.
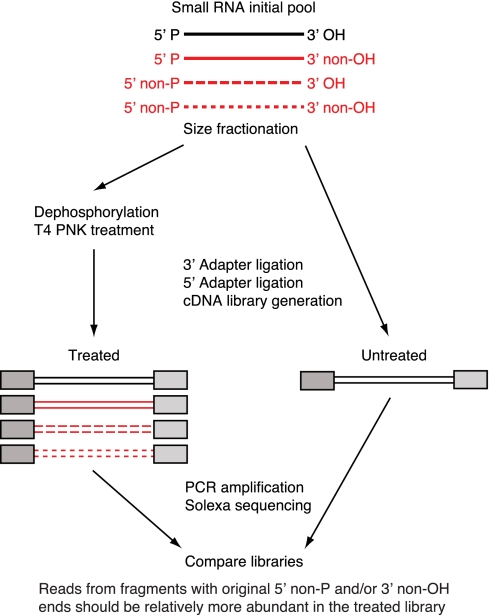
FIGURE 2.
Schematic representation of the differential RNA sequencing methodology. Short RNA molecules with original 5′P and 3′OH ends are sequenced from both treated and untreated libraries, while RNA molecules with original 5′ non-P ends (5′OH or 5′PPP ends) or 3′ non-OH ends (2′P, 3′P, or <2′,3′P) are expected to be preferentially sequenced from the treated library. We compared the two libraries and searched for reads that are far more abundant in the treated library compared with the untreated one and therefore represent putative RNA fragments with 5′ non-P and/or 3′ non-OH ends.
Treated and untreated RNA samples were subjected to sequential 3′ and 5′ adapter ligations, followed by cDNA reverse transcription, PCR amplification, and Solexa sequencing (Hafner et al. 2008). The maximum sequencing read length was 36 nt for the short-size library and 100 nt for the medium- and long-size libraries. Since the 5′ adapter can only be ligated to RNAs having a 5′P and the 3′ adapter can only be ligated only to RNAs having a 3′OH, RNAs starting with 5′OH, 5′PP, or 5′PPP or ending with 2′P, 3′P, or <2′,3′P were expected to be absent or reduced in the untreated cDNA library.
Sequence read mapping to the T. thermophilus HB8 genome
Sequence reads were mapped to the T. thermophilus HB8 genome using the SolexaMatch algorithm (Supplemental Table S1; Yassour et al. 2009). The fraction of mapped reads (allowing up to two mismatches and/or insertions/deletions), ranged from 71% (untreated long library) to 93% (treated short library), and the fraction of reads that mapped uniquely to the genome ranged from 18% (untreated short library) to 46% (untreated long library). The fraction of reads that were mapped to CRISPR-annotated regions (annotations follow CRISPRdb) (Grissa et al. 2007) ranged from 1% (untreated long library) to 17% (treated long library) (Supplemental Table S1). Only 0.5% of the nonuniquely mapped reads corresponded to CRISPR regions, and they constituted 4.5% of all reads (uniquely and nonuniquely) mapped to CRISPR regions. A large proportion of reads corresponded to annotated rRNAs, tRNAs, and mRNAs; 92% of the nonuniquely mapped reads are annotated as rRNAs and originated from two copies of each of the T. thermophilus HB8 rRNA genes.
The fraction of CRISPR-annotated uniquely mapped reads was 8.9-, 27.2-, and 12.7-fold greater in the treated versus the untreated library for the short, medium, and long library, respectively. The CRISPR-annotated read category was the most enriched category in the treated versus untreated libraries irrespective of the length of the library (Fig. 3). Since the long treated library had the best crRNA read coverage (Supplemental Table S1), we focused on this library, unless otherwise specified. In addition, hereinafter, we only consider reads that were mapped uniquely to the genome.
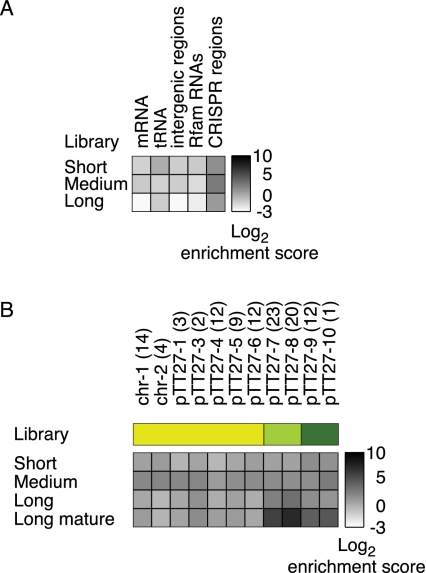
FIGURE 3.
Enrichment of different classes of RNAs in the treated libraries. (A) Global view on the enrichment of mRNA, tRNA, intergenic RNA, Rfam-annotated RNA, and CRISPR-derived RNA expression in the treated libraries. (B) Expanded view for each CRISPR region. Enrichment was calculated as the ratio between the fractions of uniquely mapped reads in the treated and untreated libraries for each gene category (displayed as log2 of the ratio). The rRNA category is omitted as it is not uniquely mapped because there are two copies of each rRNA gene. Number of spacers appears in parentheses. Although individual CRISPRs show enrichment variation both within and between libraries (short, medium, long), the CRISPR-annotated read category as a whole is the most enriched category in the treated versus untreated libraries.
As T. thermophilus HB8 encodes an Argonaute protein (Wang et al. 2008, 2009), we also searched for miRNA- or siRNA-like RNAs carrying 5′P and 3′OH ends that were expected to be enriched in the untreated short library (Supplemental Table S2). Although we identified over 20 unique reads with more than 100-fold enrichment, the read length was variable (17–36 nt), and they all corresponded to either tRNA or mRNA fragments, often representing their 5′ termini. Furthermore, we could not identify clusters of reads with uniform size distribution that were specifically enriched within the short but not the longer libraries (data not shown).
CRISPRs of T. thermophilus HB8 are constitutively expressed
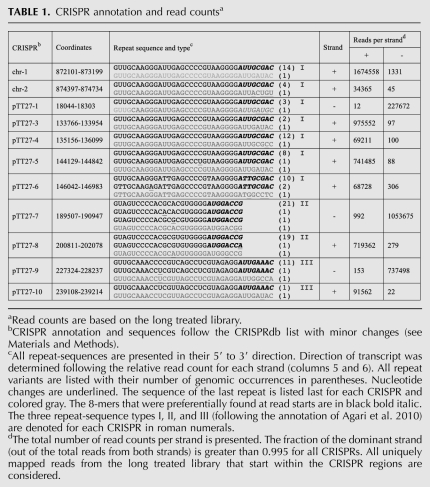
Most of the CRISPR loci listed in CRISPRdb were identified computationally (Grissa et al. 2007), and their actual expression was only demonstrated in a few bacteria. CRISPRdb lists 11 CRISPRs for T. thermophilus HB8: two on the chromosome (chr-1, chr-2) and nine on the plasmid pTT27 (pTT27-1, pTT27-3 to pTT27-10). The 11 CRISPRs can be classified into three repeat-sequence types, referred to as I, II, and III (Agari et al. 2010), as shown in Figure 1 and Table 1. We found that all T. thermophilus HB8 CRISPRs annotated in CRISPRdb are expressed (Table 1). A single-repeat once referred to as pTT27-2 (Agari et al. 2010) recovered less than 40 uniquely mapped reads (when counting also reads mapped over its flanking regions), which is a negligible number in respect to the average read count of all other CRISPRs. Furthermore it contains no spacer and does not represent an array as it is only a single-repeat. Therefore pTT27-2 was not included in further analysis. The majority of reads corresponding to the 11 CRISPR regions (>99%) were mapped unidirectionally (Table 1).
TABLE 1.
CRISPR annotation and read countsa
crRNA 5′ ends correspond to the 3′ terminal 8-nt segment of their preceding repeats
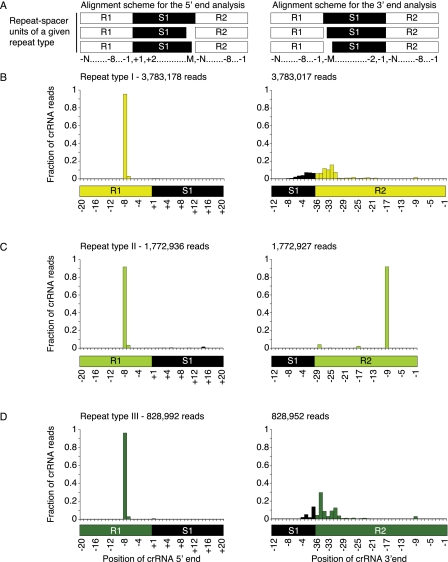
Analysis of the start positions of reads mapped to CRISPR regions revealed that they were not uniformly distributed along the CRISPR regions but preferentially mapped to the 3′ end region of the CRISPR repeats, 8 nt upstream to the spacer first position independent of specific crRNAs or the CRISPR repeat-sequence type. In accord with the study by Hale et al. (2009), there is also a small but detectable fraction of read starts mapped 7 nt upstream of the spacer (Fig. 4A–D, left panel; Supplemental Fig. S2A). There are three dominant 8-mer sequences present: AUUGCGAC, AUGGACCG, and AUUGAAAC, each corresponding to the 3′ terminal 8-nt segment of one of the three repeat-sequence types present in the T. thermophilus HB8 CRISPRs (Table 1; Supplemental Table S3). Additionally, a mutated 8-mer AUGGACCA repeat-sequence was also detected. This 8-mer corresponds to the repeat-sequence type II preceding the 12th spacer of pTT27-8, which has a G-to-A substitution. Other mutated 8-nt segments in T. thermophilus HB8 CRISPRs are only found in the last repeats of the CRISPR arrays, consistent with previous reports (Deveau et al. 2010; Touchon and Rocha 2010). Most point mutations in the last repeat reside within positions −4 to −2 (Table 1; Supplemental Table S3). Since we detected expression of the last crRNA for some of the CRISPR arrays and since we also detected reads starting in the 8-nt segment of the last repeat, it is possible that the full 8-nt segment is not required for the cleavage mechanism but is conserved for other reasons, such as assembly into Cas proteins (Haurwitz et al. 2010; Wang et al. 2011).
FIGURE 4.
Distribution of read start and end positions along the CRISPR regions. (A) A schematic of the repeat-spacer/spacer-repeat unit alignment. (Left) For the analysis of the start positions, the repeat-spacer unit is aligned using the spacer first position as an anchor. (Right) For the analysis of the end positions, the spacer-repeat unit is aligned using the spacer end as an anchor. (B–D) Start and end positions of reads by repeat-sequence type of CRISPRs. The fraction represents the relative frequency of start and end positions for each CRISPR repeat-sequence type, defined by dividing the number of reads at that position by all reads corresponding to that type. For clarity only a sub-range of the repeat-spacer/spacer-repeat unit is displayed. All omitted positions had negligible number of reads. (R) Repeat. (S) Spacer. The serial number (e.g., R1, S1, R2) denotes the relative order of the CRISPR elements. The number of reads in the analysis is indicated at the top left corner.
Different 3′ end cleavage patterns of crRNAs
Sequence analysis demonstrated that more than 99% of the mapped reads in the various libraries included the 3′ adapter (Supplemental Table S1) and that we identified the actual 3′ ends of nearly all mapped sequenced RNA fragments. Based on the analysis of all the uniquely mapped reads, we observed two major 3′ end processing patterns, which coincided with the different CRISPR repeat-sequence types (Fig. 4B–D, right panel; Supplemental Fig. S2B). The first pattern was characteristic for repeat-sequence type II CRISPRs and showed read ends that predominantly mapped to position −9 of the repeats, perfectly matching the cleavage at position −8 of the immediate downstream crRNA. In contrast, the majority of reads for repeat-sequence type I and III showed variable and truncated 3′ ends, indicative of further 3′ end processing.
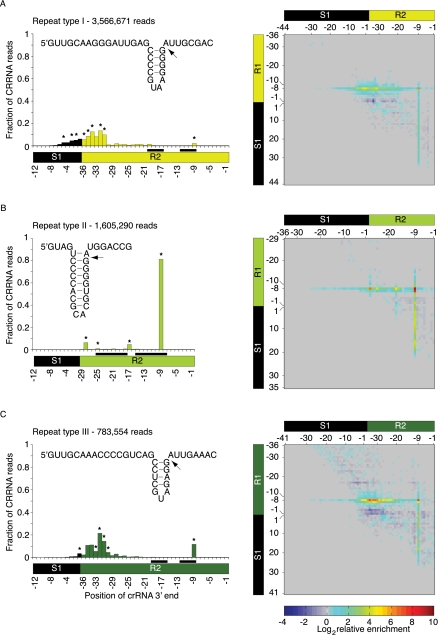
To define the 3′ end boundaries of the mature crRNAs we performed a repeat-sequence-type–specific analysis using only crRNAs with 5′ ends initiating at position −8 upstream to the spacer start (−8 reads). All −8 reads of the same CRISPR repeat-sequence type were aligned by their spacer-repeat boundary to compensate for different spacer lengths, and the average fractions of crRNAs ending at each position were plotted (Fig. 5, left panel). Most repeat-sequence type II reads end at the −9 positions of the immediate downstream repeat. The signal for reads terminating at the −9 position was independent of the spacer length of the individual crRNAs, indicative of a primary processing site located between positions −9 and −8 of the repeats (Fig. 5B, left panel). Two additional weak 3′ end signals, both independent of the spacer length, were observed at the repeat positions −16 and −28, within regions predicted to be single-stranded, and likely result from hydrolysis rather than nucleolytic cleavage. The 3′ end of repeat-sequence type I and III mature crRNAs is less well defined. A small fraction of reads shows a sharp −9 position processing event, but the majority of reads were truncated by another 20–30 nt from the −9 position (Fig. 5A,C, left panel).
FIGURE 5.
Boundaries of the mature crRNAs can be revealed from the deep-sequencing data. (A–C, left) Average fraction of reads ending at each position of the aligned CRISPR units (for the alignment scheme and the numbering of the aligned positions, see Materials and Methods; Fig. 4A, right). All reads start at the −8 of their respective repeat, annotated as R1. Positions to which a negligible number of read ends map are not displayed. (Inset) Predicted secondary structure models of the three repeat-sequence types. Models were built by identifying the longest and most stable contiguously base-paired region and supporting it by nucleotide covariation that maintains the putative stem (Supplemental Fig. S3). (Arrow) Cleavage site (−8). In repeat-sequence type II, the long stem–loop involves pairing of the cleavage site. Interestingly, this base pair was found to be less stable when complexed with T. Thermophilus HB8 Cse3 (Sashital et al. 2011). Positions predicted to base pair in the secondary structure models (insets) are marked by a pair of black rectangles on the spacer-repeat unit scheme. The number of reads in the analysis is indicated at the top left corner. (A–C, right). Heatmap representation of the average enrichment scores (log2 values) of all the reads starting at the position denoted by the y-axis and ending at the position denoted by the x-axis. Position annotation follows the Figure 4B description except for the spacer start position in the y-axis that is numbered from 1 to N and not from −N to −1. Highly enriched end positions starting at R1–8 (enrichment score is among the top 10 scores of all end positions for each type) are marked in black asterisks in the corresponding left panel graph. For all three repeat-sequence types, the highest average enrichment score was found for reads starting at R1–8 and ending at the R2–9 positions, where the enrichment score of repeat-sequence type II>type III>type I (9.27, 7.36, and 6.48, respectively). High enrichment scores were also observed for fragments starting at −7 positions, predominantly in CRISPRs of repeat-sequence type II fragments starting at R1–7 and ending at R2–9.
crRNAs have 5′OH and/or 3′ non-OH termini
The Cas endonuclease responsible for the −8 processing step of crRNA was shown to generate RNA fragments with 5′OH and <2′,3′P termini (Carte et al. 2008; Haurwitz et al. 2010; Jore et al. 2011b). We computed an enrichment score for crRNA-characteristic 5′ and 3′ ends from the ratio between the frequencies of reads in the treated and untreated libraries. All reads were ranked according to their enrichment scores. Consistent with the relative high abundance of CRISPR-derived reads in the treated compared to untreated library described above, we found that 54% (257/476) of the most enriched reads (enrichment score ≥ 200) are indeed located within CRISPR-annotated regions. Seventy-five percent of the top-ranked annotated CRISPR regions corresponded to CRISPRs pTT27-7, 8, and 9. Other highly enriched reads were annotated as tRNAs (38%, 179/476), protein-coding mRNAs (4%, 19/476), and other ncRNAs (2%, 10/476: tmRNA, seven; RNaseP, two; SRP_bact, one), and 2% (11/476) were mapped to unannotated regions (Supplemental Table S4).
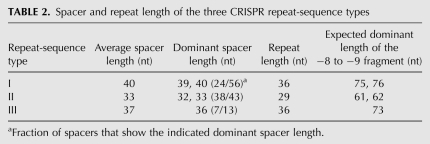
To better characterize the enriched reads that were mapped to CRISPR regions, we determined the relative start/end positions of each read within the corresponding CRISPR element (repeat or spacer) to which it was mapped. For this analysis, we used the following terminology: The CRISPR repeat and spacer are denoted by R and S, respectively. A repeat and its downstream spacer are assigned the same serial number i, defining a Ri-Si unit. Taking into account the sizes of the shortest spacer (31 nt) and repeat (29 nt) (Table 2; Supplemental Table S5) and the maximal read length (100 nt), a read could theoretically span, at most, two Ri-Si units. While there were reads spanning Si-R(i+2), their fraction was rather small (0.04%). Most reads were of the forms Ri-R(i+1) (70%), Ri-Si (17%), and Si-R(i+1) (10%) (Supplemental Table S6). Reads of these forms along with Si-Si were further analyzed.
TABLE 2.
Spacer and repeat length of the three CRISPR repeat-sequence types
For each of the three CRISPR repeat-sequence types, we computed separate average enrichment scores. The values were plotted in an N × N matrix, where N is the repeat length + maximal spacer length of the analyzed repeat-sequence type (Fig. 5A–C, right panel). Positions with high average enrichment scores are indicative of transcripts that have a 5′ non-P and/or 3′ non-OH ends. The highest enrichment scores are obtained for fragments starting at position −8 of Ri (regardless of their end position) or ending at positions −9 of R(i+1) (regardless of their start positions). This implies that the cleavage between the −8/−9 positions generated two products: one carrying a non-OH group at its 3′ end and the other carrying a non-P at its 5′ end. The 5′ non-P end could be 5′PPP, 5′PP, or 5′OH. 5′PPP and 5′PP (a hydrolysis product of 5′PPP) are found in bacteria only at transcript start sites and cannot be products of endonucleolytic cleavage, leaving 5′OH termini as the most plausible interpretation. The highest average enrichment score for each CRISPR type was detected for reads starting at the −8 position of the Ri repeat and ending at position −9 of the successive R(i+1) repeat, suggesting that these −8 to −9 fragments carry both 5′OH and 3′ non-OH ends. The enrichment score of the −8 to −9 fragments was the highest for repeat-sequence type II, supporting the conjecture that the mature repeat-sequence type II crRNAs retain their 5′OH and 3′ non-OH ends. For repeat-sequence type I and III the score was lower (especially for repeat-sequence type I), and an additional cluster of enriched positions starting at the −8 position of the R1 repeat and ending at the start positions of the R2 repeat could be observed (Fig. 5A,C). The high enrichment score of these shorter fragments most likely indicates a secondary processing by an additional nuclease, resulting in 5′OH and/or 3′ non-OH fragments.
Assuming 5′OH and/or 3′ non-OH crRNAs ends, we would not expect to find mature crRNA reads in the untreated libraries. In practice, a larger fraction than expected was found in the long untreated library, although in all cases it was substantially (greater than 50-fold) smaller than in the treated library (data not shown). This result can be attributed to potential RNA turnover processes or competing enzymatic activities that involve RNA dephosphorylation and phosphorylation events (Condon 2007).
Comparative Northern blot analyses of T. thermophilus HB8 crRNA expression
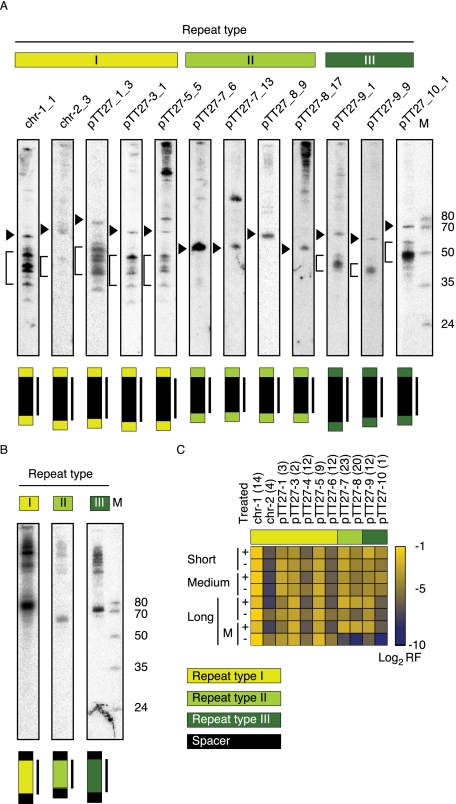
To probe for crRNA fragments longer than the size range selected for cloning, we applied Northern blotting analysis using probes either specific for the spacer elements (five, four, and three probes for spacers associated with repeat-sequence type I, II, III, respectively) (Fig. 6A) or specific for repeat elements of each of the three CRISPR repeat-sequence types (Fig. 6B). For all spacers, a strong signal for the −8 to −9 precursor fragment could be detected: 70–80 nt long for repeat-sequence type I and III and ∼60 nt long for repeat-sequence type II spacers as expected by their different spacer and repeat lengths (Fig. 6A; Supplemental Table S5). For type II, no other shorter dominant band was detected, consistent with the observation that the −8 to −9 precursor comprises the mature crRNAs for this type. For repeat-sequence types I and III, at least one stronger signal was detected for a ∼50-nt fragment, consistent with additional cleavage of the −8 to −9 fragment, yielding a shorter mature crRNA. Furthermore, for the majority of the probed repeat-sequence type I and III spacers, we observed additional bands within the 35–50 nt range, consistent with the deep sequencing analysis, where we found multiple enriched short fragments in the range of the spacer 3′ end ±6/7 positions (Fig. 5). The additional bands were sometimes with equal intensity to the dominant band (e.g., chr-1_1 and pTT27-3_1) and sometimes with weaker intensity. Probing for the repeat region revealed strong signals for the pre-crRNA transcript, weaker signals for the processing intermediates, and a strong signal for the −8 to −9 fragment (Fig. 6B). No signals shorter than the −8 to −9 fragment could be detected.
FIGURE 6.
Northern blot analyses of CRISPR spacer and repeat-sequences. Northern blots were performed with 5 μg total RNA using oligodeoxynucleotide probes designed to detect the CRISPR spacer (A) or repeat (B) sequences. The region probed against is indicated above the lane. The −8 to −9 precursor sequence is indicated by an arrowhead on the left side of each spacer panel. For type II, no other dominant band is detected, consistent with the observation that the −8 to −9 precursor comprised the mature crRNA for this type. For repeat-sequence type I and III, a bar is depicted along the mature crRNA bands. Total RNA was loaded next to a size marker cocktail (M). The sizes are indicated next to the gel. For sequences, see the Supplemental Materials and Methods. (C) Expression profile of CRISPRs in the treated (+) and untreated (−) libraries. The heatmap represents the log2 of the relative frequency of reads that were mapped to a specific CRISPR out of the total number of CRISPR reads for each corresponding library. For the long library, the values of the mature form (M) of the crRNA is also displayed. For the determination of reads corresponding to mature crRNA reads, see Materials and Methods. For an extended view of crRNA precursor and mature expression, see Supplemental Figure S4B. Interestingly, we observe a common consistent expression pattern for the three paralogous pairs of adjacent CRISPRs: chr-1:chr-2, pTT27-3:pTT27-4, and pTT27-5: pTT27-6, where the upstream CRISPR in each pair has a much higher expression level compared with the downstream CRISPR.
Previous studies showed that the whole CRISPR array is transcribed as one primary transcript from the leader to the last repeat (for review, see Marraffini and Sontheimer 2010). Therefore, one might expect similar read frequencies for cistronic members. However, we observed that crRNAs originating from the same CRISPR locus were sequenced with varying sequence read counts (Supplemental Fig. 4A,B; Supplemental Table S7). For example, the read counts across the 20 mature crRNAs derived from CRISPR pTT27-8 varied almost 400-fold. To assess whether biases in the library preparation protocol or differences in crRNA stability or processing contributed to this variation, we selected three pairs of crRNAs originating from the same CRISPR and probed those by Northern analysis: pTT27-7_6 and pTT27-7_13; pTT27-9_1 and pTT27-9_9; and pTT27-8_17 and pTT27-8_9. The differences in their estimated expression level by their mature crRNA read counts are 34.5-, 7.6- and 1.4-fold, respectively. In accord, the dominant band intensity for the three pairs is pTT27-7_6 ≪ pTT27-7_13; pTT27-9_1 < pTT27-9_9; and pTT27-8_17 ≃ pTT27-8. This confirmed that read count is related to RNA abundance, supporting the deduction of the relative expression levels of the CRISPRs and the crRNAs from the read counts and suggesting that indeed crRNAs differ in their stability or processing.
The pre-crRNA, as well as the estimated crRNA, expression is consistent across the three treated libraries (short, medium, and long) (Fig. 6C; Supplemental Fig. S4B) with correlation coefficients >0.75 between library counts. While the processing mechanism pattern could only be fully revealed by the long library, the read fraction consistency across libraries indicated that the short libraries reads still serve as a good approximation of mature crRNA abundance.
DISCUSSION
CRISPR-based immunity depends on the expression and maturation of crRNAs and their assembly into functional active complexes with Cas proteins (Al-Attar et al. 2011; Makarova et al. 2011b; Terns and Terns 2011). We analyzed T. thermophilus HB8 crRNA biogenesis and expression by differential deep sequencing with read length of up to 100 nt. Previously, microarray profiling reported the expression of 10 of 11 candidate T. thermophilus HB8 CRISPRs upon phage infection (Agari et al. 2010). Our deep sequencing approach used uninfected bacteria cultured in our laboratory and demonstrated constitutive expression of all CRISPRs, including the 11th CRISPR that was not interrogated by the array. Furthermore, we obtained reads from each spacer-repeat unit confirming their biogenesis into mature crRNAs. Constitutive CRISPR expression is consistent with the proposed role of crRNAs acting as an early response system to previously encountered phage infections (Deveau et al. 2010).
One of the advantages of RNA cDNA library sequencing over array analysis is the precise definition of the ends of the RNA molecule. Furthermore, methods for cDNA library preparation can discriminate between various 5′ and 3′ end modifications of the input RNA. By using this approach, we found that over 90% of the long treated library reads that uniquely mapped to the CRISPR region showed homogenous 5′ ends starting in the repeat region, precisely 8 nt upstream of the respective spacer, independent of the repeat-sequence type. In contrast, the 3′ ends of crRNAs varied by repeat-sequence type. While over 90% of the reads corresponding to repeat-sequence type II crRNAs terminated at the anticipated downstream −9 position, only 2% of the reads corresponding to repeat-sequence type I and III crRNAs terminated at the −9 positions, and >87% were 21- to 33-nt shorter with substantial 3′ heterogeneity. Differential processing of repeat-sequence type I and III, but not of type II, crRNAs suggests that at least two independent crRNA biogenesis mechanisms are active in T. thermophilus HB8.
The three CRISPR types differ not only in their repeat-sequence but also in the average length of their spacers with type II ≪ type III < type I (Table 2; Supplemental Table S5). Apparently, the length of the repeat-sequence type II primary fragments (from −8 to −9) is on average shorter by >10 nt compared with repeat-sequences types I and III due to their shorter spacers. The repeat-sequence type II primary fragment length resembles that of E. coli crRNAs, as the lengths of both their repeat and dominant spacers are similar (29 and 31–32 nt, respectively). Furthermore, the T. thermophilus HB8 Cas proteins encoded adjacent to repeat-sequence type II CRISPRs resemble those of E. coli. Consistently, the biogenesis of CRISPRs of repeat-sequence type II with a single −9/−8 cleavage resembles that of E. coli (Jore et al. 2011b). It is also interesting to note that the entire CRISPR-Cas module of the type II repeat of T. thermophilus HB8 is absent from its close relative T. thermophilus HB27, further supporting its independent biogenesis mechanism promoted by its module-specific Cas proteins. In turn, the fact that T. thermophilus HB27 encodes CRISPRs with repeat-sequence types I/III strongly supports the existence of an additional processing mechanism.
The biogenesis of repeat-sequence type I and III crRNAs is not as well characterized as that of repeat-sequence type II. The repeat-sequences of the stem of type I and III CRISPRs in T. thermophilus are similar with a single base-pair covariation, suggesting that they may share processing by the same Cas protein. The best candidate endonuclease implicated in type I and III crRNA biogenesis in T. thermophilus is Cas6, encoded adjacent to the repeat-sequence type III CRISPRs. Cas6 was shown to endonucleolitcally cleave CRISPRs in P. furiosus and is a core Cas protein commonly found in organisms with Tneap, Hmari, Aperm, and Mtube CRISPR-Cas modules (Wang et al. 2011), the modules that are found also in T. thermophilus HB8 (Fig. 1). Cas6 is conserved between HB8 and HB27 as well as in several other bacteria containing CRISPRs with repeat-sequence type I or III (Supplemental Table 8). Furthermore, the two most-conserved Cas proteins among all these bacteria are Cas1, which is a universal Cas protein (Makarova et al. 2011a), and Cas6 (Supplemental Table S8). Taken together, it is conceivable that Cas6 plays a role in the processing of repeat-sequence type I and III CRISPRs.
The 3′ ends of repeat-sequence type II crRNAs are likely protected from 3′ end trimming by a complex similar to the CASCADE complex present in E. coli (Jore et al. 2011b), which binds very tightly to both ends and also covers the body of the crRNA, making it inaccessible to nucleases after the initial cut between −9 and −8 by CasE (ortholog of HB8 Cas6e (Cse3), 31% identity, absent from HB27). In order to facilitate 3′ end trimming of repeat-sequence type I and III crRNAs, their 3′ ends must be accessible to either endo- and/or exonucleases. It is conceivable that the 4-bp stem–loop of the type I and III repeat is less tightly bound by its corresponding Cas6 endonuclease compared to the 8-bp stem–loop of the type II repeat and its cognate Cas6e (Cse3) protein, thereby facilitating access to 3′ exonucleases. Potential protein binding to the crRNA 5′ end can eventually block the exonucleolytic trimming of the 3′ end. This model is supported by recent findings in P. furiosus showing that Cas6 interacts with positions at the 5′ end of the CRISPR repeat (Wang et al. 2011). In particular, four repeat positions 3, 5, 6, and 8, which are conserved in all P. furiosus CRISPRs, were found to be in contact with the Cas6 proteins. Interestingly, three of these positions (3, 5, 6) are also conserved between P. furiosus and T. thermophilus HB8 and HB27 as well as in several other bacteria containing CRISPR with repeat-sequence types I or III (Supplemental Table S8; Supplemental Fig. 3). Of note, additional exonucleolytic cleavage was also suggested for the additional trimming observed for crRNAs in P. furiosus (Hale et al. 2008). Considering, however, the possibility of an endonucleolytic processing step cutting upstream of the 4-bp stem, we searched in the short treated library for multimapping sequence reads (data not shown). While we found reads corresponding to the region from the crRNA 3′ end to the successive −9 position, the number of these reads was very small. Thus, this finding can neither refute nor support an additional endonuclease mechanism.
One of the initial focuses of this study was the search for 5′ phosphorylated small RNAs performing si/miRNA-like function in T. thermophilus, given that it expresses an Ago protein. Since miRNAs and siRNAs in eukaryotic species are found constitutively expressed (although with some cell-type specificity), their complete absence, despite the expression of Ago, was somewhat surprising. It is conceivable that only when challenged with pathogens, the function of bacterial Ago proteins may be revealed.
In summary, our RNA sequencing approach revealed the constitutive expression of all the predicted T. thermophilus HB8 CRISPRs and provided new insights into different crRNA processing mechanisms associated with the repeat type. crRNAs of repeat-sequence type II retained their original 3′ end while those of type I and III underwent additional 3′ end processing. The enrichment of reads starting at the repeat −8 positions and ending at the −9 position of the successive repeat in the treated versus untreated library indicated that the primary processing intermediates and mature crRNAs have 5′ non-P and 3′ non-OH termini (Carte et al. 2008; Haurwitz et al. 2010). Taken together, our results strongly support the generality of the CRISPR cleavage mechanism across different species and suggest that various CRISPR-Cas modules containing different repeat-sequence types are operational within a single bacterial strain.
MATERIALS AND METHODS
Growth of bacteria and small RNA library preparation
T. thermophilus HB8 cells were grown in LB media supplemented with 1 mM magnesium chloride and 1 mM calcium chloride shaking at 220 rpm at 75°C. At an OD600 of 0.7, cells were collected by centrifugation (6000g, 10 min). Total RNA was prepared as described previously (Chomczynski and Sacchi 1987) with modifications described (Pfeffer et al. 2005). After gel fractionation of the small RNAs from 100 μg total RNA, the sample was split. One half was treated with alkaline phosphatase, and subsequently, a 5′P was added using T4 polynucleotide kinase and ATP. Both, treated and untreated small RNAs were subjected to adapter ligation, cDNA preparation, and Solexa sequencing as previously described (Hafner et al. 2008). For details, see the Supplemental Materials and Methods. Sequencing data were deposited at the Gene Expression Omnibus database repository under accession no. GSE34328.
Northern analysis
Northern analyses were performed as previously described (Hale et al. 2008). Briefly, 5 μg total RNA was separated on a 15% denaturating gel and probed with radiolabeled oligodeoxynucleotide anti-sense probes against various spacer and repeat regions. Hybridized blots were visualized using a PhosphorImager. For details, see the Supplemental Materials and Methods.
Genome mapping of the reads
T. thermophilus HB8 whole-genome sequence was extracted from the NCBI bacterial genome database (http://www.homd.org/download/NCBI_bacteria_genome/). The genome contains one circular chromosome (NC_006461, 1,849,742 nt) and two circular plasmids (pTT27 NC_006462, 256,992 nt; pTT8 NC_006463, 9322 nt). For annotation of the mapped reads, we followed the NCBI annotation. For the rRNA genes, we expanded slightly the genes' boundaries based on the reads mapped to the region. CRISPR annotation and sequences followed CRISPRdb (Grissa et al. 2007), with the exception of pTT27-9 and pTT27-10, for which our analysis strongly supports small modifications of the repeat definition. For pTT27-9 we redefined the first position of the repeats as the last position of the respective spacers. pTT27-10 repeats were extended by three additional nucleotides at their 3′ end, and the respective spacer was shortened accordingly. Also, for three CRISPRs (chr-1, pTT27-3, and PTT27-5) the last coordinate of the CRISPR differs in one position from the coordinate listed in the CRISPRdb. There is no pTT27-2 in CRISPRdb.
Reads were mapped to the T. thermophilus HB8 genome using the in house program SolexaMatch, which implements a mapping algorithm reported previously (Yassour et al. 2009). The SolexaMatch algorithm identified reads that match the genome from read start to read end (termed here full length reads) and reads that partially match the genome but also match the corresponding 5′ or 3′ adapter sequences (termed A5 or A3 reads, respectively). For details on the algorithm parameters, see the Supplemental Materials and Methods. As expected for Illumina RNA sequencing, the fraction of A5 reads in all libraries is very small (at most 0.7% of all uniquely mapped reads) (Supplemental Table S1).
Annotation of read positions within the CRISPR units
For the unified annotation of the read positions along the CRISPR units, we use the following terminology: The CRISPR repeat and spacer are denoted by R and S, respectively. The repeat and spacer lengths are denoted by N and M. Within the repeat, the positions are numbered from –N to −1 (from the repeat 5′ end to the position preceding the spacer). Positions within the spacer are similarly numbered from –M to −1 for analyses of read ends, and from 1 to M for analyses of the read starts. A repeat and its downstream spacer have the same serial number Ri-Si. The first repeat and spacer of a CRISPR are determined as R1 and S1, and subsequent repeat and spacer units are assigned sequential serial numbers accordingly. Note that for analyses involving alignment of crRNAs, the original positioning of reads along the CRISPR is ignored, and reads matching corresponding positions in different repeat/spacer units of the same type are aligned.
Analysis of the distribution of the 5′ and 3′ end positions of reads per CRISPR repeat-sequence type
For the analysis of the distribution of the 5′ and 3′ end positions within the same repeat-sequence type CRISPR, we aligned repeat-spacer/spacer-repeat units of the same repeat-sequence type, using the start or end of the spacer as an anchor, respectively (Fig. 4A,B). The fraction of reads was estimated by dividing the total number of reads for each aligned position within a repeat-sequence type by the total number of reads for that repeat-sequence type. Note the number of aligned sequences for most of the positions would be equal to the number of spacers in the specific repeat-sequence type. However, as spacers within a type have different lengths, the number of aligned sequences at the “edge” of the spacer alignment (i.e., end spacer–aligned position for start analysis and start spacer–aligned position for end analysis) would have less aligned sequences.
Analysis of the end positions of reads that start at the −8 positions per CRISPR repeat-sequence type
For each of the CRISPR spacers (Si), we defined the genomic coordinate of the −8 position within the preceding repeat (Ri). For each of these genomic positions, we applied the following procedure: (1) all uniquely mapped reads (A3, A5, and full length) starting at the analyzed −8 genomic position were grouped, and their total count was summed up (denoted hereafter as K). Note that although all reads start at the same −8 position, they can differ in length and consequently in their end position coordinate. (2) Each position of each read end was annotated by its relative placement along the CRISPR units as described above. (3) For each of the annotated positions, the fraction of reads ending at the same annotated position i (denoted hereafter as fi(end)) was calculated as the sum of the counts of all reads ending at that position divided by K.
To examine the relative counts of the end positions of all spacers, we applied the following steps: (1) all annotated reads were divided into three groups according to the CRISPR repeat-sequence type (I, II, or III); (2) all reads in a group were aligned using the spacer end position as a reference point; and (3) each annotated end position was denoted in reference to the spacer end, and its average fraction was calculated as ((sum of fi(end) over all the aligned annotated reads)/number of aligned spacers). To avoid high signals from positions shared by only few spacers, we divided all aligned positions by the same total number of aligned spacers.
Read enrichment score
The enrichment score of a read was calculated as the ratio between its fraction in the treated and untreated libraries. For this analysis, same-length reads that were mapped to the same genomic locations were grouped. The fractions were computed out of the total number of uniquely mapped reads in each library (two strands of the chromosome and the two plasmids). The enrichment scores described in the text were derived from the analysis of all uniquely mapped reads from the long library. For the ranking of the reads by their enrichment scores, we only considered the subset of genomic positions with at least one uniquely mapped read in at least one of the two compared libraries. In the computations of ratios, zero counts were treated as counts of 1. The average enrichment score of the grouped read fragments (Fig. 5A–C, right panel) was calculated separately for each CRISPR repeat-sequence type. The average was calculated over the total number of spacers in each repeat-sequence type. Zero values were assigned if no related fragment was observed in both treated and untreated libraries.
Expression of mature crRNAs
To estimate the expression level of the individual repeat-sequence type II crRNAs, we strictly counted reads that spanned the whole region starting at R1–8 position and ending at R2–9 position. The crRNAs of repeat-sequence type I and III were less well defined. To estimate their level, we summed over all reads that were mapped to start at R1–8 position and end at one of the last six spacer's positions or the first six positions of the successive repeat. For all counts, we used the long treated library, considering all uniquely mapped reads.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank W. Zhang, S. Dewell, and C. Zhao at the Rockefeller University Genomics Resource Center for SOLEXA sequencing. We thank A. Shinkai for sharing his data and helpful advice, Kira Makarova for her help in annotating T. thermophilus HB8 Cas proteins, L. Marraffini for useful comments on the manuscript, J. Belasco for helpful advice, and S. Altuvia, I. Rosenshine, and N. Jarrous for valuable discussions. We thank M. Yassour and T. Kaplan for their help in the initial steps of the analysis and Guy Eyal for technical assistance. This study was supported by the US-Israel Bi-national Science Foundation (H.M. and T.T.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031468.111.
REFERENCES
- Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol 395: 270–281 [DOI] [PubMed] [Google Scholar]
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 392: 277–289 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Condon C 2007. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol 10: 271–278 [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64: 475–493 [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172 doi: 10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60 doi: 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP 2008. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14: 2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329: 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Jore MM, Brouns SJJ, van der Oost J 2011a. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. 2011b. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol 8: R61 doi: 10.1186/gb-2007-8-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV 2011a. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 6: 38 doi: 10.1186/1745-6150-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011b. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Lagos-Quintana M, Tuschl T 2005. Cloning of small RNA molecules. Curr Protoc Mol Biol 26: 26.4.1–26.4.18 [DOI] [PubMed] [Google Scholar]
- Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC 2011. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol 8: 517–528 [DOI] [PubMed] [Google Scholar]
- Sashital DG, Jinek M, Doudna JA 2011. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 18: 680–687 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P 2008. CRISPR: a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6: 181–186 [DOI] [PubMed] [Google Scholar]
- Sturino JM, Klaenhammer TR 2004. Bacteriophage defense systems and strategies for lactic acid bacteria. Adv Appl Microbiol 56: 331–378 [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM 2011. CRISPR-based adaptive immune systems. Curr Opin Microbiol 14: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Rocha EP 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS ONE 5: e11126 doi: 10.1371/journal.pone.0011126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34: 401–407 [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ 2008. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Preamplume G, Terns MP, Terns RM, Li H 2011. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19: 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. 2009. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci 106: 3264–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]