The transient existence of small RNAs that are not associated with the RNA chaperone Hfq is part of the normal dynamic lifecycle of a sRNA. Small RNAs are extremely labile when not associated with Hfq, but the mechanism by which Hfq stabilizes sRNAs was still elusive. Here, the authors have found that polynucleotide phosphorylase (PNPase) is the major factor involved in the rapid degradation of small RNAs, especially those that are Hfq free. Overall, the data highlight the impact of 3′-exonucleolytic RNA decay pathways and re-evaluate the degradation mechanisms of Hfq-free small RNAs.
Keywords: Hfq, MicA, PNPase, small noncoding RNAs, RNase E
Abstract
The transient existence of small RNAs free of binding to the RNA chaperone Hfq is part of the normal dynamic lifecycle of a sRNA. Small RNAs are extremely labile when not associated with Hfq, but the mechanism by which Hfq stabilizes sRNAs has been elusive. In this work we have found that polynucleotide phosphorylase (PNPase) is the major factor involved in the rapid degradation of small RNAs, especially those that are free of binding to Hfq. The levels of MicA, GlmY, RyhB, and SgrS RNAs are drastically increased upon PNPase inactivation in Hfq− cells. In the absence of Hfq, all sRNAs are slightly shorter than their full-length species as result of 3′-end trimming. We show that the turnover of Hfq-free small RNAs is growth-phase regulated, and that PNPase activity is particularly important in stationary phase. Indeed, PNPase makes a greater contribution than RNase E, which is commonly believed to be the main enzyme in the decay of small RNAs. Lack of poly(A) polymerase I (PAP I) is also found to affect the rapid degradation of Hfq-free small RNAs, although to a lesser extent. Our data also suggest that when the sRNA is not associated with Hfq, the degradation occurs mainly in a target-independent pathway in which RNase III has a reduced impact. This work demonstrated that small RNAs free of Hfq binding are preferably degraded by PNPase. Overall, our data highlight the impact of 3′-exonucleolytic RNA decay pathways and re-evaluates the degradation mechanisms of Hfq-free small RNAs.
INTRODUCTION
The bacterial Hfq is a member of the Sm/Lsm superfamily of proteins involved in RNA metabolism (Wilusz and Wilusz 2005). It is a global regulator of cell physiology with particular impact on stress responses and affects the virulence traits of many pathogens (Tsui et al. 1994; Chao and Vogel 2010). Hfq plays a relevant role as a mediator of small noncoding RNA−mRNA interactions (Valentin-Hansen et al. 2004; Waters and Storz 2009). Base-pairing of small RNAs with their target mRNAs can alter mRNA translation and/or stability. The majority of small RNAs act as inhibitors of translation, usually triggering mRNA decay, although some other sRNAs act as positive regulators (Massé et al. 2003; Vecerek et al. 2007; Soper et al. 2010).
Hfq forms a stable hexamer with a ring-shaped structure displaying two distinct RNA-binding surfaces (Brennan and Link 2007). Biochemical and structural data support that the Hfq hexamer can bind simultaneously the sRNA on its proximal face and mRNA on its distal face, increasing the probability of RNA–RNA interactions in order to form a heteroduplex (Schumacher et al. 2002; Lease and Woodson 2004; Mikulecky et al. 2004; Link et al. 2009). In agreement, it has been suggested that the Hfq hexamer forms a ternary complex with oligo A18 and the small RNA DsrADII with a stoichiometry of 1:1:1 (Updegrove et al. 2011). Hfq can also work as a RNA chaperone and induce structural rearrangement of the RNA molecules to enable the contact between the two partner RNAs (Moll et al. 2003b; Geissmann and Touati 2004; Afonyushkin et al. 2005; Arluison et al. 2007).
Bacterial small RNAs that act as repressors bind at or near the ribosome binding site (RBS) of the target mRNA blocking its translation (Morita et al. 2006; Bouvier et al. 2008). Most of the time, this promotes cleavages in the mRNA, not only on the vicinity of the duplex (as happens with ompA mRNA/MicA) (Udekwu et al. 2005), but also downstream into the coding region (as reported for sodB mRNA/RyhB) (Prévost et al. 2011). Ribonuclease (RNase) III is an important endonuclease in the degradation of sRNA coupled to their target mRNAs (Afonyushkin et al. 2005; Deltcheva et al. 2011). However, other sRNAs mediate the destabilization of the target mRNA in an RNase E-dependent manner (Massé et al. 2003; Afonyushkin et al. 2005; Morita et al. 2005; Udekwu et al. 2005). RNase E is a single-stranded RNA endonuclease involved in mRNA decay in Escherichia coli (Arraiano et al. 2010). Hfq can associate with RNase E and sRNA in ribonucleoprotein complexes that are thought to make the degradation of target mRNAs more efficient (Aiba 2007).
Hfq is also found to interact with other proteins involved in mRNA decay. One of these proteins is the poly(A) polymerase I (PAP I), responsible for the majority of polyadenylation in E. coli cells (Régnier and Hajnsdorf 2009). Hfq is suggested to regulate polyadenylation by stimulating PAP I activity on mRNA (Hajnsdorf and Régnier 2000; Folichon et al. 2005). In the absence of Hfq, the poly(A) levels are reduced and the poly(A) tails are suggested to become smaller (Le Derout et al. 2003; Mohanty et al. 2004). Hfq was also shown to interact with the polynucleotide phosphorylase (PNPase) (Mohanty et al. 2004), a major 3′–5′ exonuclease involved in RNA degradation (Andrade et al. 2009b). PNPase responds to environmental stimuli, and its activity is modulated by metabolites such as ATP, citrate, and cyclic di-GMP (Del Favero et al. 2008; Nurmohamed et al. 2011; Tuckerman et al. 2011). We have previously shown that PNPase is a key factor in the turnover of small RNAs controlling the expression of outer membrane proteins in the stationary phase of growth (Andrade and Arraiano 2008). It was recently suggested that PNPase can also have a protector role for some sRNAs in exponentially growing cells (De Lay and Gottesman 2011). However, the details of the interplay between PNPase and Hfq in the function of sRNA are still not clear.
The interaction of Hfq with small RNAs is dynamic. Small RNAs compete for access to Hfq, and it was shown that the expression of unrelated sRNAs can dissociate Hfq–sRNA complexes already formed (Fender et al. 2010; Hussein and Lim 2011). The transient existence of small RNAs free from Hfq binding is thus part of the normal dynamic lifecycle of a sRNA. In addition, variations in the Hfq expression levels or in the availability of the free pool of Hfq can result in the reduction of Hfq–sRNA complexes. A small RNA that is not associated with Hfq is rapidly degraded, although the mechanism by which Hfq stabilizes small RNAs is not yet fully understood. RNase E was considered to be responsible for the rapid degradation of the small RNAs and was shown to compete with Hfq for access the same RNA sequences (Massé et al. 2003; Moll et al. 2003a). However, the activity of RNase E may not be as generalized; for example, RNase E was not found to be important for the in vivo degradation of OxyS upon Hfq inactivation (Basineni et al. 2009).
In this work we have characterized the degradation of small RNAs that are unassociated with Hfq. We have constructed multiple hfq mutants defective in RNases or in the poly(A) polymerase, and studied the impact of these factors in the expression of several small RNAs. We have shown that small RNAs in their Hfq-free state are rapidly degraded by PNPase, particularly in the stationary phase of growth. Moreover, PNPase was found to be more relevant than RNase E or RNase III in the degradation of small RNAs when these were not associated to Hfq. PNPase-mediated degradation of small RNAs is also found to be an active regulatory pathway in the cells expressing Hfq. Together, results show that PNPase has a predominant role in the degradation of Hfq-free small RNAs.
RESULTS
Inactivation of PNPase markedly increases the levels of small RNAs not associated with Hfq
It is commonly believed that the RNA chaperone Hfq protects regulatory RNAs from RNase E endonucleolytic cleavages (Massé et al. 2003; Moll et al. 2003a). Nevertheless, we have previously showed that 3′–5′ exonucleolytic activity can be a determinant for the degradation of small RNAs, even in the presence of Hfq (Andrade and Arraiano 2008). In this work, we wanted to characterize the role of 3′–5′ exonucleases in the degradation of small RNAs that are not associated with Hfq. Therefore, we have performed most of our studies in strains lacking Hfq.
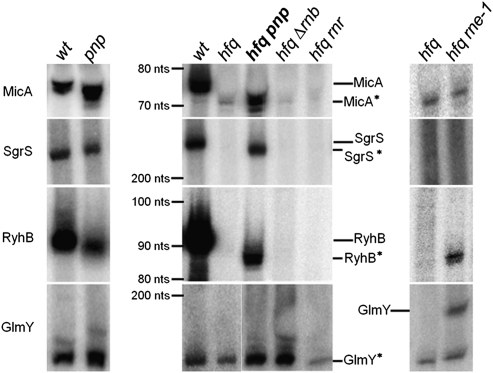
In this work we have analyzed four well-characterized E. coli sRNAs: MicA, SgrS, RyhB, and GlmY. Most of these small RNAs are induced under specific conditions of stress and in the stationary phase. Therefore, we decided to focus our work on this growth phase. Total RNA was extracted from stationary-phase cultures, and the steady-state levels of these small RNAs were analyzed by Northern blotting. Inactivation of Hfq resulted in the high reduction of all of the small RNAs analyzed when compared with the wild-type strain (Fig. 1, middle). Only GlmY levels seemed not to be so strongly affected by the absence of Hfq in the conditions analyzed. Subsequently, a set of multiple mutants lacking both Hfq and one of the main 3′–5′ exonucleases (PNPase, RNase II, or RNase R) was constructed. RNase II and RNase R had a reduced impact on this regulation; RNase II mutant was only shown to change GlmY levels, and the absence of RNase R did not affect the levels of any of these small RNAs. Upon inactivation of PNPase in cells lacking Hfq, it was possible to detect a very strong signal for all of the small RNAs analyzed. These results indicate that PNPase is a major factor controlling the levels of small RNA that are not associated with Hfq.
FIGURE 1.
PNPase strongly affects the levels of several small RNAs that are not bound to Hfq. Small RNA expression was analyzed by Northern blot. (Left) The levels of MicA, SgrS, Ryhb, and GlmY were analyzed in the wild-type (hfq+ pnp+) and a PNPase mutant (hfq+ pnp−). Total RNA was extracted from stationary-phase cultures grown at 37°C as mentioned in the Materials and Methods. (Middle) Hfq mutants lacking one of the 3′–5′ exoribonucleases PNPase (pnp), RNase II (rnb), and RNase R (rnr) were compared with wild-type (wt) and hfq single mutant. (Right) To study the impact of the essential RNase E (rne), the double hfq rne-1 mutant was grown at 30°C until it reached stationary phase and then shifted to the nonpermissive temperature of 44°C for inactivation of the thermosensitive RNase E. Samples were withdrawn after 5 min of incubation. For comparison, the single hfq mutant was treated in the same conditions. Specific [32P]-labeled probes were used to detect the small RNAs. Full-length small RNAs are clearly detected on wild type (except for GlmY), showing the expected sizes: MicA (74 nt), RyhB (90 nt), GlmY (180 nt), and SgrS (227 nt), as estimated from markers run along the gels. Small RNAs detected on hfq mutants (namely, in the hfq pnp) are slightly shorter than the corresponding full-length sRNAs; these shorter small RNAs are designated by an asterisk (*). The positions of both the full-length and the shorter small RNAs are indicated. 5S RNA or tmRNA were used as loading controls.
In Hfq− PNPase− cells we observed the accumulation of a slightly shorter form of all of the small RNAs (here designated, respectively, by MicA*, SgrS*, RyhB*, and GlmY*) (Fig. 1, middle). These shorter sRNA are not detected or are barely perceptible in the wild-type strain. The shorter GlmY* (∼140 nt) is an exception, being the predominant RNA detected in the wild type. This has been previously observed as the full-length GlmY (∼180 nt) is rapidly processed in the 3′-end to originate the GlmY* species (Reichenbach et al. 2008; Urban and Vogel 2008). Overexpression of PNPase in the hfq pnp strain was shown to reverse the accumulation of MicA* (Supplemental Fig. S1). This result confirmed that PNPase is responsible for the higher levels of small RNAs found in the Hfq− PNPase− cells.
We had previously demonstrated the involvement of PNPase in the degradation of MicA in stationary-phase cells harboring Hfq (Andrade and Arraiano 2008). To check the impact of PNPase in the regulation of all of these small RNAs in the presence of Hfq, we have analyzed the levels of the same small RNAs in the pnp single mutant and compared it with the wild-type strain (Fig. 1, left). Inactivation of PNPase in cells expressing Hfq resulted in higher levels of some of these sRNAs, namely, GlmY* and MicA. In contrast, other small RNAs such as RyhB and SgrS showed decreased amounts in the PNPase mutant strain. The reduction in the levels of other sRNAs following PNPase inactivation in exponential phase was recently observed and may potentially reflect an increase in the activity of other RNase(s) (De Lay and Gottesman 2011), but the genetic pathways involved in this regulation have not yet been elucidated. Here we show that in cells without Hfq the inactivation of PNPase (Hfq– PNPase– cells) results in increasing levels of all of the sRNAs analyzed, but that this regulation is not universal in the presence of Hfq (Hfq+ PNPase– cells). These results suggest that the binding of Hfq may impair the PNPase-dependent regulation of at least some small RNAs.
We also analyzed the effect of RNase E in the control of these regulatory RNAs in the absence of Hfq (Fig. 1, right). Only RyhB* and both GlmY/GlmY* levels were increased in the Hfq− RNase E− mutant, while MicA* and SgrS* levels did not change. This greatly contrasted with the strong stabilization of all of the small RNAs obtained in the Hfq− PNPase− mutant. Hence, our results indicated that when these small RNAs are not associated with Hfq, they are clearly more vulnerable to degradation by PNPase than to cleavages by RNase E.
PNPase is a major factor in the rapid decay of the Hfq-free MicA*
Taking into account these results and our previous data on MicA (Andrade and Arraiano 2008), we decided to use this sRNA as the main model for further investigation. MicA (previously SraD) is an antisense RNA that down-regulates the expression level of outer membrane proteins OmpA (Rasmussen et al. 2005; Udekwu et al. 2005) and LamB (Bossi and Figueroa-Bossi 2007), as well as the members of the PhoPQ regulon (Coornaert et al. 2010).
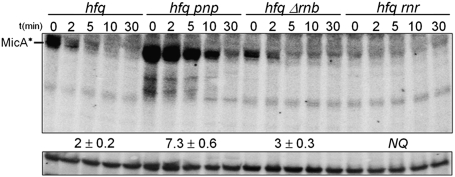
To evaluate whether the higher MicA* levels in the absence of PNPase were the consequence of increased stability, we next analyzed the decay rates of MicA* in the Hfq– PNPase– cells (Fig. 2). We also tested the potential role of RNase II and RNase R in the degradation of small RNAs in cells without Hfq. Stability measurements indicated that PNPase was found to be the only exoribonuclease significantly involved in the exo-degradation of MicA RNAs in stationary-phase cells lacking Hfq (Fig. 2). Neither RNase II nor RNase R was shown to significantly affect this decay. The hfq pnp double mutant showed a nearly fourfold stabilization of MicA* when compared with hfq single mutant. Accordingly, the increasing levels of MicA* in Hfq− PNPase− cells are a consequence of its longer stability due to the inactivation of PNPase. These results indicate that PNPase has a major role in turning over MicA species that are not associated with Hfq.
FIGURE 2.
PNPase is the major exoribonuclease involved in the degradation of MicA*. Samples from stationary-phase cultures of hfq and its derivative exoribonuclease mutants (hfq pnp, hfq Δrnb, and hfq rnr) grown at 37°C were withdrawn after inhibition of transcription (timepoints are shown in minutes) and total RNA was analyzed by Northern blot. A specific riboprobe for MicA was used. A nonspecific band that cross-hybridized with the antisense MicA probe was used as loading control. This band migrates above MicA and disappears with a more stringent washing step of the membrane without affecting MicA signal (Andrade and Arraiano 2008). Hybridization with a 5S RNA riboprobe gave identical results. Only the MicA* RNA species is detected in the absence of Hfq. Half-lives were determined after PhosphorImager densitometry quantification showing that PNPase is the major exoribonuclease involved in the degradation of the Hfq-unprotected MicA*. (NQ) Not quantifiable.
Poly(A) polymerase I promotes the degradation of MicA*
Polyadenylation can promote RNA degradation by facilitating the exonucleolytic attack of an RNA substrate (Régnier and Hajnsdorf 2009). Therefore, we decided to analyze the impact of polyadenylation in the degradation of MicA and compare it with PNPase.
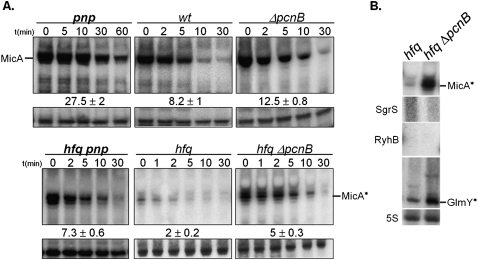
In stationary-phase cells expressing Hfq, the lack of poly(A) polymerase I (PAP I/pcnB) resulted in a modest increase in MicA half-life (only a 1.5-fold up-regulation) from 8.2 to 12.5 min, as detected by Northern blotting (Fig. 3A). On the other hand and in the same conditions, inactivation of PNPase resulted in a stronger stabilization of MicA (from 8.2 min in the wild type to 27.5 min in the pnp mutant). This indicates that in the presence of Hfq, PNPase activity against MicA surpasses the effect of PAP I polyadenylation-dependent pathways.
FIGURE 3.
Lack of poly(A) polymerase I results in increasing levels of MicA*. (A) Impact of poly(A) polymerase I (pcnB) in the degradation of the small MicA RNA in Hfq+ or Hfq− cells. Stationary-phase cultures of wild type and its derivatives pnp, ΔpcnB, hfq pnp, hfq, and hfq ΔpcnB strains were treated with rifampicin, and total RNA was analyzed by Northern blot. MicA was detected by use of a specific riboprobe. Only the shorter MicA* RNA is visible in the Hfq− cells. A nonspecific band cross-reacting with MicA probe was used as loading control. (B) The steady-state levels of several small RNAs from stationary-phase cultures of hfq and hfq ΔpcnB mutants were evaluated by Northern blot.
MicA is very unstable in the absence of Hfq; its half-life decreases from 8.2 min in the wild type to 2 min in the hfq mutant (Fig. 3A). To check whether PAP I could be involved in the rapid degradation of the MicA* in the absence of Hfq, we constructed the double hfq pcnB mutant. Two different pcnB mutations were used in this study: either a deletion ΔpcnB (O'Hara et al. 1995) or the pcnB80 allele (Hajnsdorf et al. 1995) was transferred to the hfq mutant strain; measurement of MicA half-lives gave identical results for both strains. The double-mutant hfq pcnB was found to have a significant 2.5-fold more stable MicA* when compared with the single hfq mutant (increasing half-life from 2 to 5 min). In the absence of Hfq, the MicA* RNA is more susceptible to poly(A)-mediated decay in stationary-phase cells. This was surprising, as a Hfq− mutant was reported to have low levels of polyadenylation (Hajnsdorf and Régnier 2000; Le Derout et al. 2003; Mohanty et al. 2004). Notwithstanding the higher impact that poly(A) polymerase I displays in the degradation of the MicA* in the absence of Hfq, inactivation of PNPase still renders a more stable sRNA (Fig. 3A, bottom). Altogether, these results show that PNPase has a stronger effect than poly(A) polymerase I in the degradation of MicA RNAs, irrespective of the presence of Hfq.
We also determined the relative levels of other small RNAs in the hfq ΔpncB double mutant compared with the hfq single mutant (Fig. 3B). The levels of GlmY* were also increased by the lack of PAP I in the absence of Hfq. This was also confirmed to be a consequence of the higher stabilization of GlmY* in the hfq pcnB mutant (data not shown). GlmY* is known to be highly polyadenylated in cells harboring Hfq (Reichenbach et al. 2008; Urban and Vogel 2008). We have now shown that the lack of poly(A) polymerase I is an important factor affecting the sRNA decay in the absence of Hfq in stationary-phase cells. However, inactivation of PNPase in cells devoid of Hfq resulted in higher levels of all of the small RNAs analyzed, even the ones that were not affected by the lack of PAP I (namely RyhB* and SgrS*) (Figs. 1, 3B). Hence, PNPase activity against a small RNA that is not bound to Hfq does not necessarily require an active polyadenylation-dependent pathway.
In the absence of Hfq, MicA* is a substrate for PNPase, but not for RNase E or RNase III
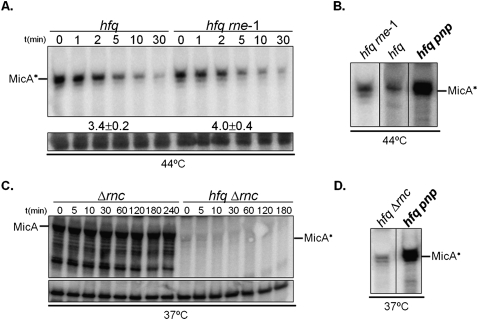
RNase E was thought to be responsible for the rapid degradation of small RNAs not protected by Hfq (Massé et al. 2003; Moll et al. 2003a). Surprisingly, we found that MicA* levels did not change substantially between the hfq single mutant and the hfq rne-1 double mutant (Fig. 1, right). To analyze this observation further, we assayed MicA decay rates in both strains. As RNase E (rne) is essential in E. coli, we used a thermolabile allele (rne-1) and performed this set of experiments at the nonpermissive temperature (Fig. 4A).
FIGURE 4.
PNPase, but not RNase E or RNase III, degrades the Hfq-free MicA* RNA. (A) Northern blot detection of MicA RNA in Hfq− cells harboring or not harboring the rne-1 allele. Stationary-phase cultures were treated at 44°C for inactivation of the thermosensitive RNase E (as mentioned before). MicA RNA stability was analyzed by Northern blot with a specific riboprobe. (B) Northern blot analysis of MicA in Hfq− cells deficient in RNase E or PNPase. The double hfq rne-1 mutant was grown at 30°C until stationary phase and then incubated at 44°C to inactivate RNase E. For comparison, the hfq and hfq pnp were treated in the same conditions. (C) Northern blot detection of MicA in stationary-phase cultures of Hfq− cells harboring or not harboring RNase III (rnc), respectively. A loading control corresponding to a nonspecific band that cross-reacted with MicA probe is shown in below. (D) Comparison of MicA* RNA steady-levels in Hfq− stationary-phase cells deficient in RNase III or PNPase grown at 37°C.
From previous work, we have identified that RNase E is involved in the degradation of MicA in cells producing Hfq (Andrade and Arraiano 2008). Surprisingly, our results revealed that MicA* is not stabilized significantly when RNase E is inactivated in the absence of Hfq (Fig. 4A). This indicates that RNase E is not able to efficiently degrade MicA* unless Hfq is present in the cell. A similar RNase E/Hfq dependency was observed in OxyS turnover (Basineni et al. 2009).
To better assess the relative impact of RNase E and PNPase, we treated the culture of the hfq pnp mutant in the same conditions used to inactivate the thermosensitive RNase E (Fig. 4B). No significant changes were detected when Hfq and RNase E were inactive, but MicA steady-state levels are substantially higher upon inactivation of both Hfq and PNPase (an eightfold increase in the hfq mutant). This result clearly showed that in the absence of Hfq, PNPase is more important than RNase E in the degradation of this sRNA. This result is also substantiated by data from Figure 1.
The other main endonuclease involved in RNA degradation is RNase III (Arraiano et al. 2010). In vitro studies showed that Salmonella RNase III can cleave MicA when bound to its target ompA mRNA (Viegas et al. 2011). To further analyze the role of RNase III in cells without Hfq, we constructed and analyzed double mutants lacking both Hfq and RNase III. RNA extracted from stationary-phase cultures from the deletion mutant of E. coli RNase III (Δrnc) and a double mutant lacking Hfq and RNase III (hfq Δrnc) was analyzed by Northern blotting (Fig. 4C). Inactivation of RNase III in the presence of Hfq appeared to block the degradation of MicA (this RNA apparently did not decay even 240 min after transcription blocking). This clearly showed that E. coli RNase III is important in the control of MicA stability. However, MicA RNAs were barely detected in the double-mutant hfq Δrnc, reflecting the results obtained with the single hfq mutant. To confirm this result we also tested another allele of RNase III, the rnc105; the double-mutant hfq rnc105 displayed identical results (data not shown). The strong decrease in MicA levels typically found in the absence of Hfq obviously reduce the number of duplexes formed between this sRNA and its target mRNAs, probably impairing RNase III activity against MicA. The MicA* levels found in the hfq pnp strain were higher (about a sixfold increase in the hfq single mutant) than the MicA* levels found in the hfq Δrnc mutant (Fig. 4D). These results clearly indicated that PNPase was more important than RNase III in the elimination of MicA* from the cell. Overall, when MicA is not associated with Hfq, the 3′–5′ exoribonucleolytic degradation pathway mediated by PNPase is found to be more important in this degradation than any of the main endoribonucleases involved in RNA turnover.
Hfq is required for the optimal expression of the full-length MicA
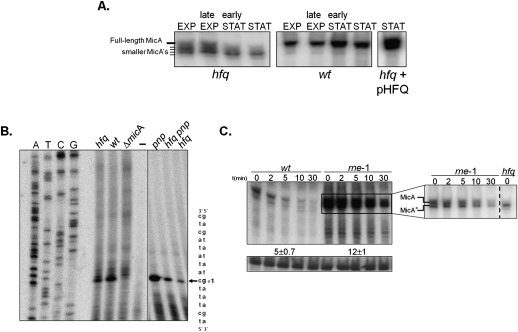
The slightly smaller MicA* is the predominant form in the hfq mutant in stationary-phase cultures, but it is barely detected in the wild-type (Fig. 1). To examine whether this RNA pattern was dependent on a growth phase-specific regulation, we analyzed both the wild-type and the hfq strain along the growth curve (Fig. 5A; Supplemental Fig. S2). In exponential phase, the hfq mutant exhibited the full-length MicA, as well as additional shorter bands of similar intensity, apparently differing a few nucleotides in size. This pattern was growth dependent. In stationary phase there was a decrease in the amount of full-length MicA and what appeared to be a concomitant accumulation of the smaller MicA*. This greatly contrasted with the wild-type strain, where the full-length MicA was the most prominent band, irrespective of the growth phase analyzed. Moreover, supplying Hfq in trans from a plasmid complemented hfq deficiency on MicA expression and resulted in the strong accumulation of the full-length RNA and in the elimination of the shorter sized RNAs (Fig. 5A). These results indicated that Hfq determines MicA full-length expression along growth, particularly in the stationary phase.
FIGURE 5.
Hfq is required for the expression of the full-length MicA RNA. (A) Steady-state levels of MicA RNA along the growth curve. Culture samples of wild-type or hfq mutant bacteria were collected at exponential (EXP), late exponential, early stationary, and stationary phase (STAT) (corresponding to OD600 values of ∼0.3, ∼1.7, ∼2.5, and ∼5.5 for the wild-type and ∼0.3, ∼0.8, ∼1.6, and ∼2.3 for the hfq mutant, respectively). The growth curves for the wild-type and the hfq mutant strain are given in Supplemental Figure S2. A specific antisense MicA riboprobe was used to detect MicA. Stationary-phase cultures of the hfq mutant transformed with the overexpressing pHFQ plasmid show complementation and do not exhibit the heterogeneous population of MicA's typically found in the hfq single mutant. (B) Determination of the 5′-end of MicA. Total RNA from stationary-phase cells of wild-type, hfq, pnp, and hfq pnp strains was analyzed by primer extension with the [32P]-labeled primer MicA-PE. The same primer extension product (indicated by an arrow) is detected on all strains and absent from the deletion micA strain (ΔmicA) and the negative control reaction (−) done without RNA. Part of the DNA sequence is indicated on the right. The transcription start site of MicA is indicated (+1) and is identical to the site described by Udekwu et al. (2005). The intensity of the primer extension product obtained is higher in the wild-type rather than the hfq mutant, in agreement with the higher amount of MicA detected in the wild-type strain (see Fig. 5A). (C) Northern blot detection of MicA in stationary-phase cultures of Hfq+ cells upon inactivation of RNase E. Cultures of wild-type and an RNase E mutant strain were grown at 30°C until they reached stationary phase, and then shifted to the nonpermissive temperature of 44°C. After 5 min, transcription was blocked with the addition of rifampicin, and samples were withdrawn at times indicated. A specific riboprobe was used to detect MicA RNA. A nonspecific band that cross-hybridized with the antisense MicA probe was used as loading control. The inset corresponds to a shorter exposure of the membrane in which it is visible that both the full-length MicA and the shorter MicA* RNA are detected and stabilized upon inactivation of RNase E in Hfq+ cells. The hfq mutant was used here as a control to clearly identify MicA* RNA.
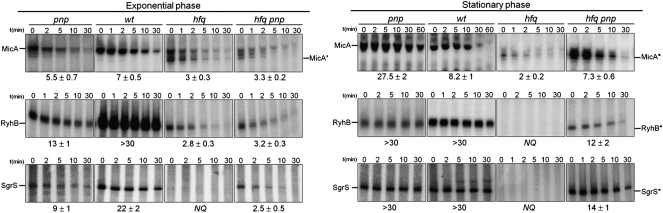
The difference in size between the full-length MicA and MicA* is small, apparently in the range of from 3 to 4 nt, visible on Northern blotting. Such small variation must lie at one of the RNA extremities. Primer extension analysis was performed to evaluate which extremity was shortened (Fig. 5B). Stationary-phase cultures of both the wild-type (which expresses full-length MicA) and the hfq mutant strain (where MicA* is detected) showed accumulation of a band that matches the start of the MicA sequence. Furthermore, this same band was identified when testing either the pnp mutant or the hfq pnp double mutant, showing that the MicA RNAs that accumulate upon PNPase inactivation retain the same 5′ end as the wild-type MicA. An additional experimental approach using nuclease S1 mapping also determined the same 5′-end for both MicA species (Supplemental Fig. S3). Altogether, these findings supported that full-length MicA and MicA* have the same 5′-end, and that the difference in size is located at the 3′-end. This suggests that the smaller RNA species probably arises from 3′-end processing of the full-length MicA.
The MicA* RNA is expressed at very low levels in the wild type. This suggests that Hfq acts in order to prevent MicA* production or to ensure its rapid elimination. We decided to analyze the kinetics of decay and found that RNase E affected MicA* levels (Fig. 5C). Inactivation of RNase E (in cells harboring Hfq) resulted in the strong elevation of MicA levels in stationary phase. However, a shorter exposure of this gel revealed the detection not only of the full-length MicA, but as well, the smaller MicA* (inset in Fig. 5C). Both RNAs showed a twofold stabilization in the absence of RNase E. This indicated that even in the presence of Hfq, the shorter MicA* RNA fragment is produced in the cell.
Growth-phase regulation of small RNAs by PNPase
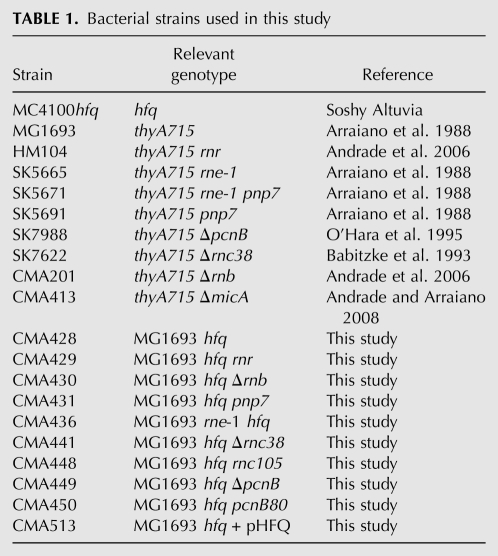
In the absence of Hfq, small RNAs are typically unstable and PNPase was found to be a major enzyme involved in the extensive degradation of MicA in stationary-phase cells. To check whether this could be generalized to other small RNAs, we extended this analysis to RyhB and SgrS. Since the RNA pattern of MicA changes along with growth in Hfq− cells (Fig. 5A), it is reasonable that different RNA degradation pathways might be involved in different stages of growth. To further analyze this, we decided to compare the small RNA stability between exponential and stationary-phase cultures.
In the absence of Hfq, all of the small RNAs analyzed were highly unstable, regardless of the growth phase that was analyzed (Fig. 6). As consequence of the extensive degradation occurring in the absence of Hfq, the MicA, RyhB, and SgrS levels were strongly reduced in the hfq mutant when compared with wild type, both in exponential and stationary-phase cultures. In contrast, all of these small RNAs were markedly stabilized in the stationary-phase cultures of the hfq pnp double mutant compared with the hfq strain. Interestingly, this regulation is not as common in exponentially growing cells. In fact, only SgrS was found to be stabilized in exponential-phase cultures of the hfq pnp mutant strain compared with the hfq strain (although this is significantly lower than the stabilization observed in stationary-phase cells). These results confirm PNPase as a major enzyme involved in the degradation of Hfq-free small RNAs in the cell.
FIGURE 6.
Growth-phase regulation of Hfq-free small RNAs by PNPase. Northern blot determination of MicA, RyhB, and SgrS RNA stabilities between the wild-type and its isogenic pnp, hfq, and hfq pnp mutants either in exponential-phase or stationary-phase cultures. Total RNA was extracted from culture samples withdrawn after inhibition of transcription with rifampicin (timepoints are shown in minutes). MicA, RyhB, and SgrS RNAs were detected by the use of specific radiolabeled probes and quantified by PhosphorImager analysis. The full-length small RNAs or their respective shorter forms (where detected) are indicated on the gels. (NQ) Not quantifiable.
Hfq deficiency resulted in the detection of shorter small RNAs that are stabilized upon further inactivation of PNPase (Figs. 1, 6). In exponential-growing cells without Hfq, only MicA was found to exhibit a heterogeneous-sized population (Figs. 5A, 6). From these fragments, MicA* is shown to be the most resistant and is even the predominant RNA species found in hfq mutants in the stationary phase. The smaller RyhB* and SgrS* RNAs were only detected in stationary-phase cells. Together, these results suggested a protection of the full-length sRNA by Hfq, which seems particularly important for sRNA expression in the stationary phase of growth.
To analyze whether PNPase is affecting the stability of small RNAs independently of Hfq, we further analyzed the decay rates of MicA, RyhB, and SgrS in the pnp single mutant. Inactivation of PNPase in exponential-phase cells producing Hfq resulted in reduced levels and decreased stability of the small RNAs. Similar results were reported with other small RNAs, suggesting that PNPase may somehow protect some sRNA in exponential phase (De Lay and Gottesman 2011). In fact, this is also observed in stationary-phase cultures, as inactivation of PNPase is also found to reduce RyhB and SgrS levels under this condition. Only MicA was shown to be a substrate for PNPase either in the absence or presence of Hfq. On the other hand, RyhB and SgrS were found to be preferably degraded by PNPase in the absence of Hfq and were greatly stabilized in the hfq pnp double mutant, particularly in the stationary phase of growth. Altogether, these results suggest that PNPase degrades small RNAs more efficiently in the absence of Hfq, although this turnover pathway is clearly active in cells with Hfq. The fact that this occurs in cells expressing Hfq may reflect the action of PNPase against small RNAs that are transiently in their Hfq-free state, a result of the dynamics of interaction with Hfq. PNPase-mediated degradation of small RNAs is suggested to be predominant in stationary-phase cells, as this regulation apparently is not so common in exponential-phase cells. Therefore, these results demonstrated that the degradation pathways of a same small RNA can be different between exponential and stationary phases and highlighted the role of PNPase in the growth-phase regulation of small RNAs.
DISCUSSION
This work demonstrated that the pool of small RNAs that are not associated with Hfq is preferably degraded by PNPase. Overall, our data highlight the impact of 3′–5′ exonucleolytic RNA decay pathways and re-evaluates the degradation mechanisms involved in the rapid decay of the Hfq-free small RNAs. The reduced levels of small RNAs typically found in the Hfq− strain were strongly increased upon inactivation of PNPase in stationary-phase cells (Fig. 1). This seems to be a general feature, since PNPase inactivation resulted in increasing levels of at least the MicA, SgrS, RyhB, and GlmY sRNAs. We only detected the accumulation of slightly shorter sRNAs rather than the full-length species, and this was shown to be the consequence of the higher stability of these fragments (Fig. 2).
The lack of poly(A) polymerase I was also found to impact the levels of small RNAs in the absence of Hfq, although to a lesser extent than PNPase (Fig. 3). These results were unexpected, as no significant differences in mRNA stability were detected between hfq and hfq ΔpcnB mutants (Mohanty et al. 2004). In contrast, our results clearly showed that in the absence of Hfq, the small RNA turnover can be affected by the lack of poly(A) polymerase I. The sRNAs found to be highly affected by polyadenylation (MicA* and GlmY*) were also found to be excellent substrates for PNPase. Nevertheless, PNPase activity against Hfq-unprotected small RNAs is not necessarily dependent on poly(A) polymerase I activity. RyhB* and SgrS* RNAs are not affected by polyadenylation, although their levels were highly increased upon PNPase inactivation, as observed in the double hfq pnp mutant (Figs. 1, 3B). Poly(A)-dependent pathways may thus not explain all the extraordinary impact of PNPase on sRNA turnover in the Hfq− cells.
Pioneer work on PNPase revealed its ability to synthesise RNA (Grunberg-Manago et al. 1955). Interestingly, it has been proposed that in the absence of Hfq, there is an increase in the biosynthetic activity of PNPase with heteropolynucleotide tails promoting RNA decay (Mohanty et al. 2004; Slomovic et al. 2008). Addition of these polynucleotide tails can potentially be responsible for PNPase notable impact on the degradation of sRNA in the absence of Hfq. RNase II (Marujo et al. 2000) and RNase R (Andrade et al. 2009a) are also major poly(A)-dependent exoribonucleases, but they were not found to be involved in the degradation of MicA (Fig. 2; Andrade and Arraiano 2008). Similar results were obtained regarding the degradation of RyhB (data not shown). Surprisingly, despite RNase R intrinsic ability to easily degrade structured RNAs on its own and its affinity to poly(A) tails, RNase R was not shown to be part of these decay pathways. The absence of RNase R resulted in the reduction of MicA* levels in cells without Hfq (Fig. 2). This might be a result of an indirect effect in which the activity of a MicA repressor is increased when RNase R is not functional. Although the protection of RNA by a ribonuclease seems paradoxical, a similar effect has been described either for RNase II or PNPase (Marujo et al. 2000; De Lay and Gottesman 2011). A major advantageous feature of PNPase in the degradation of small RNAs might be its ability to form complexes with other proteins, which can be particularly helpful in the elimination of such structured RNAs. However, we have already shown that PNPase activity on MicA can be independent of the degradosome assembly (Andrade and Arraiano 2008).
RNase E has a role in sRNA degradation (Massé et al. 2003; Morita et al. 2005; Suzuki et al. 2006; Viegas et al. 2007; Andrade and Arraiano 2008). However, our results demonstrated that its impact on Hfq− cells may not be as general as previously believed. RNase E depletion did not affect the levels of SgrS and MicA RNA. While in the presence of Hfq, both the full-length MicA and the MicA* RNAs are substrates for RNase E (Fig. 5C); this regulation is lost when Hfq is absent (Fig. 4A). This indicates that RNase E requires Hfq in order to degrade MicA. A similar RNase E dependency of Hfq to act on sRNA turnover was also reported in the growth-phase degradation of OxyS (Basineni et al. 2009). It is suggested that RNase E/Hfq cooperation (as observed in the mRNA decay mediated by sRNA) (Morita et al. 2005) can also be critical for the degradation of some small noncoding RNAs, like MicA. Nevertheless, we recognize that RNase E can also impact the levels of some sRNAs independently of Hfq. As observed, both RyhB* and GlmY are RNase E substrates, even in cells lacking Hfq (Fig. 1).
A possible RNase III/Hfq pathway was also analyzed. RNase III inactivation results in extremely long-lived MicA; however, this is strictly dependent on the presence of Hfq, as this stabilization is completely lost in the hfq Δrnc mutant (Fig. 4C). The low levels of MicA found in hfq mutants strongly decrease the probability of base-pairing with target mRNAs. The down-regulation in sRNA-target mRNA duplexes probably explains the impairment in RNase III activity on MicA, in agreement with in vitro studies (Viegas et al. 2011). Data suggest that the degradation of small RNAs that are not associated with Hfq mainly occurs in a target-independent pathway, in which RNase III has a reduced impact. The free pool of small RNAs is then preferably degraded by PNPase.
Hfq was thought to mainly protect sRNA from RNase E cleavages, as both proteins showed in vitro affinity for the same A/U-rich sequences in RNA (Moll et al. 2003a). However, it has been recently demonstrated that Hfq actually prefers to bind U-rich sequences at the 3′-end of small RNAs over internal A/U-rich sequences (Otaka et al 2011; Sauer and Weichenrieder 2011). Small RNAs, like MicA, usually display a short U-rich 3′-end sequence immediately downstream from a stem–loop as a consequence of Rho-independent transcription termination (Rasmussen et al. 2005; Udekwu et al. 2005). The physiological meaning of the high affinity of Hfq to this U-rich sequence can be the protection of the 3′-end of the RNA against degradation. Interestingly, our results showed that the 3′-ends of the small RNAs are shortened in the absence of interaction with Hfq (Figs. 1, 5B). Even though PNPase is observed to be the main exoribonuclease involved in the degradation of these shorter small RNAs, it does not seem to be the main reason for the initial 3′-end attack, as this is not prevented in a pnp background. RNase II and RNase R inactivation also did not suppress the shortening of MicA. Data suggested that other (exo)nucleases would be responsible for the 3′-end trimming of the small RNAs when they are Hfq free. The transcriptional terminator stem–loop of the small RNAs may function as a physical barrier against exoribonucleases. PNPase may be favored in this action and progress to degradation of the sRNA body, while other RNases may be inhibited, and therefore could only degrade a few nucleotides before releasing the sRNA. In the presence of Hfq, the shorter sRNAs are barely detected, probably because Hfq protects the 3′-ends of the small RNAs.
Our results also indicate that small RNAs are subject to different degradation pathways, depending on growth (Fig. 6). In the stationary phase, PNPase is shown to be the main enzyme in the degradation of small RNAs (Andrade and Arraiano 2008; this work). On the other hand, it has been proposed that in exponential phase, PNPase can actually protect small RNAs from rapid degradation by other ribonucleases, namely, from RNase E activity (De Lay and Gottesman 2011). The growth-phase regulation of sRNA turnover pathways may help to explain why RNase E was shown to affect sRNA decay in previous studies in which the exponential phase of growth was analyzed (Massé et al. 2003), whereas it is not found to be the predominant degradative enzyme in the stationary phase (this work). PNPase responds to environmental stimuli and has been suggested to be responsible for the addition of heteropolymeric tails to the 3′-end of RNAs in the stationary phase of growth (Cao and Sarkar 1997; Mohanty and Kushner 2000). PNPase could then use those tails to initiate RNA degradation. Accordingly, the growth-phase regulation of PNPase activities may thus help in explaining the growth-phase regulation of small RNAs driven by PNPase.
Variations in the levels of Hfq can most probably influence the degradation pathways of the small RNA. Interestingly, Hfq was reported to vary along the growth, and decreased levels of this protein were found in the entry to stationary phase (Ali Azam et al. 1999). Not only changes in the Hfq expression level, but also variations in the pool of free Hfq can result in low amounts of this protein and, consequently, affect the sRNA-based regulatory pathways. Hfq binds the RNA molecules very tightly and this can result in the sequestration of Hfq. A model in which an increasing concentration of a competitor RNA promotes the dissociation of the Hfq–RNA complexes has recently been proposed to explain how it is possible to cycle the Hfq pool within the cell (Fender et al. 2010). In agreement, it was shown that induction of a sRNA without the concomitant overexpression of its target mRNA (or vice versa) can sequester Hfq and abolish the function of unrelated sRNAs (Hussein and Lim 2011). Hence, Hfq depletion is likely to occur if transcription of sRNA and its target mRNAs is not coordinated. The rapid degradation of sRNA in the absence of interaction with Hfq may thus recycle any small RNAs that are produced in excess over Hfq. This reinforces the importance of studying the degradation of small RNAs when they are not associated with Hfq. Most of our work was performed in stationary-phase cells deleted for Hfq. However, we have shown that PNPase-mediated degradation of small RNAs is also an active regulatory pathway in cells expressing Hfq. This fact may reflect the action of PNPase against small RNAs that do not have their 3′-ends protected by Hfq. Our results are in agreement with in vitro data showing that Hfq can protect an mRNA from the exonucleolytic activity of PNPase (Folichon et al. 2003).
A similar phylogenetic distribution may reflect functionally linked proteins (Pellegrini et al. 1999). A large number of bacteria encode both Hfq and PNPase in their genomes (Zuo and Deutscher 2001; Sun et al. 2002), while the presence of E. coli RNase E homologs is far more restricted (Condon and Putzer 2002; Danchin 2009). Interestingly, eukaryotes lack an RNase E, but possess functional homologs of both PNPase and Hfq. The eukaryotic exosome adopts an PNPase-like conformation and is implicated in the processing and degradation of several RNAs, namely, the small nucleolar RNAs (snoRNAs) and the small nuclear RNAs (snRNAs) (Houseley et al. 2006). The exosome activity is suggested to be modulated by the Lsm1–7 complex (whose subunits are homologous to bacterial Hfq), although this interplay is still unclear (Wilusz and Wilusz 2008). The fact that Hfq and PNPase are more widespread than RNase E supports the interesting hypothesis that Hfq protection of sRNA against degradation by PNPase is far more common than was previously envisioned.
MATERIALS AND METHODS
Growth conditions, strains, and plasmids
Bacteria were grown at 37°C unless stated otherwise, with shaking at 180 rpm in Luria-Bertani (LB) medium supplemented with thymine (50 μg mL−1). SOC medium was used to recover cells after heat shock in plasmid transformation steps. When required, antibiotics were present at the following concentrations: chloramphenicol, 50 μg mL−1, kanamycin, 50 μg mL−1; tetracycline, 20 μg mL−1; ampicillin, 100 μg mL−1. The E. coli strains used in this work are listed in Table 1. Strain MC4100 hfq::cat (kindly provided by S. Altuvia) was used as donor to move the mutant hfq allele into MG1693 (wild type) and its derivative isogenic strains. Introduction of mutant alleles to different genetic backgrounds was done by P1 transduction, and positive colonies were checked by PCR. A DNA sequence of Hfq was PCR-amplified with primers hfq–EcoRI (5′-GTGACGAAGaATTcCAGGTTGTTG-3′) and hfq–HindIII (5′-CGGTCAAACAAGCtTATAACCC-3′), and following enzyme restriction it was cloned into pBAD24, yielding the overexpression pHFQ plasmid. Hfq expression is obtained even without addition of the arabinose inducer, as the cloned DNA retains hfq's own promoters. For plasmid pMicA, primers MicA–PstI (5′-TTTTCGCCACCCGAACTGCAGGC-3′) and MicA–HindIII (5′-GGCTGGAAAAACAaGCtTGACAGAAAAGAAAAAGG-3′) were used to amplify the micA gene. Following enzyme restriction, the insert was ligated into pWSK29 in sites PstI and HindIII. DNA polymerases and restriction enzymes were obtained from Fermentas, and T4 DNA Ligase from Roche. All primers were obtained from StabVida (Portugal).
TABLE 1.
Bacterial strains used in this study
RNA extraction and Northern blot analysis
Overnight cultures from isolated colonies were diluted in fresh medium to an initial OD600 ∼ 0.03 and grown to exponential (OD600 ∼ 0.3) or stationary phase (OD600 ∼ 5.5 to wild-type or OD600 ∼ 2.3 to hfq mutants). The growth curves for the wild-type strain and the hfq mutant are provided in Supplemental Figure S2. For decay experiments, blocking of transcription was obtained by adding rifampicin to a final concentration of 500 μg mL−1. Culture samples were withdrawn at defined timepoints and mixed with an equal volume of RNA stop buffer (10 mM Tris at pH 7.2, 5 mM MgCl2, 25 mM NaN3, and 500 μg mL−1 chloramphenicol). RNA was isolated following cell lysis and phenol:chloroform extraction. After a precipitation step in ethanol and 300 mM sodium acetate, RNA was resuspended in MilliQ-water. The integrity of RNA samples was evaluated by agarose gel electrophoresis. When necessary, DNase RQ (Promega) treatment following a new phenol:chloroform step was used to remove contaminant DNA. Next, 10–40 μg of total RNA was used to analyze small RNA expression on 6%–12% polyacrylamide/7 M urea gels in TBE 1x. RNA was transferred onto Hybond-N+ membrane (Amersham Biosciences) using TAE 1x as transfer buffer. RNAs were UV cross-linked to the membrane with a UVC 500 apparatus (Amersham Biosciences). DNA templates carrying a T7 promoter sequence for in vitro transcription were generated by PCR using genomic DNA of MG1693 and primers listed in Table 2. GlmY was detected by 5′-end labeling of an antisense primer (Table 2). Radiolabeled probes were purified on G25 Microspin columns (GE Healthcare). Hybridizations were carried out overnight at 42°C–68°C with the PerfectHyb Plus Hybridization Buffer (Sigma). RNA Decade markers (Ambion) or the 10-bp Step Ladder (Promega) were used for detection of small RNAs up to 150 nt; for longer transcripts, the 100–1000 bp Ladder (Biotools) was used. T7 RNA polymerase and T4 polynucleotide kinase were from Promega. All radiochemicals were purchased from Perkin-Elmer.
TABLE 2.
Oligonucleotides used in radiolabeling reactions
RNA half-life determination
Northern blot signals were visualized on PhosphorImager STORM 860, and bands intensities were quantified using the IMAGEQUANT software (Molecular Dynamics). Half-lives of RNA were determined by linear regression using the logarithm of the percentage of RNA remaining versus time, considering the amount of RNA at 0 min as 100%. A minimum of two independent RNA extractions from each strain were tested and half-lives correspond to average of at least three experiments.
Primer extension analysis
The MicA RNA was analyzed by primer extension analysis using the MicA-PE primer (5′-CGTGAGTGGCCAAAATTTCATCTCTG-3′). A total of 10 μg of each RNA sample was incubated with 1 pmol of 5′-end [γ-32P]ATP-labeled primer. Sample denaturation was done for 5 min at 80°C, immediately followed by the annealing step (30 min at 65°C and 30 min at 48°C). cDNA synthesis was obtained using 200 units of SuperScript III Reverse Transcriptase, following the manufacturer's instructions (Invitrogen). Incubation proceeded for 60 min at 55°C and was terminated by heat inactivation of the samples for 15 min at 70°C. The cDNA products were then ethanol precipitated with the addition of glycogen for 15 min in a −80°C freezer. The cDNA pellet was dissolved in 2 μL of 0.1 M NaOH/1 mM EDTA and 4 μL of formamide loading buffer. Prior to loading, samples were denatured for 5 min at 80°C, and then fractionated on 6% polyacrylamide/7 M urea gels. Plasmid pMicA was used in sequencing reactions with primer MicA-PE following the instructions of the Sequenase Version 2.0 DNA Sequencing Kit (USB). The gel was exposed on a PhosphorImager screen and the signal was detected on a PhosphorImager STORM 860.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Soshy Altuvia (The Hebrew University, Jerusalem, Israel) for providing the MC4100 hfq strain. We are grateful to Hiroji Aiba (Suzuka University of Medical Sciences, Japan) for insightful comments. This work was supported by grants from Fundação para a Ciência e a Tecnologia (FCT), Portugal, including grant PEst-OE/EQB/LA0004/2011. J.M.A. is a recipient of a FCT Postdoctoral Fellowship and V.P. is recipient of a FCT Doctoral Fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029413.111.
REFERENCES
- Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR 2005. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res 33: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol 10: 134–139 [DOI] [PubMed] [Google Scholar]
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181: 6361–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade JM, Arraiano CM 2008. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade JM, Cairrão F, Arraiano CM 2006. RNase R affects gene expression in stationary-phase: regulation of ompA. Mol Microbiol 60: 219–228 [DOI] [PubMed] [Google Scholar]
- Andrade JM, Hajnsdorf E, Régnier P, Arraiano CM 2009a. The poly(A)-dependent degradation pathway of rpsO mRNA is primarily mediated by RNase R. RNA 15: 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM 2009b. The role of 3′-5′ exoribonucleases in RNA degradation. Prog Mol Biol Transl Sci 85: 187–229 [DOI] [PubMed] [Google Scholar]
- Arluison V, Hohng S, Roy R, Pellegrini O, Régnier P, Ha T 2007. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res 35: 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano CM, Yancey SD, Kushner SR 1988. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol 170: 4625–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34: 883–923 [DOI] [PubMed] [Google Scholar]
- Babitzke P, Granger L, Olszewski J, Kushner SR 1993. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol 175: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basineni SR, Madhugiri R, Kolmsee T, Hengge R, Klug G 2009. The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol 6: 584–594 [DOI] [PubMed] [Google Scholar]
- Bossi L, Figueroa-Bossi N 2007. A small RNA downregulates LamB maltoporin in Salmonella. Mol Microbiol 65: 799–810 [DOI] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32: 827–837 [DOI] [PubMed] [Google Scholar]
- Brennan RG, Link TM 2007. Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Cao GJ, Sarkar N 1997. Stationary-phase-specific mRNAs in Escherichia coli are polyadenylated. Biochem Biophys Res Commun 239: 46–50 [DOI] [PubMed] [Google Scholar]
- Chao Y, Vogel J 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13: 24–33 [DOI] [PubMed] [Google Scholar]
- Condon C, Putzer H 2002. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res 30: 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M 2010. MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol 76: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin A 2009. A phylogenetic view of bacterial ribonucleases. Prog Mol Biol Transl Sci 85: 1–41 [DOI] [PubMed] [Google Scholar]
- De Lay N, Gottesman S 2011. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17: 1172–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Favero M, Mazzantini E, Briani F, Zangrossi S, Tortora P, Dehò G 2008. Regulation of Escherichia coli polynucleotide phosphorylase by ATP. J Biol Chem 283: 27355–27359 [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG 2010. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev 24: 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E 2003. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res 31: 7302–7310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folichon M, Allemand F, Régnier P, Hajnsdorf E 2005. Stimulation of poly(A) synthesis by Escherichia coli poly(A) polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J 272: 454–463 [DOI] [PubMed] [Google Scholar]
- Geissmann TA, Touati D 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J 23: 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M, Oritz PJ, Ochoa S 1955. Enzymatic synthesis of nucleic acidlike polynucleotides. Science 122: 907–910 [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E, Régnier P 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci 97: 1501–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P 1995. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci 92: 3973–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Hussein R, Lim HN 2011. Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci 108: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Derout J, Folichon M, Briani F, Dehò G, Régnier P, Hajnsdorf E 2003. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res 31: 4017–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Woodson SA 2004. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol 344: 1211–1223 [DOI] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennan RG 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci 106: 19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Régnier P 2000. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA 6: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11: 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR 2000. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci 97: 11966–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol 54: 905–920 [DOI] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U 2003a. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9: 1308–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Leitsch D, Steinhauser T, Blasi U 2003b. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep 4: 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H 2005. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci 103: 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmohamed S, Vincent HA, Titman CM, Chandran V, Pears MR, Du D, Griffin JL, Callaghan AJ, Luisi BF 2011. Polynucleotide phosphorylase activity may be modulated by metabolites in Escherichia coli. J Biol Chem 286: 14315–14323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR 1995. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci 92: 1807–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka H, Ishikawa H, Morita T, Alba H 2011. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci 108: 13059–13064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci 96: 4285–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E 2011. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P 2005. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol Microbiol 58: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Régnier P, Hajnsdorf E 2009. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci 85: 137–185 [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B 2008. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res 36: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer E, Weichenrieder O 2011. Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci 108: 13065–13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J 21: 3546–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Portnoy V, Yehudai-Resheff S, Bronshtein E, Schuster G 2008. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochim Biophys Acta 1779: 247–255 [DOI] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA 2010. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci 107: 9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhulin I, Wartell RM 2002. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res 30: 3662–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev 20: 2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 13: 35–49 [DOI] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol 407: 633–639 [DOI] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 19: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM 2011. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA 17: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6: e64 doi: 10.1371/journal.pbio.0060064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol 51: 1525–1533 [DOI] [PubMed] [Google Scholar]
- Vecerek B, Moll I, Blasi U 2007. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J 26: 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM 2007. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res 35: 7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM 2011. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res 39: 2918–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J 2005. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol 12: 1031–1036 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J 2008. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev 22: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res 29: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]