Abstract
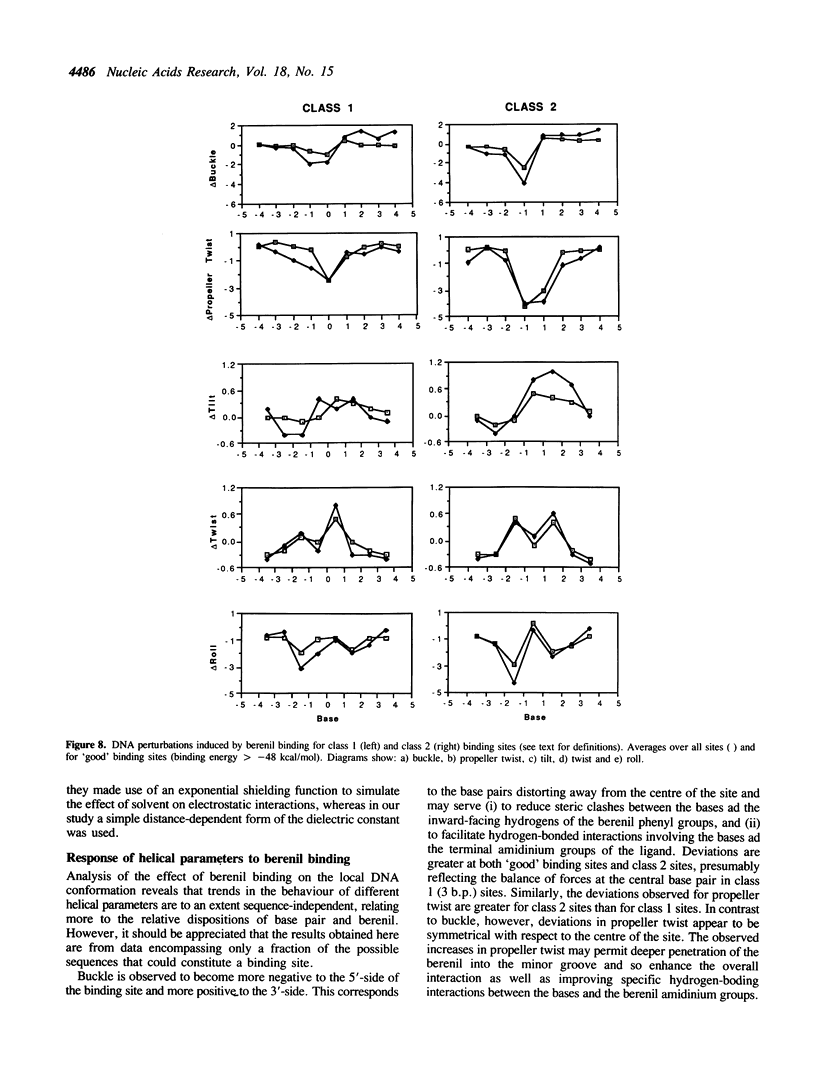
We have developed a technique of partially-restrained molecular mechanics enthalpy minimisation which enables the sequence-dependence of the DNA binding of a non-intercalating ligand to be studied for arbitrary sequences of considerable length (greater than = 60 base-pairs). The technique has been applied to analyse the binding of berenil to the minor groove of a 60 base-pair sequence derived from the tyrT promoter; the results are compared with those obtained by DNAse I and hydroxyl radical footprinting on the same sequence. The calculated and experimentally observed patterns of binding are in good agreement. Analysis of the modelling data highlights the importance of DNA flexibility in ligand binding. Further, the electrostatic component of the interaction tends to favour binding to AT-rich regions, whilst the van der Waals interaction energy term favours GC-rich ones. The results also suggest that an important contribution to the observed preference for binding in AT-rich regions arises from lower DNA perturbation energies and is not accompanied by reduced DNA structural perturbations in such sequences. It is therefore concluded that those modes of DNA distortion favourable to binding are probably more flexible in AT-rich regions. The structure of the modelled DNA sequence has also been analysed in terms of helical parameters. For the DNA energy-minimised in the absence of berenil, certain helical parameters show marked sequence-dependence. For example, purine-pyrimidine (R-Y) base pairs show a consistent positive buckle whereas this feature is consistently negative for Y-R pairs. Further, CG steps show lower than average values of slide while GC steps show lower than average values of rise. Similar analysis of the modelling data from the calculations including berenil highlights the importance of DNA flexibility in ligand binding. We observe that the binding of berenil induces characteristic responses in different helical parameters for the base-pairs around the binding site. For example, buckle and tilt tend to become more negative to the 5'-side of the binding site and more positive to the 3'-side, while the base steps at either side of the centre of the site show increased twist and decreased roll.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. G., Sanderson M. R., Skelly J. V., Jenkins T. C., Brown T., Garman E., Stuart D. I., Neidle S. Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding. EMBO J. 1990 Apr;9(4):1329–1334. doi: 10.1002/j.1460-2075.1990.tb08242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J., Kollman P. A molecular mechanical study of netropsin-DNA interactions. Biopolymers. 1986 Feb;25(2):249–266. doi: 10.1002/bip.360250207. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Carrondo M. A., Coll M., Aymami J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Binding of a Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry. 1989 Sep 19;28(19):7849–7859. doi: 10.1021/bi00445a047. [DOI] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cons B. M., Fox K. R. High resolution hydroxyl radical footprinting of the binding of mithramycin and related antibiotics to DNA. Nucleic Acids Res. 1989 Jul 25;17(14):5447–5459. doi: 10.1093/nar/17.14.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago F., Reynolds C. A., Richards W. G. The binding of nonintercalative drugs to alternating DNA sequences. Mol Pharmacol. 1989 Feb;35(2):232–241. [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Goodsell D., Dickerson R. E. Isohelical analysis of DNA groove-binding drugs. J Med Chem. 1986 May;29(5):727–733. doi: 10.1021/jm00155a023. [DOI] [PubMed] [Google Scholar]
- Gresh N., Pullman B. A theoretical study of the nonintercalative binding of berenil and stilbamidine to double-stranded (dA-dT)n oligomers. Mol Pharmacol. 1984 May;25(3):452–458. [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Nature. 1986 May 22;321(6068):449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- Jayaram B., Sharp K. A., Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989 May;28(5):975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- Kissinger K., Krowicki K., Dabrowiak J. C., Lown J. W. Molecular recognition between oligopeptides and nucleic acids. Monocationic imidazole lexitropsins that display enhanced GC sequence dependent DNA binding. Biochemistry. 1987 Sep 8;26(18):5590–5595. doi: 10.1021/bi00392a002. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985 Jun 25;183(4):553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Larsen T. A., Goodsell D. S., Cascio D., Grzeskowiak K., Dickerson R. E. The structure of DAPI bound to DNA. J Biomol Struct Dyn. 1989 Dec;7(3):477–491. doi: 10.1080/07391102.1989.10508505. [DOI] [PubMed] [Google Scholar]
- Lavery R., Zakrzewska K., Pullman B. Binding of non-intercalating antibiotics to B-DNA: a theoretical study taking into account nucleic acid flexibility. J Biomol Struct Dyn. 1986 Jun;3(6):1155–1170. doi: 10.1080/07391102.1986.10508492. [DOI] [PubMed] [Google Scholar]
- Leupin W., Chazin W. J., Hyberts S., Denny W. A., Wüthrich K. NMR studies of the complex between the decadeoxynucleotide d-(GCATTAATGC)2 and a minor-groove-binding drug. Biochemistry. 1986 Oct 7;25(20):5902–5910. doi: 10.1021/bi00368a010. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Origins of netropsin binding affinity and specificity: correlations of thermodynamic and structural data. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4359–4363. doi: 10.1073/pnas.84.13.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J. A., Palecek E., Lilley D. M. (A-T)n tracts embedded in random sequence DNA--formation of a structure which is chemically reactive and torsionally deformable. Nucleic Acids Res. 1986 Dec 9;14(23):9291–9309. doi: 10.1093/nar/14.23.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L. H., Skelly J. V., Hudson B. D., Neidle S. The crystal structure of the DNA-binding drug berenil: molecular modelling studies of berenil-DNA complexes. Nucleic Acids Res. 1987 Apr 24;15(8):3469–3478. doi: 10.1093/nar/15.8.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peticolas W. L., Wang Y., Thomas G. A. Some rules for predicting the base-sequence dependence of DNA conformation. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2579–2583. doi: 10.1073/pnas.85.8.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjura P. E., Grzeskowiak K., Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol. 1987 Sep 20;197(2):257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Comparison of binding sites in DNA for berenil, netropsin and distamycin. A footprinting study. Eur J Biochem. 1987 Sep 1;167(2):281–289. doi: 10.1111/j.1432-1033.1987.tb13334.x. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Hydroxyl radical footprinting of the sequence-selective binding of netropsin and distamycin to DNA. FEBS Lett. 1987 Dec 10;225(1-2):195–200. doi: 10.1016/0014-5793(87)81156-0. [DOI] [PubMed] [Google Scholar]
- Randrianarivelo M., Zakrzewska K., Pullman B. Theoretical modeling of DNA-monocationic lexitropsin complexation: influence of ligand binding on DNA curvature. J Biomol Struct Dyn. 1989 Feb;6(4):769–779. doi: 10.1080/07391102.1989.10507736. [DOI] [PubMed] [Google Scholar]
- Rehfuss R., Goodisman J., Dabrowiak J. C. Quantitative footprinting analysis. Binding to a single site. Biochemistry. 1990 Jan 23;29(3):777–781. doi: 10.1021/bi00455a027. [DOI] [PubMed] [Google Scholar]
- Sarai A., Mazur J., Nussinov R., Jernigan R. L. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989 Sep 19;28(19):7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Seibel G. L., Singh U. C., Kollman P. A. A molecular dynamics simulation of double-helical B-DNA including counterions and water. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6537–6540. doi: 10.1073/pnas.82.19.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A. R., Torres R., Clark W., Olson W. K. Base sequence effects in double helical DNA. I. Potential energy estimates of local base morphology. J Biomol Struct Dyn. 1987 Dec;5(3):459–496. doi: 10.1080/07391102.1987.10506409. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidor B., Irikura K. K., Brooks B. R., Karplus M. Dynamics of DNA oligomers. J Biomol Struct Dyn. 1983 Oct;1(1):231–252. doi: 10.1080/07391102.1983.10507437. [DOI] [PubMed] [Google Scholar]
- Tilton R. F., Jr, Weiner P. K., Kollman P. A. An analysis of the sequence dependence of the structure and energy of A- and B-DNA models using molecular mechanics. Biopolymers. 1983 Mar;22(3):969–1002. doi: 10.1002/bip.360220316. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gunsteren W. F., Berendsen H. J., Geurtsen R. G., Zwinderman H. R. A molecular dynamics computer simulation of an eight-base-pair DNA fragment in aqueous solution: comparison with experimental two-dimensional NMR data. Ann N Y Acad Sci. 1986;482:287–303. doi: 10.1111/j.1749-6632.1986.tb20962.x. [DOI] [PubMed] [Google Scholar]
- Vorlícková M., Kypr J. Conformational variability of poly(dA-dT).poly(dA-dT) and some other deoxyribonucleic acids includes a novel type of double helix. J Biomol Struct Dyn. 1985 Aug;3(1):67–83. doi: 10.1080/07391102.1985.10508399. [DOI] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Goodisman J., Dabrowiak J. C. Determination of netropsin-DNA binding constants from footprinting data. Biochemistry. 1988 Feb 23;27(4):1198–1205. doi: 10.1021/bi00404a020. [DOI] [PubMed] [Google Scholar]
- Weiner P. K., Langridge R., Blaney J. M., Schaefer R., Kollman P. A. Electrostatic potential molecular surfaces. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3754–3758. doi: 10.1073/pnas.79.12.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska K., Lavery R., Pullman B. Theoretical studies of the selective binding to DNA of two non-intercalating ligands: netropsin and SN 18071. Nucleic Acids Res. 1983 Dec 20;11(24):8825–8839. doi: 10.1093/nar/11.24.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kitzing E., Diekmann S. Molecular mechanics calculations of dA12.dT12 and of the curved molecule d(GCTCGAAAAA)4.d(TTTTTCGAGC)4. Eur Biophys J. 1987;15(1):13–26. doi: 10.1007/BF00255031. [DOI] [PubMed] [Google Scholar]



