Summary
In bacteria, many small regulatory RNAs (sRNAs) are induced in response to specific environmental signals or stresses and act by base pairing with mRNA targets to affect protein translation or mRNA stability. In Escherichia coli, the gene for the sRNA IS061/IsrA, here renamed McaS, was predicted to reside in an intergenic region between abgR, encoding a transcription regulator and ydaL, encoding a small MutS-related protein. We show that McaS is a ~95 nt transcript whose expression increases over growth, peaking in early-to-mid stationary phase, or when glucose is limiting. McaS uses three discrete single-stranded regions to regulate mRNA targets involved in various aspects of biofilm formation. McaS represses csgD, the transcription regulator of curli biogenesis and activates flhD, the master transcription regulator of flagella synthesis leading to increased motility, a process not previously reported to be regulated by sRNAs. McaS also regulates pgaA, a porin required for the export of the polysaccharide poly β-1,6 N-acetyl-D-glucosamine. Consequently, high levels of McaS result in increased biofilm formation while a strain lacking mcaS shows reduced biofilm formation. Based on our observations, we propose that, in response to limited nutrient availability, increasing levels of McaS modulate steps in the progression to a sessile lifestyle.
Keywords: small RNA, Hfq, CRP, curli, flagella
Introduction
In the last ten years it has become apparent that base-pairing small RNAs (sRNAs) regulate a number of processes in bacterial cells. These sRNAs fall into two categories, trans-encoded and cis-encoded (or antisense), based on the location of the sRNA gene relative to the target mRNA gene (reviewed in (Storz et al., 2011)). While the majority of sRNAs characterized to date are trans-encoded sRNAs, and only share limited complementarity with their targets, increasing evidence suggests there are numerous cis-encoded sRNAs, which share extensive complementarity with the gene encoded on the opposite strand (reviewed in (Thomason & Storz, 2010)). Both cis and trans-encoded sRNAs regulate gene expression by base pairing with their targets, resulting in degradation of the mRNA or occlusion of the ribosomal binding site, preventing translation of the mRNA. In some cases, base pairing with an sRNA can increase access to the ribosome binding site or increase mRNA stability. Cis-encoded sRNAs also can modulate expression of their mRNA targets by interfering with transcription initiation or inducing premature transcription termination.
Many of the E. coli trans-encoded sRNAs, which require the RNA chaperone Hfq for function, turn off the expression of proteins that are not needed under a particular stress condition. For example, anaerobic induction of FnrS blocks the expression of proteins not required for growth under limiting oxygen conditions (Durand & Storz, 2010, Boysen et al., 2010), while GcvB, whose expression is high in rich media, represses the synthesis of amino acid uptake systems that are superfluous when nutrients are plentiful (Urbanowski et al., 2000, Sharma et al., 2007). The action of some trans-encoded sRNAs also leads to increased synthesis of beneficial proteins. For instance, DsrA, RprA and ArcZ increase translation of the stationary phase sigma factor σS in response to various stresses (Majdalani et al., 1998, Majdalani et al., 2002, Mandin & Gottesman, 2010).
A few sRNAs regulate targets connected to bacterial group behavior, including the sensing of bacterial density, termed quorum sensing, and the formation of biofilms, structurally complex communities of bacteria attached to biotic or abiotic surfaces. For example, in E. coli, CyaR negatively regulates luxS, encoding the autoinducer-2 synthase, resulting in decreased levels of the quorum signal molecule (De Lay & Gottesman, 2009). In Vibrio cholerae, the homologous Qrr sRNAs repress the synthesis of HapR, the master transcription regulator of quorum sensing, thus impacting expression of virulence and biofilm-associated genes (Lenz et al., 2004). Recently, two E. coli sRNAs, OmrA and OmrB, were found to repress the synthesis of CsgD, the transcription regulator of curli biogenesis (Holmqvist et al., 2010). Curli are extracellular proteinaceous structures extending from the surface of enteric bacteria that are used for attachment during biofilm formation (reviewed in (Barnhart & Chapman, 2006)). CsgD activates the csgBAC operon, encoding the structural components of curli filaments and adrA, encoding a diguanylate cyclase involved in regulating cellulose synthesis (Brombacher et al., 2003, Hammar et al., 1995, Zogaj et al., 2001). Synthesis of CsgD is modulated by multiple transcription factors, the stationary phase sigma factor σS, variations in c-di-GMP levels, in addition to the aforementioned OmrA and OmrB sRNAs (Pesavento et al., 2008, Ogasawara et al., 2010, Holmqvist et al., 2010). Here we describe another E. coli small RNA that regulates CsgD as well as other aspects of biofilm formation.
Interestingly, curli biogenesis, controlled by CsgD, is inversely correlated with flagellar synthesis. This inverse expression occurs by competition for core RNA polymerase, direct regulation of flagellar genes by CsgD, and opposing responses to low and high c-di-GMP levels (high levels repress motility) (Ogasawara et al., 2011, Pesavento et al., 2008). Much of this regulation is exerted on FlhD2C2, the master transcription regulator of flagellar synthesis. FlhD2C2 activates expression of a cascade of ~50 genes involved in motility and chemotaxis, including fliE and fliFGHIJK, two operons known to be repressed by CsgD, and yhjI, a gene encoding a phosphodiesterase that modulates c-di-GMP levels (Soutourina & Bertin, 2003, Ogasawara et al., 2011, Pesavento et al., 2008).
Extracellular structures like curli and flagella have been shown to be required for biofilm formation in enteric bacteria. However, to generate the mushroom shaped architecture associated with mature biofilms, an extracellular matrix comprised of proteins, nucleic acids and exopolysaccharides is also needed (reviewed in (Lopez et al., 2010, O’Toole et al., 2000, Stoodley et al., 2002). Different E. coli isolates synthesize various exopolysaccharides including cellulose (Bokranz et al., 2005), lipopolysaccharides (LPS), K antigen (reviewed in (Whitfield, 2006)), colanic acid (reviewed in (Majdalani & Gottesman, 2005)) and poly β-1,6-N acetyl D-glucosamine (PGA). PGA is a cell bound exopolysaccharide adhesin that is synthesized by the pgaB and pgaC-encoded enzymes and is exported through the pgaA-encoded porin (Wang et al., 2004).
The decision to maintain a planktonic lifestyle or to commit to the formation of a biofilm impacts the synthesis of several complex molecules such as curli, flagella and PGA. Not surprisingly, the decision is regulated on multiple levels and can vary depending on strain background and environmental signals. We report that an sRNA, IS061/IsrA, initially predicted in a computational screen (Chen et al., 2002), regulates three aspects of biofilm formation in E. coli and is thus renamed McaS (multi-cellular adhesive sRNA). McaS directly base pairs with the 5′ UTR of the csgD mRNA and represses CsgD expression while activating translation of FlhD and PgaA. We propose McaS thus helps regulate the progression from a planktonic lifestyle to a sessile state in a biofilm and/or shifts the timing of the transition.
Results
McaS RNA induction during the transition into stationary phase
The E. coli McaS (IS061/IsrA) RNA was previously predicted in a search for orphan promoter and terminator sequences and suggested to be ~158 nt in length (Chen et al., 2002). The sRNA is encoded in an intergenic region between the gene encoding a LysR-type transcription regulator, AbgR, a regulator of the divergently encoded p-aminobenzoyl glutamate catabolism operon abgABT (Hussein et al., 1998), and a gene encoding a small MutS-related protein, YdaL (Gui et al., 2011) (Fig. 1A and 1B). Based on sequence conservation, McaS is present in other E. coli and Shigella strains, but is absent from most enteric bacteria such as Salmonella (Zhang et al., 2003) (data not shown).
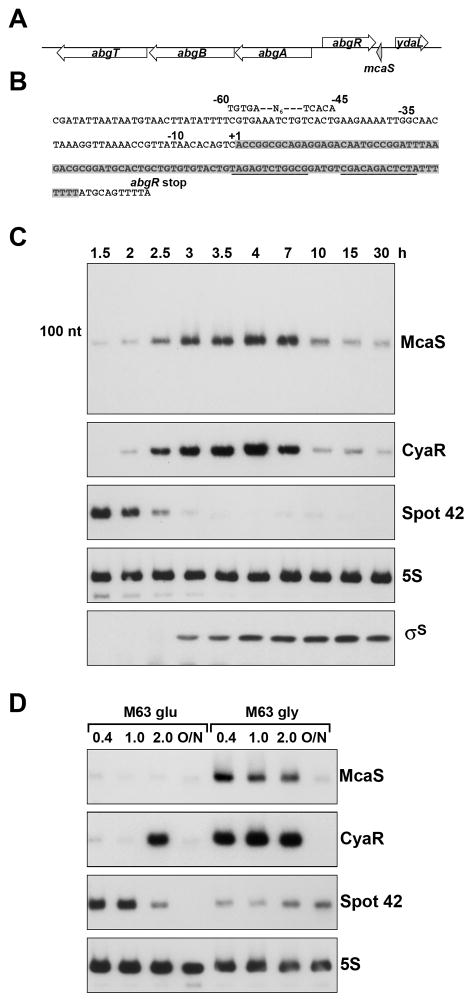
Fig. 1. McaS expression is induced during entry into stationary phase and by non-preferred carbon sources.
A. Genomic context of McaS. The gene encoding McaS (shaded grey) is located downstream of abgR, encoding a transcription regulator and upstream of ydaL, encoding a small MutS-related protein.
B. Sequence of McaS. The sequence of the sRNA is shaded grey. The RNA terminates within the poly(T) stretch of the rho-independent terminator (underlined sequence). The −10 and −35 promoter elements are indicated along with a putative CRP binding site. The abgR stop codon is also indicated.
C. Levels of the McaS, CyaR and Spot 42 RNAs and σS throughout growth. McaS, CyaR and Spot 42 levels were assayed by northern analysis of RNA taken from wild type MG1655 grown in LB media at 37°C for 30 h. At the indicated times (corresponding to OD600 of ~0.1, 0.2, 0.6, 0.9, 1.8, 2.5, 4.7, 5.5, 6, and 6), total RNA was extracted and 10 μg was separated on an 8% polyacrylamide-7M urea gel, transferred to a membrane and probed with a 32P-labeled oligonucleotide specific for McaS, CyaR, Spot 42 or 5S as a control. σS levels were assayed by western blot analysis of cells simultaneously.
D. Levels of McaS, CyaR and Spot 42 under conditions of carbon limitation. Wild type cells were grown in M63 minimal media with 0.2% glucose or 0.4% glycerol at 37°C for 30 h (O/N). RNA was extracted at the indicated OD600 and northern analysis was performed as in C. Upon longer exposure, some McaS expression is observed for cells grown in minimal media with glucose.
To validate McaS expression, we probed total RNA isolated from wild type MG1655 grown in LB media over a period of 30 h (Fig. 1C and Fig. S1). We detected a single predominant band of ~95 nt that was low during exponential phase and increased during the transition from exponential to stationary phase. Maximal McaS expression was reached around ~4–7 h of growth (OD600 ~2.5–4.0) with levels decreasing as the cells entered late stationary phase at ~30 h. A strain carrying 227 nt of the McaS promoter fused to a lacZ reporter (PmcaS(227)-lacZ) also showed an increase in β-galactosidase activity during the transition into stationary phase (Fig. S2). However, the activity did not decrease later in stationary phase, most likely due to the stability of the β-galactosidase protein.
End mapping by RACE analysis showed McaS has a single 5′ end (Fig. 1B indicated by the +1, Table S1) and terminates within the poly-T stretch of the rho-independent terminator, ~8 nt from the abgR stop codon (Table S1). These boundaries correspond to the ~95 nt transcript detected by northern analysis. McaS is not derived from a longer RNA, since the ~95 nt transcript also was detected by northern analysis of samples treated with terminator 5′ phosphate-dependent exonuclease, which preferentially degrades processed transcripts (Sharma et al., 2010) (Fig. 1B and data not shown). We were not able to detect the predicted ~158 nt RNA by northern analysis, even using oligonucleotides specific to the previously predicted 5′ end (data not shown). It is possible a longer RNA is transcribed from an upstream promoter under different conditions.
The proximity and orientation of mcaS to the upstream gene, abgR, is reminiscent of the location and orientation of gadY to gadXW, where the antisense GadY RNA base pairs with the gadXW transcript resulting in cleavage of the dicistronic message and accumulation of the gadX and gadW mRNAs (Opdyke et al., 2004, Opdyke et al., 2011). To determine if McaS is antisense to abgR, we placed abgR under the control of the PBAD promoter on the chromosome. The abgR 3′ ends in this strain mapped to multiple sites, some of which overlap mcaS sequences (Fig. S3). However, we did not observe an effect of increased or decreased McaS expression on the levels of the abgR mRNA under the conditions tested (data not shown).
CRP-dependent induction of McaS
Along with the σ70 promoter elements for McaS, we were able to identify a putative binding site for the transcription regulator CRP (cAMP receptor protein) upstream of the mapped McaS 5′ end (Fig. 1B). In E. coli, CRP is the global regulator of catabolite repression and previously was shown to negatively regulate Spot 42 RNA and positively regulate CyaR RNA expression (De Lay & Gottesman, 2009, Johansen et al., 2008, Polayes et al., 1988, Papenfort et al., 2008). To determine if McaS is also subject to catabolite repression, wild type MG1655 was grown in M63 minimal media containing either glucose or glycerol (Fig. 1D). Expression of McaS decreased when glucose was the sole carbon source compared to glycerol. The same membrane was probed for Spot 42 and CyaR, confirming that Spot 42 levels decrease and CyaR levels increase under glucose limiting conditions. We did see an increase in CyaR, but not McaS expression, in the presence of glucose as the cells reached an OD600 of ~2.0.
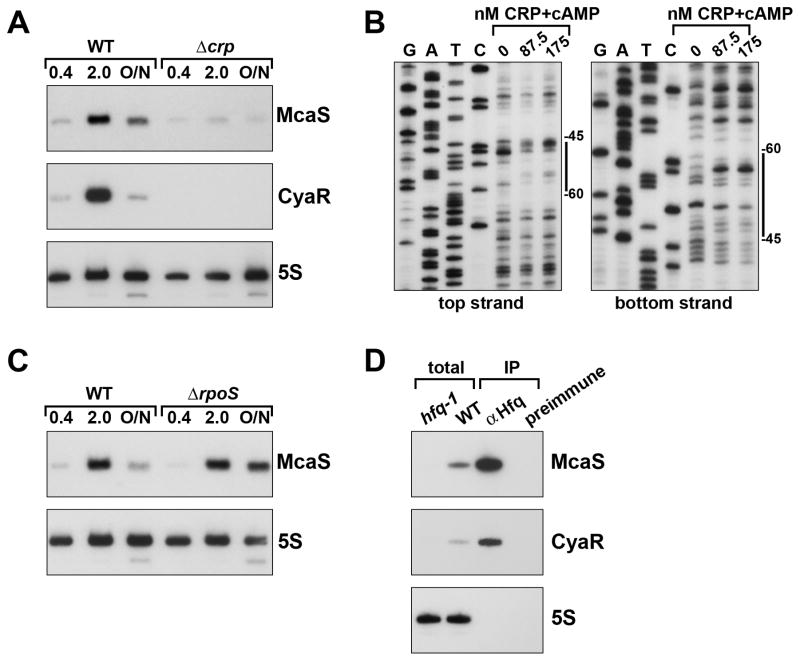
To further test the role of CRP in regulating mcaS, we created a Δcrp mutant strain and examined the levels of both McaS and CyaR by northern analysis. The amounts of both sRNAs were decreased relative to the wild type strain (Fig. 2A), although expression of McaS was not completely abolished in the Δcrp mutant. Expression of the PmcaS(227)-lacZ fusion was also reduced in the Δcrp deletion (Fig. S2). To test CRP binding to the mcaS promoter, a fragment of the promoter, radiolabeled on either the top or bottom strand, was incubated with increasing amounts of purified CRP with cAMP. Following DNase I digestion in the presence of CRP, altered cleavage patterns are evident on both strands in the region of the fragment corresponding to the predicted CRP binding site (Fig. 2B).
Fig. 2. McaS levels are regulated by CRP and Hfq.
A. Effect of Δcrp on McaS and CyaR levels. Wild type MG1655 and the isogenic Δcrp::cat mutant (GSO549), were grown for 20 h at 37°C in LB media. At the indicated OD600 samples were taken and RNA was processed for northern analysis as in Fig. 1.
B. DNase I footprinting of CRP binding at the mcaS promoter. The ~300 nt fragment labeled on the 5′ end of the top and bottom strands were incubated with increasing amounts of CRP+cAMP and then cleaved with DNase I. Sequencing ladders generated with the same labeled oligonucleotides were run along side the footprinting reactions. The positions indicated are relative to the +1 of McaS transcription.
C. Effect of ΔrpoS on McaS levels. Wild type MG1655 and the isogenic ΔrpoS::kan mutant (GSO548), were grown for 20 h in LB media at 37°C. At the indicated OD600 samples were taken and RNA was processed for northern analysis as in Fig. 1.
D. Immunoprecipitation with Hfq. Cells extracts were prepared from wild type MG1655 grown in LB to early stationary phase (OD600 ~1) and subject to immunoprecipitation with α-Hfq or preimmune serum. Northern analysis was carried out on the immunoprecipitated samples (0.5 μg RNA loaded) as well as on total RNA isolated from wild type and the isogenic hfq-1 mutant (GSO550) (5 μg loaded) as in Fig. 1.
Given that McaS levels peak in stationary phase we sought to determine if expression was also dependent on σS. As shown in Fig. 2C, levels of McaS in the ΔrpoS mutant were similar to the wild type strain in exponential (OD600 ~0.4) and early stationary (OD600 ~2.0) phases. However, in late stationary phase (~20 h), expression of McaS was elevated in the ΔrpoS mutant. No difference in β-galactosidase activity was seen in the PmcaS(227)-lacZ fusion between wild type and the ΔrpoS mutant strain, indicating the apparent repressive effects of σS in late stationary phase are most likely to be indirect and may be at a post-transcriptional level (Fig. S2).
McaS binding to Hfq
Many E. coli sRNAs bind to the RNA chaperone protein Hfq and are generally unstable in the absence of Hfq (reviewed in (Vogel & Luisi, 2011)). To assess if Hfq binds McaS, lysate from wild type MG1655 was subject to immunoprecipitation with either α-Hfq or preimmune serum. Immunoprecipitated RNA (0.5 μg) as well as total RNA (5 μg) samples from wild type and hfq-1 mutant cells were then probed for McaS and the known Hfq-binding sRNA CyaR (Fig. 2D). Similar patterns of expression and enrichment are observed for McaS and CyaR. The stabilities of both sRNAs are strongly dependent on Hfq, as the sRNAs are undetectable in the hfq-1 strain. In addition, both are enriched upon α-Hfq precipitation. Together these results indicate McaS binds Hfq.
McaS repression of curli biogenesis
The binding of McaS to Hfq indicated the potential to act as a trans-encoded base pairing sRNA. The algorithm TargetRNA (Tjaden et al., 2006) predicted 16 nt of uninterrupted base pairing between McaS and the ~148 nt 5′ UTR of the transcription regulator csgD, previously found to be regulated by the OmrA and OmrB RNAs (Holmqvist et al., 2010). To determine if McaS is an additional sRNA regulator of csgD and curli production, we examined the effects of overexpressing McaS using Congo red indicator plates. Strains proficient in curli production bind the hydrophobic dye, resulting in red-colored colonies. Strains unable to produce functional curli, and thus unable to bind the dye, appear white or light pink. The mcaS gene placed under the control of an IPTG inducible promoter on the plasmid pBRplac (pBR-McaS) was transformed into wild type and ΔcsgD strains and grown on Congo red indicator plates. As shown in Fig. 3A, the wild type strain containing the control vector appeared red. In contrast, colonies from the ΔcsgD strain as well as the wild type strain with McaS overexpression were light pink, consistent with the lack of functional curli. This assay was not sufficiently sensitive to detect a difference between the wild type strain and a strain lacking mcaS (data not shown).
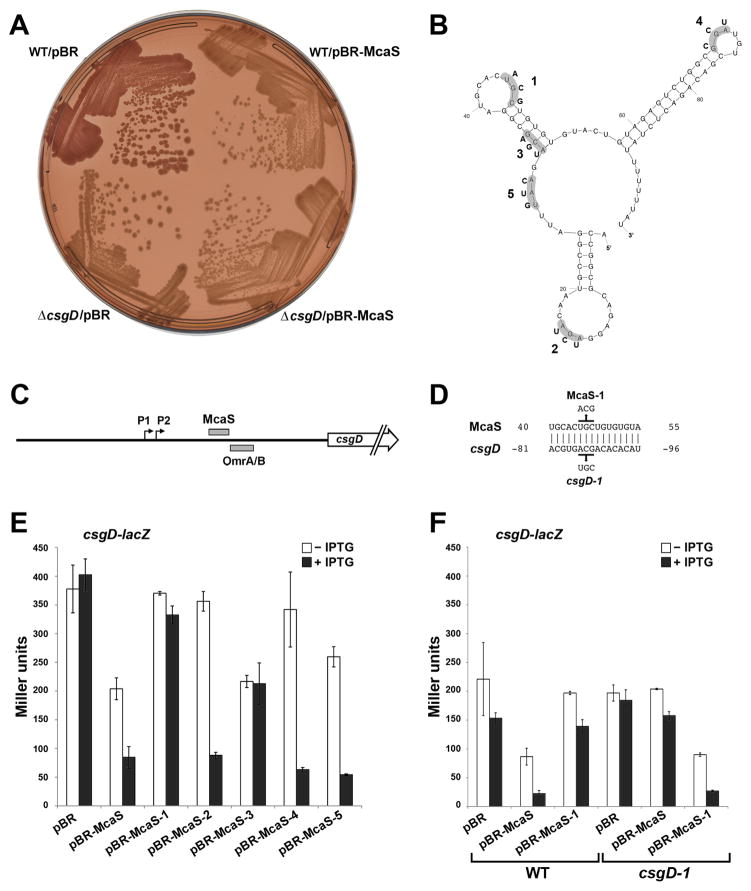
Fig. 3. McaS negatively regulates expression of the transcription regulator CsgD.
A. Congo red indicator plates show decreased curli production upon McaS overexpression. The wild type NM525 and NM525 ΔcsgD::cat (GSO554) strains were transformed with the control vector or pBR-McaS. Strains were grown on Congo red indicator plates with 1 mM IPTG for 48 h at 25°C to assay curli formation. The image shown is representative of three independent plates.
B. A predicted structure of the McaS sRNA as determined by the Mfold (Zuker, 2003) RNA folding algorithm. There is an alternative prediction for the central stem-loop structure. The bases mutated in the different McaS derivatives are indicated.
C. Schematic of the csgD 5′ UTR showing the location of the predicted McaS base pairing and the region of OmrA/B base pairing (Holmqvist et al., 2010).
D. Base pairing between McaS and csgD predicted by TargetRNA (Tjaden et al., 2006) using full length McaS and modified parameters. Residues mutated in McaS and the csgD-lacZ fusion are indicated. The numbers indicate the position relative to the start codon for csgD and +1 for McaS.
E. Negative regulation of csgD-lacZ by McaS. The reporter strain PM1205 ΔabgR-ydaL::kan csgD-lacZ (GSO560) was transformed with the control vector, pBR-McaS or plasmids containing various McaS mutant derivatives. The levels of β-galactosidase activity of the csgD-lacZ fusion was assayed after 1 h of induction with 0.2% arabinose and either 1 mM IPTG (black bars) or no IPTG (white bars). The average values from three independent assays are shown and error bars correspond to the standard deviation of those values.
F. Disruption and restoration of base pairing between McaS and csgD. Plasmids carrying wild type McaS or McaS-1 mutant derivative were transformed into strains PM1205 ΔabgR-ydaL::kan csgD-lacZ (GSO560) and PM1205 ΔabgR-ydaL::kan csgD-1-lacZ (GSO562) which carries compensatory mutations to restore base pairing with McaS-1. β-galactosidase activity was assayed as in E.
To test the predicted effect of McaS on csgD expression (Fig. 3B-D), we created a translational fusion of the entire 148 nt 5′ UTR of csgD through the 10th codon of the open reading frame fused to lacZ under the control of a PBAD promoter. A deletion of the abgR-ydaL region was also moved into this background to eliminate chromosomal McaS expression. As shown in Fig. 3E, when McaS was induced from pBR-McaS, expression from the csgD-lacZ fusion was reduced by ~4.5 fold compared to the vector control strain. There also was a ~1.9 fold reduction in csgD-lacZ expression in strains carrying pBR-McaS in the absence of IPTG, likely due to leaky expression from the Plac promoter (Fig. 3E, black versus white bars).
To determine whether there was a direct interaction between McaS and csgD, we generated a series of mutations in the single stranded regions of McaS (Fig. 3B) including McaS-1, which interrupts the predicted McaS base pairing with csgD (Fig. 3D). Northern analysis of the McaS mutants showed that the levels of McaS-3 were similar to wild type McaS, while the levels of McaS-2, McaS-4 and McaS-5 levels were slightly lower (Fig. S4). McaS-1 and McaS2+4, though detectable, were the least abundant. The McaS-1 mutant no longer repressed the csgD-lacZ fusion, restoring β-galactosidase activity to the levels with the control vector (Fig 3E). The McaS-3 mutant also showed reduced repression of the fusion in the presence of IPTG, although not to the extent of McaS-1. The effect of the mcaS-3 mutation is not completely understood, but could indicate additional base pairing at a second site in the csgD 5′ UTR or be due to alterations in the McaS structure that could impact Hfq binding or indirectly affect base pairing. The McaS-2, McaS-4 and McaS-5 mutants did not repress under non-inducing conditions, possibly due to lower basal levels, but were able to repress the csgD-lacZ fusion upon induction (Fig. 3E).
We next introduced compensatory mutations into the csgD 5′ UTR (csgD-1) to restore base pairing with the McaS-1 mutant (Fig. 3D). As shown in Fig. 3F, regulation of the mutant csgD-1-lacZ fusion was lost with wild type McaS but was restored with the McaS-1 mutant. Taken together these results indicate that McaS directly base pairs with the csgD 5′ UTR resulting in post-transcriptional repression of CsgD synthesis, ultimately leading to reduced curli formation.
McaS activation of flagella synthesis
Given the inverse regulation of genes involved in curli biogenesis and flagellar synthesis (Pesavento et al., 2008, Ogasawara et al., 2011), we wondered if McaS also could affect flagellar synthesis, and thus motility. We assessed the effects of overexpressing McaS on the ability of wild type and a strain lacking the master flagellar regulator, FlhD2C2, to swim using motility swim agar plates (Fig. 4A). As expected, the ΔflhDC strain showed no motility. In contrast, overexpression of McaS resulted in a clear increase in motility compared to the control vector-containing strain at both 25°C (32 mm diameter compared to 11 mm as shown in Fig. 4A) and 37°C (data not shown). However, no difference in motility was observed between a wild type and a ΔabgR-yadL strain lacking McaS at either temperature (data not shown). We also examined a ΔabgR-yadL strain carrying the control vector or pBR-McaS by electron microscopy. As shown in representative images in Fig. 4B, under the conditions tested, no flagella were visible for cells carrying the control vector (0 flagella for 15 cells), while cells overexpressing McaS all had one or more flagella (66 flagella for 26 cells).
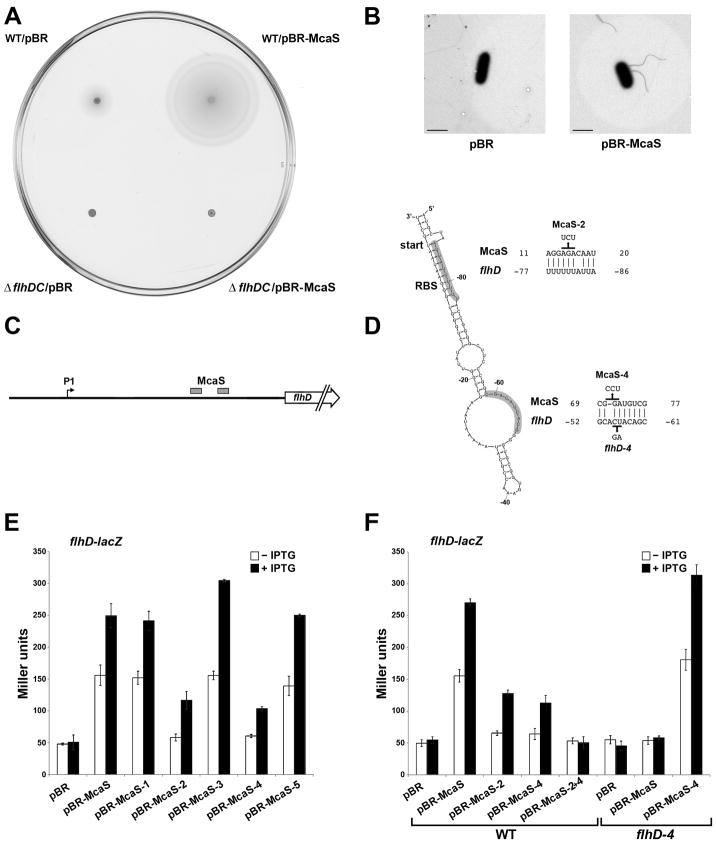
Fig. 4. McaS positively regulates expression of the transcription regulator FlhD resulting in increased motility.
A. Overexpression of McaS leads to increased motility on swim agar plates. Wild type NM525 and NM525 ΔflhDC::kan (GSO553) were transformed with the control vector or pBR-McaS and inoculated on tryptone swim agar plates containing 100 μM IPTG, which were then incubated for 16 h at 25°C. The image shown here is representative of three independent experiments.
B. Electron microscopy of flagellar staining. Strain NM525 ΔabgR-ydaL::kan (GSO552) was transformed with the control vector or pBR-McaS. Overnight cultures were diluted back in fresh LB + 1 mM IPTG and grown to an OD600 ~1.0. Cells were collected and negatively stained for flagella. The images are representative of multiple cells from two independent experiments. The bar corresponds to 2 μM. It is striking that no flagella were observed for the cells carrying pBR despite the fact that these cells were motile on swim agar plates. This discrepancy may be due to differences in culture conditions.
C. Schematic of the flhD 5′ UTR leader sequence showing two regions of predicted McaS base pairing.
D. Structure of part of the flhD 5′ UTR predicted by Mfold. Positions of the start codon and ribosome binding site (RBS) are indicated. Base pairing between McaS and flhD predicted by TargetRNA using single stranded regions of McaS as input with modified parameters and by manual inspection are shown with numbers indicating position relative to the start codon of flhD and +1 of McaS. Mutations made in either the McaS sequence or the flhD 5′ UTR are indicated.
E. Positive regulation of flhD-lacZ by McaS. The reporter strain PM1205 ΔabgR-ydaL::kan flhD-lacZ (GSO564) was transformed with the control vector, pBR-McaS or plasmids containing various McaS mutant derivatives.
F. Disruption and restoration of base pairing between McaS and flhD. Plasmids carrying wild type McaS and mutant McaS-2, McaS-4, and McaS-2+4 derivatives were transformed into strain PM1205 ΔabgR-ydaL::kan flhD-lacZ (GSO564) and plasmids carrying wild type McaS and mutant McaS-4 were transformed into strain PM1205 ΔabgR-ydaL::kan flhD-4-lacZ (GSO566) which carries compensatory mutations to restore base pairing with McaS-4. β-galactosidase activity in both D and E was assayed as in Fig. 3.
We predicted the increase in motility and flagella synthesis might be due to McaS effects on FlhD2C2 synthesis. The flhDC mRNA contains a long 5′ UTR with a σ70 promoter residing 198 nt upstream of the start codon (indicated in Fig. 4C) (Soutourina et al., 1999). The Mfold algorithm (Zuker, 2003) predicted a highly base paired secondary structure for this UTR in which the ribosome binding site is sequestered by a hairpin (Fig. 4D). This hairpin could prevent access to the ribosome binding site and reduce translation of the FlhD protein in a manner similar to the inhibitory structure of the 5′ UTR of the DsrA, RprA and ArcZ target rpoS (Majdalani et al., 1998, Soper et al., 2010),
Using the single stranded regions of McaS in TargetRNA together with manual scanning, we predicted two regions of base pairing between McaS and the flhD leader; one site ~60 nt from the start codon and a second site that maps opposite the ribosome binding site in the predicted secondary structure (Fig. 4D). We hypothesized that McaS could base pair with the flhD 5′ UTR in both places, relieving the secondary structure around the ribosome binding site, resulting in increased translation of the FlhD and FlhC proteins.
To test this hypothesis, we created an flhD-lacZ translational fusion incorporating the entire 198 nt 5′ UTR through the 10th codon fused to lacZ under the control of the PBAD promoter in the ΔabgR-ydaL background. Upon induction of both McaS and the flhD-lacZ fusion, there was a ~5 fold increase in β-galactosidase activity with pBR-McaS compared to the control vector strain (Fig. 4E). This increase is consistent with the observed increase in motility and visible flagella. We also observed ~1.4 fold higher levels of flhD-lacZ expression in a wild type background compared to a ΔabgR-yadL strain (Fig. S5) documenting that endogenous levels of McaS are sufficient to activate flhD expression.
We then assayed the flhD-lacZ fusion in strains expressing the McaS mutants, among which McaS-2 and McaS-4 were predicted to disrupt base pairing (Fig. 4D). When the two regions of McaS were mutated independently, activation of the flhD-lacZ fusion was reduced, resulting in only ~2 fold induction compared to wild type McaS (Fig. 4E). However, with a McaS double mutant (McaS-2+4), flhD-lacZ activation was completely eliminated, indicating both regions may be involved in McaS regulation of the flhD-lacZ fusion (Fig. 4F). In contrast, the McaS-1, McaS-3 and McaS-5 mutants all gave wild type levels of induction.
Compensatory mutations in the 5′ UTR of the flhD leader (flhD-4) that could restore base pairing with the McaS-4 mutant restored activation of flhD by McaS-4 but not by wild type McaS (Fig. 4F). These results indicate that McaS acts as a positive regulator of flhD by base pairing with two potential regions in the flhD leader sequence, which, in the absence of McaS sequesters the ribosome binding site, preventing translation.
Cell autoaggregation associated with elevated levels of McaS
Overexpression of an sRNA has frequently been used to identify phenotypes that give insights into the function of the sRNA (reviewed in (Storz et al., 2011)). To that end, we cloned mcaS behind the strong Ptac promoter on the multi-copy plasmid pRI. Intriguingly, we observed a distinctive aggregative phenotype in overnight cultures of strains carrying pRI-McaS (Fig. 5A). The cells appeared to stick to the walls of the culture tubes and, in the absence of shaking, immediately sedimented to the bottom of the tube. Curli and the auto-transporter adhesin Antigen-43 have been implicated in the autoaggregation of cells (Henderson et al., 1997, Vidal et al., 1998). However, deletion of neither csgD nor the flu gene, encoding Antigen-43, affected McaS-dependent autoaggregation (data not shown). It was recently reported that reduced levels of the RNA binding protein, CsrA, resulted in autoaggregation in a manner that was dependent on the polysaccharide adhesin PGA (Jin et al., 2009). Deletion of the pgaA gene, encoding the porin through which PGA is excreted (Wang et al., 2004), abolished the McaS-dependent autoaggregation phenotype (Fig. 5A), suggesting that pgaA might be a target of McaS.
Fig. 5. McaS positively regulates expression of the outer membrane porin PgaA.
A. McaS-induced autoaggregation of cells is dependent on pgaA. Wild type MG1655 or ΔpgaA::kan (GSO551) cells overexpressing McaS were grown in LB at 37°C for 20 h with shaking. The cells overexpressing McaS in the wild type pgaA background stick to the walls and sediment to the bottom of the tube in the absence of shaking.
B. Schematic of the pgaA 5′ UTR showing the region of predicted base pairing between the single stranded region in McaS mutated in McaS-3 and 100 nt upstream of the pgaA start codon.
C. Structure of a portion of the pgaA 5′ UTR predicted by Mfold. Positions of the start codon and ribosome binding site (RBS) are indicated. Base pairing between McaS and pgaA predicted by TargetRNA using single stranded regions of McaS and by manual inspection are shown with numbers indicating position relative to the start codon of pgaA and +1 of McaS. Mutations made in the McaS sequence are indicated.
D. Positive regulation of pgaA-lacZ by McaS. The reporter strain PM1205 ΔabgR-ydaL::kan, pgaA-lacZ (GSO568) was transformed with the control vector, pBR-McaS or plasmids containing various McaS mutant derivatives. β-galactosidase activity was assayed as in Fig. 3.
Again, using the single stranded regions of McaS in TargetRNA and manual scanning, we identified several regions of possible complementarity between McaS and multiple sites in the ~234 nt long 5′ UTR of the pgaABCD operon (Fig. 5B, 5C). To test for McaS regulation of pgaA and possible base pairing with the 5′ UTR, we generated a pgaA-lacZ fusion. Expression of wild type McaS resulted in an ~18 fold increase in β-galactosidase activity compared to the control vector (Fig. 5D). Induction was almost completely lost with the McaS-2 and McaS-5 mutants, consistent with the possibility that the McaS regions affected by these mutations are involved in the predicted pairing with the pgaA 5′ UTR. We also did not observe induction with the McaS-1 mutant. This could be due to base pairing that we have not yet predicted or the reduced levels of this mutant RNA (Fig. S4). Expression was somewhat reduced for the McaS-4 mutant, although not as drastically as for McaS-1, McaS-2 and McaS-5. Surprisingly, induction of the McaS-3 mutant, which, although mutated, is still predicted to pair with the sequences ~100 nt upstream of the start codon, up regulated the pgaA-lacZ fusion by ~59 fold compared to the control vector and ~3 fold compared to wild type McaS (Fig. 5D). We interpret this increase in activation to be the result of mutated residues disrupting the central hairpin structure of McaS allowing for more efficient base-pairing with pgaA by the single stranded regions of McaS. We note that no single compensatory mutation restored the expected regulation (data not shown). Conceivably, McaS acts directly through base pairing with multiple sites in the pgaA 5′ UTR. It is also possible that McaS acts indirectly by binding CsrA via GGA sequences present on McaS, thus relieving CsrA repression of pgaA. Regardless, we propose that McaS promotes a change in the structure of the leader, which leads to increased ribosome access and PgaA translation, which in turn results in increased export of PGA and autoaggregation.
Dependence of target regulation on McaS levels
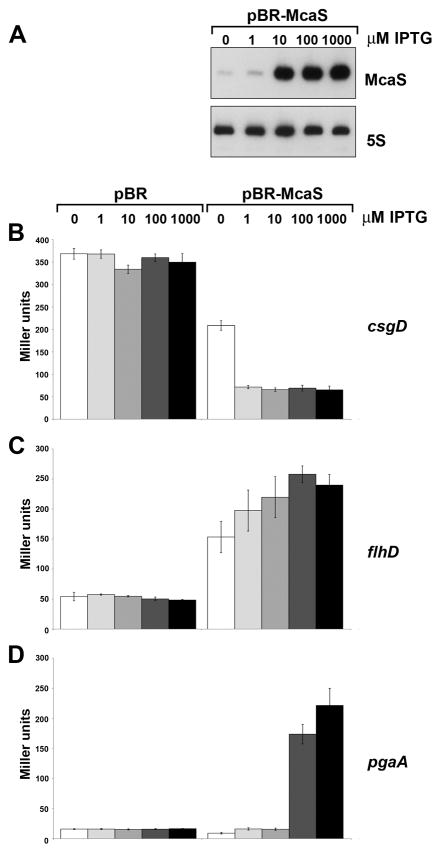
We suspected the three McaS targets might be differentially sensitive to McaS levels since cell aggregation was only observed when McaS was placed under the control of the strong Ptac promoter on pRI or when induced with high levels of IPTG from the Plac promoter on pBR. Additionally, leaky expression of McaS from the Plac promoter on pBR-McaS in the absence of IPTG, affected expression of some target lacZ fusions (csgD and flhD) but not others (pgaA). Quantitation of McaS expression by northern analysis showed that the levels of leaky McaS expression from pBR-McaS in the absence of IPTG were approximately equivalent to McaS levels in wild type cells grown in LB for 4 h (Fig. S6). Induction with 1000 μM IPTG for 1 or 4 h resulted in ~150-fold higher levels and induction for 15 h led to ~500-fold higher levels compared to the chromosomally-expressed McaS. For comparison, the McaS levels from pRI-McaS were ~3.5-fold higher than the highest levels detected for pBR-McaS (Fig. S6).
We systematically examined the effects of different McaS levels by inducing sRNA synthesis from pBR-McaS with different concentrations of IPTG in a ΔabgR-ydaL background (Fig. 6A). We found that McaS-mediated repression of the csgD-lacZ fusion can occur equally well when low levels (1 μM) or high levels (1000 μM) of IPTG are used (Fig. 6B). Similarly, even lower levels of McaS activated the flhD-lacZ fusion, as regulation occurred even with leaky McaS expression in the absence of IPTG (Fig. 6C). In contrast, strong activation of the pgaA-lacZ fusion was only seen with concentrations of IPTG above 100 μM (Fig. 6D). We note, however, that endogenous levels of McaS can impact pgaA, given that we consistently observed slightly higher pgaA-lacZ expression in the wild type strain compared to the ΔabgR-ydaL deletion strain (Fig. S5).
Fig. 6. McaS targets display varying sensitivity to McaS levels.
A. McaS levels induced with different IPTG concentrations from pBR-McaS. Overnight culture of strain NM525 ΔabgR-ydaL::kan (GSO552) with the pBR-McaS plasmid was diluted to an OD600 ~0.05 in fresh LB + amp and grown at 37°C for 1.5 h. McaS expression was induced for 1 h with the indicated final concentrations of IPTG. Cells were collected, RNA extracted and northern analysis performed as in Fig. 1.
B.–D. Varied sensitivity of (B) csgD-lacZ, (C) flhD-lacZ, and (D) pgaA-lacZ expression to different McaS levels. The reporter strains PM1205 ΔabgR-ydaL::kan csgD-lacZ (GSO562), PM1205 ΔabgR-ydaL::kan flhD-lacZ (GSO564), or PM1205 ΔabgR-ydaL::kan pgaA-lacZ (GSO568) were transformed with vectors pBR and pBR-McaS. β-galactosidase activity was assayed as in Fig. 3 except that the indicated final concentrations of IPTG were used.
Effect of McaS on biofilm formation
Given that McaS regulates the synthesis of curli, flagella and a polysaccharide, three structures involved in biofilm formation, we next tested whether McaS impacted this group behavior. We examined the biofilm formed in 96-well microtiter plates for strains carrying either pBR-McaS or the control vector grown in LB, colonization factor antigen (CFA), and YESCA medium at 25°C and 37°C in the presence of increasing IPTG. After incubation, planktonic cells were removed by washing with ddH2O and any attached cells retained in a biofilm were stained with crystal violet and quantitated by measuring OD570 and normalizing to OD600 (Wu & Outten, 2009). In the wild type background, cells carrying the control vector showed little biofilm formation. In contrast, McaS overexpression resulted in extensive biofilm formation in a manner dependent on the amount of McaS expressed (Fig. 7 and Fig. S7). The strongest effects were observed for cells grown in LB medium at 25°C (Fig. 7A). Minimal biofilm was seen in the absence of IPTG or with 1 μM IPTG. However, when the concentration of IPTG was increased to 10 μM, the amount of biofilm formation increased ~80 fold compared to the control vector strain. Higher concentrations of IPTG (100 μM, 1000 μM) led to even higher levels of biofilm formation, ~100 fold more than the control strain. Thus, increased expression of McaS correlates with increased biofilm formation.
Fig. 7. Biofilm formation is affected by different levels of McaS expression.
A. Growth assay shows increased biofilm formation in the presence of McaS overexpression. Overnight cultures of strains NM525, NM525 ΔcsgD::cat (GSO554), NM525 ΔflhDC::kan (GSO553) and NM525 ΔpgaA::kan (GSO570) with the indicated plasmids were diluted to an OD600 ~0.05 into 200 μl of fresh LB + amp + IPTG and grown at 25°C for 24 h in a 96-well microtiter plate. Planktonic cells were washed away and the amount of biofilm retained in the wells was determined by crystal violet staining and measuring absorbance at OD570 and normalizingby OD600. The image is representative of three independent experiments.
B. Growth assay shows decreased biofilm formation in strains lacking mcaS. Four independent cultures of wild type MG1655 or the isogenic ΔabgR-ydaL::kan mutant (GSO569) lacking chromosomal McaS were grown overnight in LB at 37°C. The cultures were diluted to an OD600 ~0.05 in 200 μl fresh LB, CFA or YESCA media and grown at 37°C for 24 h in a 96-well microtiter plate. The amount of biofilm formation was determined as in A. The image is representative of three independent experiments.
We sought to determine if the effects of McaS overexpression on biofilm formation were dependent on one particular McaS target and thus also examined biofilm levels for strains carrying individual ΔcsgD, ΔflhDC and ΔpgaA deletions in LB at 25°C. Biofilm formation upon McaS overexpression was not affected by deletion of csgD or flhDC, consistent with the hypothesis that the two different extracellular structures can compensate for one another. In contrast, biofilm formation was significantly reduced in the ΔpgaA deletion strain indicating that McaS activation of pgaA expression is particularly critical for this phenotype.
We also tested if endogenous levels of McaS had an affect on biofilm formation under all conditions assayed above. Despite the fact that the biofilm levels are much lower, we observed an ~3.5 fold decrease in biofilm formation in the CFA medium at 37°C for the ΔabgR-ydaL deletion strain compared to wild type. These results indicate that chromosomally-expressed McaS modulates biofilm formation under some growth conditions.
Discussion
The McaS sRNA was originally identified in a computational screen for orphan promoter and terminator sequences (Chen et al., 2002). In this work, we document that McaS is a primary transcript of ~95 nt whose expression is induced by non-preferred carbon sources and entry into stationary phase. The McaS RNA overlaps the 3′ UTR of abgR, the upstream gene, suggesting it could act as a cis-encoded sRNA. However, here we show that McaS functions as a trans-encoded base pairing sRNA that binds to the RNA chaperone Hfq and regulates expression of the global transcription regulators CsgD and FlhD and an outer membrane porin, PgaA.
CRP-dependent regulation of McaS
Expression of McaS increases under limiting nutrient conditions. Its regulation is partially dependent on σS in a manner that is not understood, as well as on the catabolite repressor protein CRP. The CRP binding site identified in this promoter region is conserved across 36 E. coli and Shigella strains examined, yet the distance between the CRP binding site and start of mcaS transcription is sub-optimal, falling between that normally found for class I and class II CRP-dependent promoters. Nevertheless, the conservation of the CRP binding site, the repression of McaS in the presence of glucose and the DNase I footprinting data provide evidence for direct CRP regulation of McaS.
In vitro studies indicated CRP also directly regulates two of the three targets of McaS: csgD and flhD (Soutourina et al., 1999, Zhao et al., 2007, Zheng et al., 2004). McaS thus has the potential to participate in two distinct regulatory circuits in conjunction with CRP. The CRP-McaS-csgD circuit forms an incoherent feedforward loop where CRP and McaS act in opposition (CRP activates csgD and McaS while McaS represses csgD), while the regulatory circuit formed by CRP-McaS- flhD is a coherent feedforward loop (CRP activates both mcaS and flhD while McaS activates flhD). The potential benefits of the two types of feedforward loops in regulating motility and biofilm formation are not yet known. A search of the pgaA promoter did not reveal a CRP binding site. However, it is conceivable that the reduced biofilm formation observed in several E. coli K-12 strains in the presence of glucose and in a CRP mutant strain (Jackson et al., 2002) might be the indirect result of decreased McaS activation of flhD and pgaA.
Overall the expression of McaS is similar to that of the CRP-activated CyaR sRNA but opposite that of the CRP-repressed sRNA Spot 42 (De Lay & Gottesman, 2009, Polayes et al., 1988). Nevertheless, some differences in McaS and CyaR expression were noted. CyaR levels are elevated in the presence of glucose during early stationary phase while the levels of McaS remain consistently low (Fig. 1D). Additionally, CyaR was not detected in the Δcrp mutant while there is still residual expression of McaS (Fig. 2A). CyaR, which modulates group behavior through repression of luxS, is conserved across a wide range of enteric bacteria, while McaS is found in fewer species and notably is absent from Salmonella. Although there is no obvious sequence similarity between CyaR and McaS, it will be interesting to explore whether CyaR and McaS have overlapping roles in the cell as has been found for the two σE–regulated sRNAs, MicA and RybB (Gogol et al., 2011), and whether CyaR can compensate for McaS in species that lack the second CRP-activated sRNA.
Deletion of crp did not completely abolish McaS expression, indicating the potential for an additional regulator. We considered a number of possibilities including the divergently-encoded AbgR (Hussein et al., 1998), the McaS target CsgD, and MlrA, a regulator of csgD (Brown et al., 2001). However, northern analysis showed individual deletions of abgR, csgD and mlrA did not alter McaS expression (data not shown). We note that Jørgensen et al. found that McaS was not specifically induced in a cya strain defective in catabolite control (Jørgensen et al., 2011). This difference between our results might be due to differences in an additional regulator(s) in our respective strain backgrounds.
Role of multiple single stranded regions in McaS base pairing with mRNA targets
The three McaS targets we have identified share a number of characteristics. Two of the targets encode highly regulated global transcription regulators (CsgD and FlhD) that initiate cascades of gene expression impacting multi-cellular behavior of E. coli. The regulation of both csgD and flhD is complex with multiple transcription factors modulating expression from these genes. At the post-transcriptional level, both flhD and pgaA are regulated by the carbon storage protein, CsrA, though in opposite ways. CsrA binds to the pgaA mRNA to repress translation of PgaA (in opposition to McaS) and to the flhD mRNA to increase expression of FlhD (in conjunction with McaS) (Wang et al., 2005, Wei et al., 2001). The 5′ UTRs of csgD, flhD and pgaA also are all unusually long, 148, 198 and 234 nt, respectively. For each of these 5′ UTRs, McaS base pairs well upstream of the ribosome binding site and start codon, unlike the case for a majority of base pairing sRNA, which base pair at or near the ribosome binding site. We propose that while McaS regulates csgD through one region of base pairing and flhD through two regions of base pairing, base pairing with the pgaA leader may be more complex, potentially involving multiple sequences in the 5′ UTR.
A 16 nt region of base pairing between McaS and the csgD mRNA appears to be responsible for repression. This base pairing is direct, as mutating residues in McaS or the csgD mRNA disrupts the regulation (Fig. 3D–F). The region of base pairing is only 1 nt away from the previously reported region of OmrA/B interaction with the csgD leader (Holmqvist et al., 2010). OmrA/B were shown to decrease translation of CsgD by limiting access of the ribosome to the translation initiation region through an unknown mechanism that does not involve occlusion of the ribosome binding site. As the binding sites for McaS and OmrA/B are located in such close proximity, it could be that McaS represses CsgD synthesis in a manner similar to OmrA/B. Jorgensen et al. have proposed that McaS base pairs with an additional region of csgD closer to the site of translation initiation, which may directly block ribosome binding (Jørgensen et al., 2011). Intriguingly, these authors showed that CsgD synthesis is also repressed by two additional sRNAs, GcvB and RprA, which are predicted to pair in the same regions as McaS.
While McaS represses synthesis of one transcription factor, CsgD, it increases synthesis of another transcription factor, FlhD and also likely the FlhC transcription factor encoded on the same transcript. The 5′ UTR of the flhD mRNA is predicted to be highly structured with the ribosome binding site occluded by a long stem loop (Fig. 4D). We propose that pairing of McaS with two regions of the flhD mRNA leads to increased translation by relieving the secondary structure around the ribosome binding site. We hypothesize that the first region of base pairing between McaS and flhD is within a single stranded region, where an initial kissing complex may form between the loop of flhD and the terminator loop of McaS. Part of this flhD loop also appears to contain an ARN Hfq binding motif (Vogel & Luisi, 2011), which may facilitate the base pairing between McaS and flhD. The initial pairing may partially relieve the secondary structure of the flhD leader allowing McaS to base pair with the region opposite the ribosome binding site. Interestingly, the flhD mRNA is also repressed by multiple sRNAs, one of which, ArcZ, base pairs within the loop of the flhD mRNA overlapping the McaS binding site (N. De Lay and S. Gottesman, manuscript in preparation). This interplay between activation of flhD by McaS base pairing with two sites within the 5′ UTR, repression through additional sRNAs, and activation by CsrA further increases the complexity of the already nuanced regulation of FlhD synthesis.
McaS regulation of pgaA, which has the longest 5′ UTR of the McaS targets, is sensitive to the levels of McaS. Compared to csgD and flhD, pgaA-lacZ expression is only slightly reduced in the abgR-ydaL deletion compared to the wild type strain (Fig. S5, Jørgensen et al., 2011), and the strongest pgaA activation only occurs with high levels of McaS (Fig. 6). This requirement for high levels of McaS expression might be explained by a requirement for multiple interactions between McaS and pgaA. Pairing predictions indicate McaS has the potential to base pair with multiple regions in the pgaA 5′ UTR. Possibly, opening of the pgaA secondary structure to allow ribosome binding requires simultaneous base pairing with more than one McaS molecule. Alternatively, the secondary structure may be more recalcitrant to base pairing interactions. Interestingly, pgaA is also regulated by CsrA, which has been shown to bind to six regions within the pgaA 5′ UTR to repress PgaA translation (Wang et al., 2005). Given that McaS contains multiple GGA sequences known to be bound by CsrA, it is also possible that McaS regulates pgaA directly through base pairing and indirectly through titration of CsrA from the pgaA mRNA. Further experiments are needed to fully understand the post-transcriptional regulation of the pgaA mRNA by both McaS and CsrA. While pgaA encodes the porin for PGA export, it is probable that synthesis of the enzymes required for PGA production and encoded in the same operon is also increased.
Network inference analysis inferred that McaS (IsrA) is involved in regulating the E. coli response to DNA damaging conditions (Modi et al., 2011). Although a ΔisrA strain showed wild type sensitivity to DNA damaging agents, a ΔisrA ΔglmZ mutant was less sensitive than the wild type strain. The effects seen in the double deletion strain conceivably could result from polar effects of the ΔisrA deletion on the downstream ydaL gene. YdaL contains a Smr-like (small MutS-related) domain and was recently shown both to bind double stranded DNA and to possess endonuclease activity typical of proteins involved in DNA repair (Moreira & Philippe, 1999, Gui et al., 2011). None of the three targets confirmed in this work were identified in the network inference analysis, highlighting limitations of this global approach.
McaS modulation of biofilm formation
We observed dramatic effects of McaS on biofilm formation, a complex process that requires temporal and spatial coordination of gene expression within and between individual bacteria. Biofilm formation progresses through a number of stages. These include initial attachment of bacteria to a surface followed by more permanent attachment and the formation of the mature mushroom like architecture, which requires an extracellular matrix (reviewed in (Lopez et al., 2010, Wood, 2009)). The exact cellular requirements for biofilm formation can vary as some extracellular structures can compensate for others. For example, when cells are grown in LB, flagella reportedly are required to mediate initial attachment and spreading during biofilm formation (Pratt & Kolter, 1998). However, when a strain containing an ompR234 mutation that constitutively activates csgD expression is grown in minimal media with glucose, flagella are not required (Prigent-Combaret et al., 2000). Instead, increased curli expression is sufficient to mediate biofilm formation. The polysaccharide PGA appears to be required for even distribution of cells across the biofilm substructure as well as for the formation of permanent attachments during biofilm establishment (Agladze et al., 2005). The McaS effects on biofilm formation, in large part, appear to be due to the activation of the pgaABCD operon as the dramatic increase in biofilm amount upon McaS overexpression was not observed in a ΔpgaA strain (Fig. 7A).
Not surprisingly, the process of biofilm formation is highly regulated with different surface appendages and patterns of gene expression required depending on the temperature, carbon source, surface and strains tested (Pratt & Kolter, 1998, Wood et al., 2006, Pruss et al., 2010, Jackson et al., 2002, Prigent-Combaret et al., 2000). Given the varied levels of McaS expression throughout growth and the finding that endogenous McaS levels have graded effects on the McaS targets, we propose differing levels of McaS impact various stages of biofilm formation. In line with this conclusion, we observed that a lack of McaS most strongly affected biofilm formation when cells were grown at 37°C in CFA media, a finding that is consistent with McaS activation of pgaA, which has previously been shown to be required for biofilm formation in CFA media (Wang et al., 2004). It is interesting to note that both McaS and the pgaABCD operon are absent from other enteric bacteria such as Salmonella highlighting key differences in the complex regulation surrounding the motile-to-sessile switch in these closely related species.
Regulatory sRNAs already have been shown to regulate various aspects of bacterial physiology. Here we report that a new sRNA regulator, McaS, through activation by CRP responds to nutrient limitation and coordinates the regulation of motility, yet another physiological response not previously known to be targeted by sRNAs, and biofilm formation. An important direction for future work will be to understand the mechanism of McaS integration into the extensive regulatory network surrounding the complex decision to switch from a motile to a sessile lifestyle.
Experimental procedures
Bacterial strains
The bacterial strains used in the study are listed in Table S2. Gene knockouts were performed in strain NM500 using λ Red-mediated recombination with fragments generated by PCR using oligonucleotides listed in Table S3 (Datsenko & Wanner, 2000, Yu et al., 2000, Court et al., 2003). The deletion alleles flanked by FRT sites, were moved into new backgrounds by P1 transduction, and where indicated, antibiotic resistance markers were removed using pCP20 (Cherepanov & Wackernagel, 1995). The method of Mandin and Gottesman was used to create the csgD-lacZ, flhD-lacZ, and pgaA-lacZ translational fusions (and mutant derivatives) by fusing the entire 5′ UTR to the 10th codon of the coding sequence to lacZ (Mandin & Gottesman, 2009). The mutant derivatives of the lacZ fusions were constructed using overlapping PCR as described previously (Ho et al., 1989). The mcaS promoter fusion was created by overlapping PCR amplifying the entire promoter region using the oligonucleotides listed in Table S3 and integrating the fragment upstream of lacZ in the PM1205 strain background. The PBAD-abgR strain was constructed by amplifying the PBAD promoter from plasmid pTM26 (Morita et al., 2004) followed by recombineering into the abgR locus of MG1655. All mutations and fusions were confirmed by sequencing.
Plasmids
The plasmids used in this study are listed in Table S4. For high expression from the Ptac promoter of plasmid pRI (pRI-McaS), the McaS fragment was PCR amplified from MG1655 genomic DNA, digested with EcoRI and HindIII and cloned into the corresponding sites of pRI (Opdyke et al., 2004). For Plac-controlled expression (pBR-McaS), McaS was PCR amplified, digested with AatII and HindIII and cloned into the corresponding sites of pBRplac (Guillier & Gottesman, 2006). pRI and pBRplac are both high copy pBR322-derived vectors. Mutant derivatives of pBR-McaS were generated by overlapping PCR as described previously (Ho et al., 1989) using the oligonucleotides listed in Table S3 and cloned into the AatII and HindIII sites of pBRplac. All cloning was performed in E. coli TOP10 cells (Invitrogen). All plasmid inserts were confirmed by sequencing.
Growth conditions
Unless indicated otherwise, strains were grown aerobically at 37°C in either LB (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter) or M63 minimal media supplemented with final concentrations of 0.001% Vitamin B1 and 0.2% glucose or 0.4% glycerol. Curli production was monitored using Congo red plates (LB agar without NaCl containing 20 μg/ml Congo red and 10 μg/ml Brilliant Blue) incubated at 25°C for 48 h. Motility was monitored using motility swim agar (10 g tryptone, 5 g NaCl, 2.5 g agar per liter) incubated at 25°C for 16 h. Biofilm formation was performed in LB, colonizaton factor antigen media (CFA) (10 g casamino acids, 5 g yeast extract, 50 mg MgSO4 and 5 mg MnCl2 per liter) (Agladze et al., 2005) or in YESCA media (10 g casamino acids and 1 g yeast extract per liter) (Epstein et al., 2009). Where indicated, IPTG was added at a final concentration of 1 mM except for the motility plates, where 100 μM was used. Antibiotics were added at the following concentrations: 100 μg/ml ampicillin (amp), 30 μg/ml kanamycin (kan), 12.5 μg/ml tetracycline (tet), or 25 μg/ml chloramphenicol (cat).
RNA extraction
For cells grown in LB, total RNA was extracted by hot acid phenol as described previously (Masse et al., 2003) with some minor modifications. Briefly, 750 μl of cells were combined with 102 μl lysis solution (320 mM sodium acetate, 8% SDS and 16 mM EDTA). The lysed cells were incubated with 500 μl of hot acid phenol choloroform (pH 4.5, Ambion) at 65°C for 10 min. For cells grown in M63 medium, RNA was extracted by hot phenol extraction as described previously with minor modifications (Kawano et al., 2002). At the indicated time points, 20 ml (exponential phase) or 10 ml (stationary phase) of cells were collected in a 50 ml conical tube by centrifugation at 4°C 5000 g for 10 min. Following centrifugation, the cell pellets were resuspended in 700 μL of solution A (0.5% SDS, 20 mM sodium acetate and 10 mM EDTA), transferred to a 1.5 ml tube containing 500 μl of hot acid phenol, and incubated at 65°C for 10 min. After the initial extractions, the samples from both growth conditions were extracted two additional times with hot acid phenol. A final extraction with phenol:chloroform:isoamyl alcohol (Invitrogen) was performed using Phase Lock Gel Heavy 2.0 ml tubes (5Prime). The supernatant was combined with 700 μl 100% ethanol to precipitate the RNA, after which the pellets were resuspended in DEPC water. Total RNA concentration was determined based on OD260.
Northern analysis
Unless indicated otherwise, total RNA (10 μg) was separated on an 8% polyacrylamide-7M urea gel in 1X TBE and transferred to Zeta-Probe membrane (Bio-Rad) for 1 h at 55V followed by 16 h at 20V in 0.5X TBE or for 5 h at 55V. Oligonucleotides were end-labeled with 32P-ATP by T4 polynucleotide kinase (NEB). Membranes were hybridized overnight at 45°C in UltraHyb (Ambion) hybridization buffer. Following hybridization, membranes were washed once with 2X SSC + 0.1% SDS followed by an incubation for 10 min at 45°C with 2X SSC + 0.1% SDS. Membranes were subsequently washed briefly 5X with 0.2X SSC + 0.1% SDS allowed to dry and exposed to KODAK Biomax XAR film at −80°C.
Western Blot analysis
Western blot analysis was performed as described previously with minor changes (Hemm et al., 2008). Briefly, samples were taken at the indicated times from the same cultures grown in LB over 30 h at 37°C used for RNA extraction. Samples were separated on a 10–20% Tris-Glycine gel (Invitrogen) and transferred to a nitrocellulose membrane (Invitrogen). Samples were blocked in 5% milk and probed with 1:4000 dilution of α-RpoS antibody in 5% milk followed by incubation with 1:20,000 dilution of HRP-anti-rabbit IgG in 5% milk. The membrane was developed using Amersham ECL Western Blot Detection Reagent (GE Healthcare) and exposed to KODAK Blue-XB film.
DNase I footprinting
Fragments of the mcaS promoter were amplified by PCR using 5′-32P-labeled MK0043 and unlabeled MK0018 for the labeled top strand and 5′-32P-labeled MK0063 and unlabeled MK0043 for the labeled bottom strand. The PCR products were purified using QiaQuick PCR purification columns (Qiagen). Increasing amounts of purified CRP were incubated with the labeled DNA fragment (0.5 pmol) for 20 min at 37°C in a binding buffer containing 10 mM Tris-Cl (pH7.4), 50 mM KCl, 0.5 mM DTT, 1 mM MgCl2, 4% glycerol, 0.05 mg/ml BSA, 0.05 mg/ml sheared salmon sperm DNA, 0.5 mM EDTA, and 2 mM cAMP in a final volume of 12 μl. To initiate DNA digestion, 12 μl of a solution containing 5 mM CaCl2 and 10 mM MgCl2 as well as DNase I at a final concentration of 120 ng/μl were added and incubated for 1 min at room temperature. The reaction was stopped by the addition of 125 μl stop solution (200 mM NaCl, 30 mM EDTA, 1% SDS and 250 μg/ml of total yeast RNA) followed by phenol:chloroform extraction and ethanol precipitation. The partially digested DNA samples were resuspended in 10 μl of water and analyzed on an 8% polyacrylamide-7M urea sequencing gel. A sequencing ladder was generated using the corresponding PCR product and radiolabeled primer (MK0043 for top strand and MK0063 for the bottom strand) and the SequiTherm EXCEL II DNA sequencing Kit (Epicentre Biotechnologies) following the manufacturer’s instructions.
Immunoprecipitation
Wild type MG1655 or an isogenic hfq-1 derivative were grown in LB to early stationary phase (OD600 ~1), and concentrated in lysis buffer [20 mM Tris-HCl (pH 8), 150 mM KCl, 1 mM MgCl2, 1mM DTT)] containing RNase inhibitor (Invitrogen). Immunoprecipitation was performed as described previously (Zhang et al., 2003). Briefly, cells were lysed by vortexing with glass beads (Sigma G1277) for 10 cycles of 30 s. The lysate (200 μl) from wild type cells was incubated for 2 h at 4°C in buffer Net2 [50mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.05% Triton X-100] with 24 mg protein A sepharose beads pre-bound with 20 μl α-Hfq or pre-immune serum. Beads were then washed 5 times with Net2 buffer. Immunoprecipitated RNA as well as total RNA from 100 μl of wild type and hfq-1 cell lysates were extracted by phenol:chloroform:isoamyl alcohol (Invitrogen) followed by ethanol precipitation. Northern analysis was performed as above, using 0.5 μg RNA from the immunoprecipitated samples and 5 μg RNA from the total RNA samples.
RACE analysis
RACE end mapping was performed as described previously (Argaman et al., 2001) using oligonucleotides listed in Table S3 and RNA isolated from strain MC4100. The 3′ and 5′ cDNA amplification products were cloned into pCRII-TOPO cloning vector (Invitrogen) and the ends were determined by sequencing.
β-galactosidase assays
Overnight cultures grown in LB were diluted into fresh LB to an OD600 ~0.05 and were grown at 37°C for 1.5 h with shaking until they reached an OD600 ~ 0.1–0.2 Expression of the lacZ fusions was induced by the addition of 0.2% arabinose. For half the samples, expression of McaS from the pBR-McaS plasmid was induced simultaneously by also adding 1mM IPTG unless indicated otherwise. After 1 h induction, the cells were lysed in Z buffer (700 μl) with 15 μl 0.1% SDS and 30 μl chloroform. β-galactosidase activity was assayed as described (Durand & Storz, 2010).
Electron microscopy
Overnight cultures of strain NM525 ΔabgR-ydaL::kan carrying either pBR or pBR-McaS grown in LB + amp, were diluted back 1:500 into fresh LB + amp containing 1 mM IPTG and grown to an OD600 ~ 1.0. Cells were collected and absorbed onto EM grids covered with formvar/carbon film for 5 min. Grids were fixed with 2% formaldehyde and negatively stained with 2% aqueous uranyl acetate and allowed to air dry. The samples were examined on Tecnai 200 transmission electron microscope (FEI, Hillsborough, OR) at 80 kV accelerating voltage and images were recorded on Gatan CCD camera (Gatan, Pleasanton, CA). The total numbers of flagella and cells were counted for each image obtained from two independent experiments.
Biofilm assay
Biofilm growth assays were performed as described previously (Wu & Outten, 2009). Briefly, overnight cultures grown in LB were diluted to a final OD600 of ~ 0.05 in fresh LB + amp + IPTG or LB, CFA or YESCA media. A 200 μl aliquot of the diluted culture was added to each well of a 96-well polystyrene microtiter plate (Costar). Plates were incubated at 25°C or 37°C without shaking for 24 h. Planktonic growth was determined by measuring OD600 using a BioTek Synergy H4 Microplate Reader. The plates were washed twice with 220 μl of ddH2O to remove planktonic cells. Any bacteria retained in the biofilm were stained with 220 μl of 0.1% crystal violet for 10 min and then washed three additional times with ddH2O to remove unbound excess dye. The plate was allowed to dry for 20 min at 37°C. The crystal violet was solubilized by addition of a solution of acetone:ethanol (1:4), and the amount of biofilm formation was determined by measuring the OD570 and normalizing by the OD600.
Supplementary Material
Acknowledgments
We thank M. Jarnik for help with the electron microscopy, S. Adhya for providing the purified CRP and N. Majdalani for providing the anti-RpoS antibody. We would like to thank S. Gottesman and current and former members of the Storz lab for comments on the manuscript. This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
References
- Agladze K, Wang X, Romeo T. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J Bacteriol. 2005;187:8237–8246. doi: 10.1128/JB.187.24.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokranz W, Wang X, Tschape H, Romling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- Boysen A, Moller-Jensen J, Kallipolitis B, Valentin-Hansen P, Overgaard M. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J Biol Chem. 2010;285:10690–10702. doi: 10.1074/jbc.M109.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher E, Dorel C, Zehnder AJ, Landini P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology. 2003;149:2847–2857. doi: 10.1099/mic.0.26306-0. [DOI] [PubMed] [Google Scholar]
- Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R., 3rd MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol. 2001;41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EA, Reizian MA, Chapman MR. Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J Bacteriol. 2009;191:608–615. doi: 10.1128/JB.01244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogol EB, V, Rhodius A, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui WJ, Qu QH, Chen YY, Wang M, Zhang XE, Bi LJ, Jiang T. Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli. J Struct Biol. 2011;174:282–289. doi: 10.1016/j.jsb.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol. 2008;70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Reimegard J, Sterk M, Grantcharova N, Romling U, Wagner EG. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein MJ, Green JM, Nichols BP. Characterization of mutations that allow p-aminobenzoyl-glutamate utilization by Escherichia coli. J Bacteriol. 1998;180:6260–6268. doi: 10.1128/jb.180.23.6260-6268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DW, Simecka JW, Romeo T. Catabolite repression of Escherichia coli biofilm formation. J Bacteriol. 2002;184:3406–3410. doi: 10.1128/JB.184.12.3406-3410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Watt RM, Danchin A, Huang JD. Use of a riboswitch-controlled conditional hypomorphic mutation to uncover a role for the essential csrA gene in bacterial autoaggregation. J Biol Chem. 2009;284:28738–28745. doi: 10.1074/jbc.M109.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and sigmaE-dependent CyaR-ompX regulatory case. J Mol Biol. 2008;383:1–9. doi: 10.1016/j.jmb.2008.06.058. [DOI] [PubMed] [Google Scholar]
- Jørgensen M, Nielsen J, Boysen A, Franch T, Møller-Jensen J, Valentin-Hansen P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2012.07976.x. in press. [DOI] [PubMed] [Google Scholar]
- Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Majdalani N, Gottesman S. Regulatory roles for small RNAs in bacteria. Curr Opin Microbiol. 2003;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- Modi SR, Camacho DM, Kohanski MA, Walker GC, Collins JJ. Functional characterization of bacterial sRNAs using a network biology approach. Proc Natl Acad Sci U S A. 2011;108:15522–15527. doi: 10.1073/pnas.1104318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Philippe H. Smr: a bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem Sci. 1999;24:298–300. doi: 10.1016/s0968-0004(99)01419-x. [DOI] [PubMed] [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology. 2010;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol. 2011;406:29–43. doi: 10.1016/j.jmb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008;22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol. 1988;170:3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Pruss BM, Verma K, Samanta P, Sule P, Kumar S, Wu J, Christianson D, Horne SM, Stafslien SJ, Wolfe AJ, Denton A. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch Microbiol. 2010;192:715–728. doi: 10.1007/s00203-010-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Wood TK. Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol. 2009;11:1–15. doi: 10.1111/j.1462-2920.2008.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, Gonzalez Barrios AF, Herzberg M, Lee J. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- Wu Y, Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol. 2009;191:1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- Zhao K, Liu M, Burgess RR. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 2007;35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Constantinidou C, Hobman JL, Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.