Abstract
It has been postulated that obligate asexual lineages may persist in the long term if they escape from negative interactions with either sexual lineages or biological enemies; and thus, parthenogenetic populations will be more likely to occur in places that are difficult for sexuals to colonize, or those in which biological interactions are rare, such as islands or island-like habitats. Ischnura hastata is the only known example of natural parthenogenesis within the insect order Odonata, and it represents also a typical example of geographic parthenogenesis, as sexual populations are widely distributed in North America, whereas parthenogenetic populations of this species have only been found at the Azores archipelago. In order to gain insight in the origin and distribution of parthenogenetic I. hastata lineages, we have used microsatellites, mitochondrial and nuclear DNA sequence data, to examine the population genetic structure of this species over a wide geographic area. Our results suggest that sexual populations of I. hastata in North America conform to a large subdivided population that has gone through a recent spatial expansion. A recent single long distance dispersal event, followed by a demographic expansion, is the most parsimonious hypothesis explaining the origin of the parthenogenetic population of this species in the Azores islands.
Keywords: Ischnura hastata, parthenogenetic reproduction, geographic parthenogenesis, apomixis, metapopulation structure
Introduction
Despite the fact that clonal reproduction is a priori expected to be reproductively advantageous (Maynard-Smith, 1978; Bell, 1982), obligate asexual organisms are evolutionarily short lived and have been traditionally regarded as evolutionary dead ends (Bell, 1982; Judson and Normark, 1996). It has been suggested that asexual organisms accumulate deleterious mutations more quickly than sexuals (Muller, 1964; Kondrashov, 1988) and that co-evolutionary arm races with parasites, predators and competitors will favor sexual reproduction (Lively, 2010). One way by which asexuals may temporarily escape these forces is through the observed differences in the distribution of closely related sexual and asexual lineages.
Asexuals tend to occupy more isolated habitats, have broader distributions and are often found in marginal habitats, relative to their sexual counterparts; a pattern that may provide insight into potential mechanisms by which asexual lineages may persist, at least in the short term. Thus, a number of hypotheses have been proposed to explain the observed patterns of distribution of sexual and asexual lineages: (1) the superior colonizing ability of parthenogens could explain their larger geographical ranges, given that every single asexual individual is able to found a new population, because finding a mate is not necessary to ensure reproduction (Cuéllar, 1994); (2) the interactions with parasites, predators and competitors will influence the distribution of asexual forms, and thus parthenogens will be more common in sparsely inhabited habitats, where these interactions are rare, and where environmental factors are dominating (Glesener and Tilman, 1978; Hamilton et al., 1990); (3) the lower fitness of asexual/bisexual hybrids might keep populations separated (Lynch, 1984); (4) heterozygosity assurance provided by clonal reproduction will favor asexual lineages in subdivided metapopulations (Vrijenhoek, 1985; Haag and Ebert, 2004). Most of these models are based on the idea that parthenogens may persist in the long term if they escape from negative interactions with either sexual lineages or biological enemies (predators and parasites).
Ischnura hastata (Zygoptera, Coenagrionidae) is the only known example of parthenogenetic reproduction within the insect order Odonata, and represents a typical example of geographic parthenogenesis: sexual populations of this species are widely distributed in the American continent, whereas only female, parthenogenetic populations have been described only in the Azores archipelago (Cordero Rivera et al., 2005). This system allows us the opportunity to study the mechanisms that maintain parthenogenesis and how they have arisen in a previously unidentified group. It also allows us to investigate the influence of geographic isolation on the evolution of parthenogenetic populations, and to contribute further to our understanding of geographic parthenogenesis.
Previous molecular genetics work strongly suggests that parthenogenetic I. hastata females are diploid and that the number of chromosomes of the unfertilized eggs is restored by an apomictic mechanism of parthenogenesis; although these results are not entirely conclusive without further cytological and karyotype analyses (see Lorenzo-Carballa and Cordero-Rivera, 2009). No evidence for bacterial involvement in the parthenogenesis has been found, which suggests that tychoparthenogenesis (that is, a small capacity for parthenogenesis in otherwise sexually reproducing females) could occur in the sexual populations of this species (Lorenzo-Carballa and Cordero-Rivera, 2009). I. hastata has been inferred to be a migratory species (Corbet, 1999) based on its widespread distribution in the American continent. It is found in temporal or permanent water bodies, which suggests that the sexual demes may have an island-like structure subject to frequent extinction and/or recolonization, as described for other odonate species (Gibbons et al., 2002; Purse et al., 2003; De Block et al., 2005; Rouquette and Thompson, 2007). This potential for long distance dispersal should favor population mixing, leading to low levels of genetic subdivision.
In order to gain insight in the origin and patterns of distribution of parthenogenetic I. hastata lineages, we have studied the population structure of this species over a wide geographic area. Here, we use microsatellite loci, mitochondrial and nuclear DNA sequence data to examine the population genetic structure of sexual and parthenogenetic populations of I. hastata. Our results suggest that the North-American demes of this species are strongly interconnected by dispersal so they can be considered a single population, and that the asexual population found in the Azores archipelago is the result of a demographic expansion after a recent single colonization event.
Materials and methods
Sample collection and DNA extraction
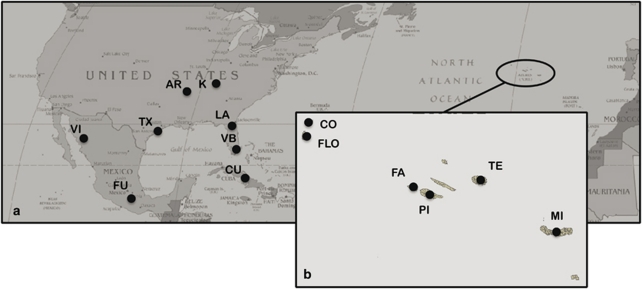
Adult individuals of I. hastata were collected from 14 locations: six parthenogenetic populations (Pico, Terceira, Faial, Flores, São Miguel and Corvo), covering most of the species' range in the Azores islands and eight sexual populations spread across North America (Arkansas; Kentucky; Texas; Florida, Lake County; Florida, Vero Beach; México, Las Fuentes and México, Vicam) and the Caribbean (Cuba) (Figure 1; Table 1). Genomic DNA from all of these samples was extracted using a CTAB protocol (modified from Doyle and Doyle, 1987) or the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany), following the manufacturer's instructions.
Figure 1.
Sampling locations of specimens of Ischnura hastata in North America (a) and the Azores islands (b). Sites are labelled according to abbreviations in Table 1.
Table 1. Ischnura hastata: geographic distribution of the analyzed specimens.
| Abr. | Lat. (N) | Long. (W) |
COI |
EF1-α |
|||
|---|---|---|---|---|---|---|---|
| n | Haplotypes | n | Haplotypes | ||||
| Arkansas | AR | 34 14 21 | 93 11 10 | 9 | 1, 2, 3, 4 | 7 | 1, 2, 7, 8, 9, 10, 11, 12, 13 |
| Kentucky | K | 37 9 3 | 86 5 50 | 12 | 1, 4, 11 | 3 | 1, 10, 30, 31, 32, 33 |
| Cuba | CU | 20 1 11 | 75 49 22 | 6 | 1 | 4 | 1, 2, 3, 4, 5, 6 |
| Texasa | TX | 30 33 15 | 95 8 2 | 4 | 1, 25, 26 | ||
| Florida, Lake Co. | LA | 29 8 17 | 81 36 50 | 10 | 1, 5 | 10 | 1, 2, 5, 7, 14, 15, 16, 17, 18, 19, 20, 21 |
| Florida, Vero Beach | VB | 27 38 45 | 80 24 49 | 21 | 1, 3, 6, 7, 8, 9, 10 | 14 | 1, 2, 6, 16, 21, 22, 23, 24, 25, 26, 27, 28, 29 |
| Mexico, Las Fuentes | FU | 18 56 10 | 99 14 2 | 16 | 1, 3, 12, 13, 14, 15, 16, 17, 18, 19, 20 | 6 | 34, 35, 36, 37, 38, 39 |
| Mexico, Vicam | VI | 27 38 37 | 110 17 46 | 19 | 1, 3, 12, 15, 21, 22, 23, 24 | 8 | 1, 35, 38, 40, 41, 42, 43, 44, 45, 46 |
| Pico | PI | 38 27 36 | 28 15 57 | 12 | 27 | 12 | 1 |
| Faial | FA | 38 35 28 | 28 42 39 | 12 | 27 | 12 | 1 |
| Flores | FLO | 39 25 20 | 25 11 3 | 10 | 27 | 10 | 1 |
| Terceira | TE | 38 41 52 | 27 9 26 | 12 | 27 | 13 | 1 |
| Sâo Miguel | MI | 37 49 33 | 25 44 28 | 10 | 27, 28 | 10 | 1 |
| Corvo | CO | 39 42 42 | 25 6 17 | 5 | 27 | 5 | 1 |
Abbreviations: COI, Cytochrome c Oxidase I; EF1-α, Elongation Factor 1-α.
For each gene are listed the number of individuals sequenced (n) and the haplotypes found in each population for each gene. Parthenogenetic populations are indicated in bold.
Only COI sequenced.
Sequencing and haplotype determination
Nucleotide variation was assessed in one mitochondrial (Cytochrome c Oxidase I, COI) and one nuclear gene (Elongation Factor 1-α, EF1-α). Genomic DNA was PCR amplified using locus-specific primers and conditions (see Supplementary information). Sequencing reactions were carried out bidirectionally either on a MegaBACE 500 sequencer, using the DYEnamic ET Dye Terminator Cycle Sequencing Kit (Amersham Biosciences, Buckinghamshire, UK) or on a 3730xl DNA Analyzer, using the BigDye Terminator Kit (Applied Biosystems, Foster City, CA, USA). Sequencing results were consistent across platforms, and only high quality traces were included in our analyses. For EF1-α data, allelic phases of heterozygous individuals were inferred using PHASE (Stephens et al., 2001). Excluding autapomorphies, all identified haplotypes had posterior probabilities >0.8.
Microsatellite genotyping
Nine polymorphic microsatellite loci (Lorenzo Carballa et al., 2007) were used to screen 144 individuals from sexual and parthenogenetic populations (Table 3). Forward primers were labelled with a fluorescent dye (HEX or 6-FAM, Eurofings MWG Operon, Ebesberg, Germany) for microsatellite typing. PCR conditions were the same as described in Lorenzo Carballa et al. (2007). Automated genotyping was performed on a MegaBACE 500 automated DNA sequencer using the ET-550 Size Standard (Amersham Biosciences). Fragment size was recorded and scored using Genetic Profiler v. 1.2 (Amersham Biosciences).
Genetic diversity analyses
Forward and reverse sequence strands were assembled and edited using SeqManII v. 5.03 (DNAstar, Inc., Madison, WI, USA) and consensus sequences were aligned using Clustal W (Thompson et al., 1994), as implemented in MEGA v. 4.0 (Tamura et al., 2007). The population genetic parameters number of haplotypes (h), number of segregating sites (S), haplotype diversity (H), nucleotide silent diversity (πS) and number of synonymous and non-synonymous substitutions for each locus and population were calculated using DNAsp v. 4.20.2 (Rozas et al., 2003). COI and EF1-α haplotypes are deposited in GenBank with accession numbers JN578143–JN578216.
For the microsatellite data set, allelic richness, observed and expected heterozygosities, and the number of private alleles for each locus and population were calculated using the software GDA (Lewis and Zaykin, 2001). Deviations from Hardy–Weinberg equilibrium for each locus in each population were calculated using Arlequin v. 3.01 (Excoffier et al., 2005). Linkage disequilibrium analyses in the sexual populations were performed using correlation-based tests for linkage disequilibrium (Zaykin et al., 2008).
Phylogenetic and phylogeographic analyses
Genealogical relationships between I. hastata haplotypes were analyzed using statistical parsimony networks (95% probability criterion) as implemented in TCS 1.21 (Clement et al., 2000). Additionally, we used multiple tree-building methods (NJ, Maximum Likelihood and Bayesian Inference) to reconstruct phylogenetic relationships among these haplotypes. The results obtained with these methods were almost identical to the reported networks (data not shown).
For the COI data set, we used the refined nesting algorithm of the nested clade phylogeographic analysis (Templeton et al., 1995) as implemented in Geodis v. 2.5 (Posada et al., 2000) to identify haplotype clades showing significant geographic associations. However, because of the severe limitations of the nested clade phylogeographic analysis to infer the multiple historical processes that may characterize a species' history (see Knowles and Maddison, 2002; Petit and Grivet, 2002; Knowles, 2004; Panchal and Beaumont, 2007; Petit, 2008a, 2008b; Beaumont et al., 2010) we did not use this analysis to make any inferences about the demographical or historical processes that could have shaped the genetic variation observed in I. hastata.
Population structure
First, we assessed population structure assuming a priori populations (that is, sampling locality=population), using hierarchical analysis of molecular variance (Excoffier et al., 1992) including (i) populations, (ii) populations within reproductive mode and (iii) reproductive mode. Analysis of molecular variance was also implemented to analyze the genetic structure of sexual populations. Analyses were conducted for all loci (COI, EF1-α and microsatellites) in Arlequin v. 3.01 (Excoffier et al., 2005). For the microsatellites we selected a locus-by-locus strategy, which is the most desirable option when missing data exist (Excoffier et al., 2005).
Pairwise Fst values were estimated with Arlequin v. 3.01 (Excoffier et al., 2005), using pairwise distances among EF1-α and COI sequences. The significance of Fst for population comparisons was assessed by 10 000 permutations.
To test for a possible scenario of isolation by distance in the sexual populations, we tested the correlation of genetic differentiation over geographical distances for all pairs of populations. Geographical distances among locations were estimated using the latitude/longitude distance calculator (http://jan.ucc.nau.edu/~cvm/latlongdist.html), and correlations between geographic and genetic distances for both microsatellites and COI were tested using a Mantel permutation procedure (Sokal and Rohlf, 1995) as implemented in IBD v. 3.15 (http://ibdws.sdsu.edu/~ibdws/; Jensen et al., 2005). The significance of the Mantel test was assessed by performing 10 000 random iterations.
To get insight into the substructuring among the sexual populations, the results with predefined populations were then compared with the Bayesian model-based clustering methods implemented in Structure v. 2.1 (Pritchard et al., 2000). We used the admixture model, and the number of clusters, K, was estimated by comparing the log-likelihood ratios in two independent runs for values of K between 1 (panmixia) and 4 (the total number of independent sites sampled for microsatellites). Each run consisted in 106 iterations, with a burn-in period of 105. All summary statistics stabilized before the end of the burn-in period and independent runs always attained the same results.
Patterns of demographic history in sexual populations
We used COI data to infer patterns of demographic history in sexual I. hastata. First, two tests based on the distribution of segregating sites (Tajima's D; Tajima, 1989) and on the haplotype distribution (Fu's FS; Fu, 1997) were applied to the data. Negative values of these tests are expected in populations that have undergone a recent expansion, because rare alleles are more numerous than expected; whereas positive values occur if rare alleles are eliminated from population following genetic bottlenecks (Tajima, 1989). Significance of D and FS statistics was tested by generating 10 000 coalescent simulations in Arlequin v. 3.01 (Excoffier et al., 2005). To further investigate the possibility of demographic expansions, we plotted the distribution of pairwise sequence differences (mismatch distribution) for all COI haplotypes using Arlequin v. 3.01 (Excoffier et al., 2005). Unimodal mismatch distributions are characteristic of haplotypes drawn from a population that has undergone a recent expansion and is the null hypothesis by which the mismatch distribution is assessed. Significant divergences from this model (multimodality) reflect the highly stochastic shape of gene trees under demographic equilibrium (Rogers and Harpending, 1992). All these analyses were carried out in each of the sexual localities sampled, as well as in the entire sexual data set.
As an independent test for demographic events, we used microsatellite data to test for the possibility of recent population size reduction and expansion in the sexual cluster. For bottleneck detection, calculations were performed with the program BOTTLENECK (Cornuet and Luikart, 1997), using the Two-Phase Model of mutation as suggested by Luikart et al. (1998). Wilcoxon's signed rank tests were used to determine whether populations exhibit significant number of loci with gene diversity excess. Signatures of population expansion were examined using the β imbalance index (Kimmel et al., 1998). This statistic is analogous to a mismatch distribution, and relies in two different estimates of the genetic diversity parameter θ (4Neμ). One estimate is based on allele size variance (θV), and the other is estimated from the variance in repeat numbers (θP). The imbalance index should be <1 for expanding populations whose pre-expansion history is stable, but is characteristically >1 in populations with a reduction in size that precedes a detectable growth phase (Kimmel et al., 1998, King et al., 2000). Here, we calculated β1, the difference in the natural logarithm of these estimators, averaged over all loci. This logarithm-based metric was chosen because of its higher sensitivity to detect historic signals of population expansion (King et al., 2000). Confidence intervals of β1 were estimated by bootstrapping over loci using R 2.9.0.
Results
Genetic diversity
Analyses of sequence polymorphisms in sexual and parthenogenetic I. hastata are summarized in Table 2. For EF1-α, 228 sequences were obtained. In the 46 inferred haplotypes 44 out of 459 sites (9.59 %) and two indels were found to be polymorphic. Only one haplotype was found in the parthenogenetic populations, which was also present in the sexual populations. The other 45 haplotypes were found only in the sexual individuals: one haplotype was present in four populations, seven haplotypes were present in two populations and three were found in different individuals from the same population. Each of the remaining (38) was found in single individuals. Nei's nucleotide diversity (π) varied from 0.006 to 0.022 in the sexual populations, with an average of 0.014. The number of haplotypes per location varied from 3 (K) to 10 (VB), and the number of private haplotypes per site fluctuated between 2 (VI) and 7 (VB), which yields an overall haplotype diversity in the sexual populations of 0.9 (ranging from 0.78 to 1).
Table 2. Polymorphism statistics for each gene.
| Locus | Population | n | P | L | Cod | S | Syn | Rep | πsa | Ha |
|---|---|---|---|---|---|---|---|---|---|---|
| EF1-α | Arkansas | 9 | 5 | 459 | 360 | 17 | 9 | 0 | 0.016 | 0.912 |
| Kentucky | 6 | 4 | 459 | 360 | 21 | 10 | 0 | 0.022 | 1.000 | |
| Texas | — | — | — | — | — | — | — | — | — | |
| Cuba | 6 | 2 | 459 | 360 | 7 | 5 | 0 | 0.006 | 0.929 | |
| Lake Co. | 12 | 6 | 459 | 360 | 19 | 11 | 0 | 0.012 | 0.900 | |
| Vero Beach | 12 | 9 | 459 | 360 | 23 | 13 | 0 | 0.008 | 0.865 | |
| Las Fuentes | 6 | 5 | 459 | 360 | 17 | 11 | 0 | 0.018 | 0.879 | |
| Vicam | 10 | 7 | 459 | 360 | 20 | 12 | 0 | 0.015 | 0.900 | |
| Pico | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| Faial | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| Flores | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| Terceira | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| Sâo Miguel | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| Corvo | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| Sexuals | 46 | 45 | 459 | 360 | 44 | 44 | 0 | 0.0146 | 0.939 | |
| Parthenogens | 1 | 0 | 459 | 360 | 0 | 0 | 0 | — | — | |
| COI | Arkansas | 4 | 1 | 550 | 550 | 4 | 4 | 0 | 0.002 | 0.583 |
| Kentucky | 3 | 1 | 550 | 550 | 2 | 2 | 0 | 0.001 | 0.318 | |
| Texas | 3 | 2 | 550 | 550 | 2 | 2 | 0 | 0.002 | 0.833 | |
| Cuba | 1 | 0 | 550 | 550 | 0 | 0 | 0 | — | — | |
| Lake Co. | 2 | 1 | 550 | 550 | 1 | 1 | 0 | 0.0004 | 0.200 | |
| Vero Beach | 7 | 5 | 550 | 550 | 7 | 7 | 0 | 0.001 | 0.500 | |
| Las Fuentes | 11 | 7 | 550 | 550 | 17 | 17 | 0 | 0.008 | 0.933 | |
| Vicam | 8 | 4 | 550 | 550 | 17 | 17 | 0 | 0.008 | 0.725 | |
| Pico | 1 | 0 | 550 | 550 | 0 | 0 | 0 | — | — | |
| Faial | 1 | 0 | 550 | 550 | 0 | 0 | 0 | — | — | |
| Flores | 1 | 0 | 550 | 550 | 0 | 0 | 0 | — | — | |
| Terceira | 1 | 0 | 550 | 550 | 0 | 0 | 0 | — | — | |
| Sâo Miguel | 2 | 1 | 550 | 550 | 1 | 1 | 0 | 0.0007 | 0.356 | |
| Corvo | 1 | 0 | 550 | 550 | 0 | 0 | 0 | — | — | |
| Sexuals | 26 | 26 | 550 | 550 | 33 | 33 | 0 | 0.004 | 0.616 | |
| Parthenogens | 2 | 2 | 550 | 550 | 1 | 1 | 0 | 0.00012 | 0.064 |
Abbreviations: COI, Cytochrome c Oxidase I; EF1-α, Elongation Factor 1-α.
Listed are number of haplotypes (n) and private haplotypes (P) analyzed; length of sequence (L) and coding region (Cod) analyzed; total number of polymorphic sites in coding and non-coding regions (S); number of synonymous (Syn) and non-synonymous (Rep) changes in coding regions; nucleotide diversity (πS) and haplotype diversity (H). Parthenogenetic populations are indicated in bold.
DNA polymorphism statistics calculated for the whole sequence (including coding and non-coding regions).
One-hundred and fifty-eight sexual and parthenogenetic I. hastata individuals were sequenced for COI. In all, 34 out of 550 sites were polymorphic (6.18%) and 28 haplotypes were identified, 2 of which were found only in the parthenogenetic populations. The most common haplotype in the parthenogens was found in 54 individuals, whereas the second, which differs only in 1 nucleotide, was only found in 2 individuals from São Miguel Island. In the sexual populations, 26 haplotypes were found: 1 was shared by all sexual populations and represented 61.85% of the total number of sexual individuals, 1 haplotype was found in 4 populations, 3 haplotypes were found in 2 populations and 2 were found in different individuals of the same population. The rest (19) were each found in single individuals. Nucleotide diversity varied from 0 to 0.01, with an average of 0.003. The number of haplotypes per location fluctuated between 11 (FU) and 1 (CU), and the number of private haplotypes between 7 (FU) and 0 (CU), which yields an average haplotype diversity of 0.51 (ranging from 0 to 0.95).
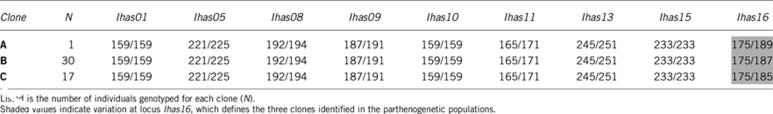
Polymorphism was analyzed across 9 microsatellite loci in 144 individuals from 10 populations (Table 3). Allelic diversity ranged from 7 to 13 alleles per locus, with 87 alleles identified across all 9 loci. All the alleles present in the parthenogenetic females were also found in the sexual populations, with the exception of one allele at microsatellite locus Ihas16, exclusive of the parthenogenetic populations, that was found in one individual from Pico Island.
Table 3. Microsatellite polymorphisms.
| Locus | Population | n | Np | He | Ho | PA |
|---|---|---|---|---|---|---|
| Ihas01 | Lake Co. | 12 | 3 | 0.366 | 0.083* | 0 |
| Vero B. | 15 | 3 | 0.301 | 0.200* | 0 | |
| Las Fuentes | 17 | 4 | 0.357 | 0.176* | 2 | |
| Vicam | 19 | 5 | 0.546 | 0.421 | 2 | |
| Pico | 20 | 1 | 0 | 0 | ||
| Faial | 17 | 1 | 0 | 0 | ||
| Flores | 10 | 1 | 0 | 0 | ||
| Terceira | 19 | 1 | 0 | 0 | ||
| Sâo Miguel | 10 | 1 | 0 | 0 | ||
| Corvo | 5 | 1 | 0 | 0 | ||
| Sexuals | 63 | 7 | 0.412 | 0.238 | 6 | |
| Parthenogens | 81 | 1 | 0 | 0 | ||
| Ihas05 | Lake Co. | 12 | 6 | 0.746 | 0.583 | 0 |
| Vero B. | 13 | 4 | 0.618 | 0.538 | 0 | |
| Las Fuentes | 17 | 6 | 0.765 | 0.706 | 0 | |
| Vicam | 19 | 7 | 0.752 | 0.632 | 1 | |
| Pico | 20 | 2 | 1 | 0 | ||
| Faial | 17 | 2 | 1 | 0 | ||
| Flores | 10 | 2 | 1 | 0 | ||
| Terceira | 19 | 2 | 1 | 0 | ||
| Sâo Miguel | 10 | 2 | 1 | 0 | ||
| Corvo | 5 | 2 | 1 | 0 | ||
| Sexuals | 61 | 8 | 0.751 | 0.683 | 6 | |
| Parthenogens | 81 | 2 | 1 | 0 | ||
| Ihas08 | Lake Co. | 11 | 6 | 0.814 | 0.636 | 0 |
| Vero B. | 15 | 9 | 0.887 | 0.800 | 1 | |
| Las Fuentes | 15 | 5 | 0.807 | 0.733 | 0 | |
| Vicam | 19 | 6 | 0.717 | 0.579 | 0 | |
| Pico | 19 | 2 | 1 | 0 | ||
| Faial | 17 | 2 | 1 | 0 | ||
| Flores | 10 | 2 | 1 | 0 | ||
| Terceira | 18 | 2 | 1 | 0 | ||
| Sâo Miguel | 10 | 2 | 1 | 0 | ||
| Corvo | 5 | 2 | 1 | 0 | ||
| Sexuals | 60 | 9 | 0.822 | 0.683 | 7 | |
| Parthenogens | 79 | 2 | 1 | 0 | ||
| Ihas09 | Lake Co. | 12 | 7 | 0.837 | 0.750 | 0 |
| Vero B. | 14 | 6 | 0.765 | 0.643 | 1 | |
| Las Fuentes | 16 | 8 | 0.768 | 0.765 | 2 | |
| Vicam | 19 | 8 | 0.824 | 0.579* | 1 | |
| Pico | 19 | 2 | 1 | 0 | ||
| Faial | 16 | 2 | 1 | 0 | ||
| Flores | 10 | 2 | 1 | 0 | ||
| Terceira | 18 | 2 | 1 | 0 | ||
| Sâo Miguel | 10 | 2 | 1 | 0 | ||
| Corvo | 5 | 2 | 1 | 0 | ||
| Sexuals | 62 | 12 | 0.804 | 0.677 | 10 | |
| Parthenogens | 79 | 2 | 1 | 0 | ||
| Ihas10 | Lake Co. | 12 | 7 | 0.822 | 0.750 | 0 |
| Vero B. | 14 | 7 | 0.817 | 0.571 | 0 | |
| Las Fuentes | 12 | 7 | 0.819 | 0.750 | 0 | |
| Vicam | 14 | 7 | 0.836 | 0.500* | 0 | |
| Pico | 20 | 1 | 0 | 0 | ||
| Faial | 16 | 1 | 0 | 0 | ||
| Flores | 8 | 1 | 0 | 0 | ||
| Terceira | 18 | 1 | 0 | 0 | ||
| Sâo Miguel | 10 | 1 | 0 | 0 | ||
| Corvo | 5 | 1 | 0 | 0 | ||
| Sexuals | 52 | 9 | 0.822 | 0.635 | 8 | |
| Parthenogens | 77 | 1 | 0 | 0 | ||
| Ihas11 | Lake Co. | 11 | 10 | 0.892 | 0.910 | 0 |
| Vero B. | 13 | 9 | 0.871 | 0.770 | 0 | |
| Las Fuentes | 12 | 7 | 0.779 | 0.500* | 0 | |
| Vicam | 16 | 10 | 0.865 | 0.688 | 2 | |
| Pico | 19 | 2 | 1 | 0 | ||
| Faial | 15 | 2 | 1 | 0 | ||
| Flores | 10 | 2 | 1 | 0 | ||
| Terceira | 17 | 2 | 1 | 0 | ||
| Sâo Miguel | 10 | 2 | 1 | 0 | ||
| Corvo | 5 | 2 | 1 | 0 | ||
| Sexuals | 52 | 13 | 0.867 | 0.712 | 11 | |
| Parthenogens | 76 | 2 | 1 | 0 | ||
| Ihas13 | Lake Co. | 12 | 7 | 0.815 | 0.917 | 2 |
| Vero B. | 15 | 7 | 0.793 | 0.533* | 0 | |
| Las Fuentes | 14 | 6 | 0.725 | 0.643 | 0 | |
| Vicam | 19 | 8 | 0.801 | 0.632* | 0 | |
| Pico | 20 | 2 | 1 | 0 | ||
| Faial | 17 | 2 | 1 | 0 | ||
| Flores | 10 | 2 | 1 | 0 | ||
| Terceira | 18 | 2 | 1 | 0 | ||
| Sâo Miguel | 10 | 2 | 1 | 0 | ||
| Corvo | 5 | 2 | 1 | 0 | ||
| Sexuals | 60 | 10 | 0.822 | 0.667 | 8 | |
| Parthenogens | 80 | 2 | 1 | 0 | ||
| Ihas15 | Lake Co. | 11 | 5 | 0.766 | 0.545 | 1 |
| Vero B. | 14 | 5 | 0.751 | 0.786 | 0 | |
| Las Fuentes | 12 | 5 | 0.725 | 0.750 | 0 | |
| Vicam | 18 | 5 | 0.640 | 0.389* | 0 | |
| Pico | 18 | 1 | 0 | 0 | ||
| Faial | 1 | 1 | 0 | 0 | ||
| Flores | 10 | 1 | 0 | 0 | ||
| Terceira | 11 | 1 | 0 | 0 | ||
| Sâo Miguel | 9 | 1 | 0 | 0 | ||
| Corvo | 1 | 1 | 0 | 0 | ||
| Sexuals | 55 | 7 | 0.731 | 0.600 | 6 | |
| Parthenogens | 50 | 1 | 0 | 0 | ||
| Ihas16 | Lake Co. | 11 | 8 | 0.771 | 0.818 | 0 |
| Vero B. | 14 | 9 | 0.767 | 0.857 | 0 | |
| Las Fuentes | 16 | 6 | 0.520 | 0.438 | 0 | |
| Vicam | 19 | 6 | 0.592 | 0.632 | 1 | |
| Pico | 20 | 3 | 1 | 1 | ||
| Faial | 17 | 2 | 1 | 0 | ||
| Flores | 10 | 2 | 1 | 0 | ||
| Terceira | 19 | 3 | 1 | 0 | ||
| Sâo Miguel | 10 | 3 | 1 | 0 | ||
| Corvo | 5 | 3 | 1 | 0 | ||
| Sexuals | 60 | 11 | 0.649 | 0.667 | 8 | |
| Parthenogens | 81 | 4 | 1 | 1 |
Listed are the number of individuals genotyped for each locus (n), number of alleles in each population (Np), expected (He) and observed (Ho) heterozygosities, and the number of private alleles (PA). An asterisk indicates significant deviation from Hardy–Weinberg equilibrium. Parthenogenetic populations are indicated in bold.
Given that apomictic parthenogenesis produces offspring that is heterozygous at all loci in which their mothers were heterozygous (Lorenzo-Carballa and Cordero-Rivera, 2009), deviations from Hardy–Weinberg equilibrium might be expected at segregating loci. Accordingly, for variable microsatellite loci (Ihas05, Ihas08, Ihas09, Ihas11, Ihas13 and Ihas16) all parthenogenetic females were heterozygous (Table 3). Variation in parthenogens was only observed at locus Ihas16, with four different alleles that defined the three highly related clones found in the asexual population (Table 4). In contrast, 40 unique multilocus genotypes were found in the sexuals. In all the sexual populations, we observed a deficit of heterozygotes across multiple loci, although this excess of homozygotes was only significant (P<0.05) in the populations from Mexico (FU and VI) and Florida (LA and VB) (Table 3). Cases of linkage disequilibrium were found between loci Ihas01-Ihas05, Ihas05-16 (LA); Ihas01-Ihas09, Ihas05-Ihas13 (VB); Ihas01-Ihas10, Ihas08-Ihas09 (VI); and Ihas01-Ihas09, Ihas01-Ihas11 (FU).
Table 4. Microsatellite multilocus genotypes found in the parthenogenetic Ischnura hastata populations.
Phylogenetic and phylogeographic analyses
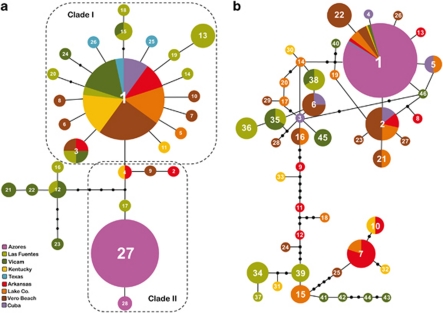
The COI and EF1-α haplotype networks are shown in Figure 2. The main characteristic of these networks is the lack of genetic structure in the sexual populations. For the COI, parthenogenetic I. hastata cluster together, whereas for the EF1-α, there is no separation among sexual and asexual populations, suggesting a relative recent divergence between both lineages. Both loci networks showed a star-like shape, with the most frequent haplotypes in the center, surrounded by several low frequency haplotypes; a pattern typically observed in expanding populations.
Figure 2.
Mutational haplotype networks based on statistical parsimony, representing genealogical relationships between 28 COI haplotypes (a) and 46 EF1-α haplotypes (b) in sexual and parthenogenetic populations of Ischnura hastata. Black dots represent missing mutational steps or non-sampled haplotypes. Haplotypes connected by a single line differ only in one mutational step. The size of the circles correlates with haplotype frequency. The dashed lines in the COI network indicate the two main clades with significant geographic association (see main text).
For the COI data, the nested clade phylogeographic analysis nesting algorithm resolved two two-step clades that showed significant geographic association: clade I, which contained the most common haplotype in sexual populations and its surrounding haplotypes and clade II, which included the haplotypes found in the parthenogenetic populations and their more closely related sexual haplotypes. Parthenogens differ from their closest sexual relative by a single mutational step (Figure 2).
Population structure
I. hastata exhibited significant partitioning of genetic variation. The analysis of molecular variance including sexual and parthenogenetic populations showed that for the nuclear markers almost all genetic variation (∼60–85%) resides within the sampled populations. In contrast, the main source of genetic variation for mtDNA (∼72%) is attributed to haplotype differences between reproductive groups (sexual vs asexual populations; Table 5). Within these groups, both the nuclear and the mitochondrial genome showed very little population structure. A complementary analysis of molecular variance of the sexual populations showed a very similar pattern. At a continental scale, I. hastata can be characterized as a large subdivided population where the vast majority of the observed variation corresponds to variation within the independently sampled populations (Table 5). Fst estimates in North-American (that is, sexual) populations were higher for EF1-α than for any other locus reflecting the existence of many population-exclusive alleles in this marker (Table 6). We found a strong correlation between pairwise Fst values and geographic distances for both the COI (r=0.5508, Mantel test P=0.0062) and the microsatellite data sets (r=0.8416, P=0.1626). However, as we were only able to genotype four of the sexual populations (see Table 3) the latter correlation is not significant.
Table 5. AMOVA and hierarchical analyses for Ischnura hastata populations.
| Source of variation (%) |
Sequences |
Microsatellites |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EF1-α | COI | Ihas01 | Ihas05 | Ihas08 | Ihas09 | Ihas10 | Ihas11 | Ihas13 | Ihas15 | Ihas16 | Average | |
| All populations | ||||||||||||
| Among groups (sexual–asexual) | 17.81 | 71.66 | 20.87 | 22.45 | 21.89 | 31.8 | 48.59 | 15.51 | 21.67 | 61.78 | 31.25 | 32.04 |
| Among populations within groups | 22.22 | 2.79 | 1.77 | 0.14 | −0.03 | −0.94 | −0.32 | −0.73 | 1.03 | 0.81 | 4.69 | 0.70 |
| Within populations | 59.97 | 25.55 | 77.36 | 77.4 | 78.15 | 69.14 | 51.73 | 85.22 | 77.31 | 87.41 | 64.06 | 67.26 |
| Fixation indices | ||||||||||||
| FSC (pop/reproductive mode) | 0.270*** | 0.099*** | 0.022 | 0.002* | −0.0004* | −0.013 | −0.006 | −0.009 | 0.013** | 0.021 | 0.068*** | 0.010*** |
| FST (pop/total) | 0.400*** | 0.745*** | 0.226*** | 0.226*** | 0.219*** | 0.309*** | 0.483*** | 0.148*** | 0.227*** | 0.626*** | 0.359*** | 0.327*** |
| FCT (reproductive mode/total) | 0.178** | 0.717*** | 0.209** | 0.224** | 0.219** | 0.318** | 0.486** | 0.155** | 0.217** | 0.618** | 0.313** | 0.320*** |
| Sexual populations only | ||||||||||||
| Among populations | 25.34 | 10.37 | 2.86 | 4.28 | 3.62 | 1.13 | 0 | 2.31 | 6.11 | 3.59 | 0.63 | 2.71 |
| Within populations | 74.66 | 89.63 | 97.14 | 95.72 | 96.38 | 98.87 | 100 | 97.69 | 93.89 | 96.41 | 99.37 | 97.29 |
| Fixation indices | ||||||||||||
| FST | 0.253*** | 0.104** | 0.028 | 0.042* | 0.036* | 0.011 | −0.004 | 0.023 | 0.061** | 0.036 | 0.0063 | 0.027*** |
Abbreviations: AMOVA, analysis of molecular variance; COI, Cytochrome c Oxidase I; EF1-α, Elongation Factor 1-α.
Negative values should be interpreted as zero.
*P<0.05.
**P<0.01.
***P<0.001.
Table 6. Pairwise Fst values between sexual Ischnura hastata populations based on COI (upper diagonal) and EF1-α (lower diagonal).
| CU | AR | LA | VB | K | FU | VI | TX | |
|---|---|---|---|---|---|---|---|---|
| CU | −0.006 | −0.059 | −0.084 | −0.069 | 0.027 | 0.095 | 0.111 | |
| AR | 0.268 | 0.052 | −0.002 | 0.002 | 0.051 | 0.106 | 0.022 | |
| LA | −0.055 | 0.213 | −0.025 | −0.004 | 0.082 | 0.147 | 0.140 | |
| VB | −0.010 | 0.337 | −0.019 | −0.026 | 0.128 | 0.198 | 0.047 | |
| K | 0.208 | −0.059 | 0.118 | 0.263 | 0.087 | 0.151 | 0.111 | |
| FU | 0.373 | 0.133 | 0.361 | 0.465 | −0.070 | 0.058 | 0.001 | |
| VI | 0.213 | 0.326 | 0.187 | 0.263 | 0.283 | 0.361 | 0.064 |
Abbreviations: COI, Cytochrome c Oxidase I; EF1-α, Elongation Factor 1-α.
Significant values are indicated in bold. Negative values should be interpreted as zero.
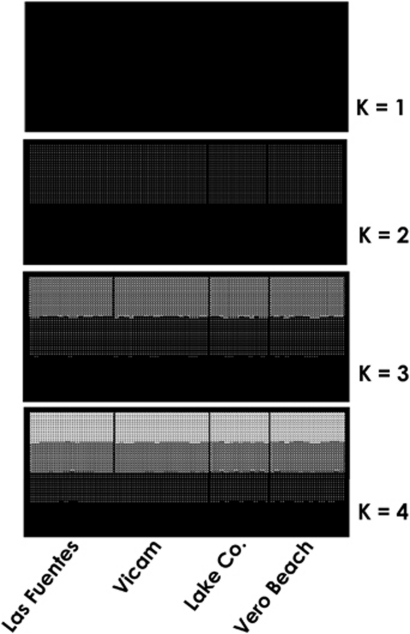
As for the phylogenetic analyses, the microsatellite-based Bayesian population assignment tests failed to detect any genetic structure among sexual populations: an increase in the number of clusters resulted in nearly the same probability for all the sexual individuals to be assigned to each of the sampled localities (Figure 3).
Figure 3.
Genetic clustering of sexual Ischnura hastata, based on nine microsatellite loci. Estimated population structure (from K=1 to K=4). Sampled localities (labelled below the figure) are separated by black lines. Each individual is represented by a narrow vertical column with the proportion of the colors indicating the genome proportion derived from each population.
Patterns of demographic history in sexual populations
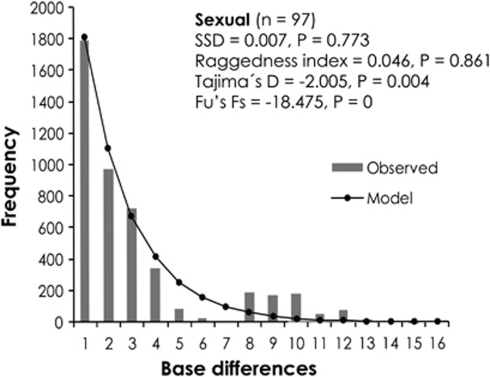
Because demographic events in continental (that is, sexual) populations might help to explain the colonization of the Azores archipelago, we conducted a series of analyses aimed to characterize the recent demographic history of I. hastata in North America. Overall sequencing data (Fu's FS, Tajima's D and mismatch statistics) rejected constant population and support the hypothesis of population expansion. When the data from each locality were analyzed separately, neutrality tests were significantly negative at Vero Beach; and significantly negative FS values were also found in Kentucky and Las Fuentes. With the exception of Vicam, a trend toward negative values of both statistics was found in all populations. According to the mismatch distributions, the hypothesis of a sudden expansion could not be rejected in most populations, thus providing further evidence of population expansion (Table 7). Similarly, the frequency distribution of pairwise differences in the sexual cluster was not significantly different from that expected under a model of demographic expansion, and neutrality tests were also significantly negative (Figure 4).
Table 7. Demographic estimates from COI for each of the sexual localities sampled.
| Locality | Fu's Fs (P) | Tajima's D (P) | SSD (P) | Raggedness index (P) |
|---|---|---|---|---|
| Arkansas | −0.928 (0.137) | −1.149 (0.141) | 0.0198 (0.479) | 0.116 (0.766) |
| Kentucky | −1.325 (0.025) | −1.451 (0.071) | 0.003 (0.308) | 0.227 (0.738) |
| Texas | −0.89 (0.092) | −0.71 (0.281) | n.a. | n.a. |
| Cuba | — | — | — | — |
| Lake Co. | −0.34 (0.156) | −1.112 (0.189) | 0.0004 (0.18) | 0.4 (0.821) |
| Vero Beach | −5.074 (0.0001) | −2.131 (0.003) | 0.0002 (0.795) | 0.087 (0.633) |
| Las Fuentes | −3.698 (0.033) | −0.57 (0.327) | 0.015 (0.256) | 0.029 (0.541) |
| Vicam | 0.334 (0.579) | −0.208 (0.459) | 0.054 (0.368) | 0.103 (0.31) |
Abbreviation: COI, Cytochrome c Oxidase I.
SSD tests the validity of a sudden expansion model based on the sum of squares deviations between the observed and expected mismatch. Non-significant values for SSD signify that the data do not deviate from that expected under the model of expansion. Raggedness index is calculated similarly, and non-significant raggedness values also indicate population expansion.
n.a., least square procedure to fit model mismatch distribution and observed distribution did not converge.
Figure 4.
Mismatch distributions for the sexual group of Ischnura hastata. Raggedness index and probability levels are given under the null distribution (model frequency) consistent with a sudden population expansion.
The microsatellite data set further confirmed this pattern of population expansion. The observed imbalance index for the sexual cluster (β1 mean: −0.492; range: −0.889 to −0.137) suggests that the sexual population has recently expanded without significant previous bottlenecks. Consistently, our bottleneck analyses did not detect a non-significant excess of heterozygosity across all loci (Wilcoxon's test, P=0.684), suggesting that the sexual population has not suffered significant bottlenecks.
Discussion
Genetic diversity, structure and demography of sexual populations
Our results suggest that in I. hastata, local populations may be so strongly interconnected that they can be better considered as a single ‘patchy population', where gene flow (that is, dispersal) exerts a strong homogenizing force over wide geographic ranges. Bayesian model-based clustering, phylogenetic and genetic differentiation analyses suggest that at a broad continental scale there is no significant population structure, and that very little genetic differentiation exists between demes. In fact, most of the total genetic variation found across North America (average 97% for microsatellite loci) is found within each of the independently sampled populations. However, some of these local populations are significantly differentiated, suggesting varying amounts of dispersal among localities. Only moderate population structure is detected over distances of 1400 km, but the vast extent of the North-American subcontinent allows significant population differentiation. Thus, demes may currently be panmictic over a regional scale with low or sporadic gene flow over greater distances. This restricted pattern of gene flow over very long distances, combined with high levels of local (or even regional) recruitment, can explain the significant association between genetic differentiation and geographic distance detected in I. hastata populations (see Palumbi, 2003). Yet, other non-equilibrium alternatives cannot be entirely excluded. For example, a very recent disruption of gene flow (for example, Pogson et al., 2001), or an ongoing turnover of local populations where colonizers tend to be from different demes (Wade and McCauley, 1988) could potentially explain the observed geographic patterns of genetic variation. The analyses of the site-frequency distribution in two different markers (that is, mitochondrial COI and nuclear EF1-α) revealed a relatively small, but consistent, excess of rare alleles within local populations. This excess could be the result of sampling admixture (Ptak and Przeworski, 2002; Hammer et al., 2003), as the genetic pool of adults flying in each local population includes both local recruitment and natal dispersal (Corbet, 1999). Accordingly, analyses of genetic variation at nine unlinked microsatellite loci revealed a significant genome-wide reduction of heterozygosity, suggesting that sexual populations are indeed comprising genetically dissimilar individuals from different neighboring locations (Freeland et al., 2000).
This pattern of population structure that we observe in I. hastata is the result not only of the present-day gene flow, but also reflects historical processes. Both slow (mitochondrial COI and nuclear EF1-α) and fast (microsatellites) evolving polymorphic markers have allowed us to infer the demographic history of this species in North America. First, the star-like shape of the nuclear and mitochondrial haplotype networks is indicative of a population expansion occurred during the recent history of the species. Neutrality tests of mitochondrial DNA provided also strong indication for the deviation from mutation-drift equilibrium, and mismatch analysis was unimodal for the sexual cluster, thus providing further indication of population expansion.
The erosion of mitochondrial polymorphism might be also due to natural selection either purifying or adaptive (Bazin et al., 2006), since the lack of recombination in mitochondrial DNA makes it act as a single locus, and thus selection acting anywhere in the mtDNA would exactly mimic a demographic bottleneck/expansion. To distinguish between these two evolutionary processes, we have used the imbalance index and bottleneck tests to infer aspects of population history and demography of sexual I. hastata using the microsatellite data. We found concordance between the results of these analyses and the ones obtained from COI data, suggesting that the sexual populations of this species have suffered a recent expansion, and that the demography of pre-expansion was relatively stable.
Genetic diversity of asexual populations: age and origin of parthenogenetic lineage
Our results confirm that sexual populations of I. hastata are connected by dispersal over a wide geographic area. The detection of a pattern of population expansion in multiple, independent, loci allows us to distinguish it from selection and to attribute it to a past demographic change, which is consistent with the low levels of genetic differentiation currently found across the species' range in North America. This range expansion is likely to be related to the biology and habitat preferences of I. hastata, a pioneer species usually found in temporary ponds where local extinctions are frequent, and adult individuals are thus forced to emigrate to new suitable habitats. Weather fronts have a major role in the dispersal of odonates, and long distance dispersal being passively transported as part of the aerial plankton is frequent in the case of weak flyers such as I. hastata. In fact, this species has been reported over water far from mainland in the Gulf of Mexico (Corbet, 1999). The possibility then exists that this species has been transported to the Azores archipelago by the surface winds associated with the Gulf Stream. Thus, the origin of the populations of this species in the Azores islands is likely to be the result of a single long distance dispersal event, rather than the result of a human introduction associated with shipping trade between the North-American continent and the Azores archipelago (Cordero Rivera et al., 2005).
As expected if the parthenogenetic populations from the Azores have their origin in the sexual populations that inhabit the American continent, microsatellite alleles and the only EF1-α allele found in the islands are a subset of those found in the sexual populations. In contrast, mtDNA revealed parthenogenetic populations as a differentiated cluster, and none of the two almost identical COI haplotypes that are present in the archipelago are found in any of the sexual populations sampled. This observed discordance between nuclear and mitochondrial markers can be potentially explained by a recent gene exchange between both reproductive modes, although this hypothesis is extremely is unlikely, given the great geographic distance that exists between both lineages. Instead, the observed discordance is better explained as the result of a recent fixation of a single diploid thelytokous lineage in the Azores archipelago (see Lorenzo-Carballa and Cordero-Rivera, 2009).
Because apomixis (that is, thelytokous parthenogenesis in which the eggs do not undergo meiosis) generates instant complete linkage disequilibrium between the mitochondrial and nuclear genome, shortly after the fixation there should be no genetic variation among clones. However, there is at least one very recently derived (that is, S=1) COI haplotype, and one derived microsatellite allele segregating in the population at frequencies of 3.5 and 0.6%, respectively, indicating that the ancestral parthenogenetic lineage has been in the archipelago long enough to start accumulating mutations.
The use of mitochondrial DNA to study sexual–asexual lineage divergence can lead to invalid age estimates, due to the extinction of sexual lineages, an inadequate sampling, or if sexual populations have diverged considerably (see Johnson, 2006; and references therein). In our case, we found a single mutational step between sexual and parthenogenetic lineages suggesting a very recent origin for the parthenogenetic lineages. Population frequencies of the Ihas16 alleles can be used to obtain maximum likelihood estimates of the age of the specific parthenogenetic clones (Slatkin and Rannala, 1997; data not shown) However, these estimates showed wide confidence intervals, and could not reject a very recent origin (<20 generations ago), again indicating the young age of the parthenogenetic lineages.
Despite the great geographic area comprised by the sexual localities sampled in this study, they do not represent the actual distribution range of sexual I. hastata, which has been recorded from Canada to South America. A more extensive sampling effort in the American continent would probably reveal higher genetic diversity in the sexual lineage and could help to better understand metapopulation dynamics in this species. Also, the fact that we have found three highly related microsatellite multilocus genotypes in the asexual populations suggests that genetic variability in the parthenogens could be higher than what has been detected in the current study. Thus, including more samples from the Azores, and increasing the resolution power of the analyses by adding more loci (for example, amplified fragment lenght polymorphisms, AFLPs) will probably reveal more diversity in the asexual populations and could also help to better date the time of divergence among both lineages.
One of the most intriguing questions that arise from this work is whether the I. hastata population found at the Azores arose from the immigration of sexual damselflies, which subsequently evolved into parthenogens, or whether parthenogens originated in America and then dispersed to the archipelago. Thus, asexuality could have arisen once in the Azores, and asexuals subsequently outcompeted and drove to extinction their sexual ancestors, given that the lower intrinsic rate of demographic growth of sexual reproduction would theoretically cause sexual individuals to be replaced by parthenogens, all else being equal (Maynard-Smith, 1978). Alternatively, the capacity for parthenogenetic reproduction could be already present in the ancestral sexual population, and therefore in the individuals that colonized the Azores. In this case, parthenogenetic reproduction could have been advantageous for the colonization of the islands. However, with the data available, it is not possible to answer this question. Future research should focus on the study of the potential for parthenogenetic reproduction in sexual females, to better understand the ecological conditions that could have favored the loss of sex in this species.
Acknowledgments
This work was supported by a research grant of the Spanish Ministry of Education and Science to ACR, which included FEDER funds (CGL2005-00122 and CGL2008-02799) and a predoctoral grant to MOLC. We thank Bernd Schierwater for providing the laboratory facilities and all the staff of the ITZ in Hannover for their help during most of the laboratory work. This work would not have been possible without the collaboration of Laura Sirot, Carl Cook, Alejandro Córdoba-Aguilar, Dennis Paulson and George L Harp who kindly provided specimens of I. hastata from most of the sexual populations. We also thank Thomas M McCleskey for writing a program, which formats sequence data for PHASE analysis and reincorporates the PHASE output into a nexus format. Mario García-París, Richard G Harrison and two anonymous referees gave useful comments in early drafts of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bazin E, Glemin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1132585. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nielsen R, Robert C, Hey J, Gaggiotti O, Knowles L, et al. In defence of model-based inference in phylogeography. Mol Ecol. 2010;19:436–446. doi: 10.1111/j.1365-294X.2009.04515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. University of California Press: Berkeley; 1982. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Corbet PS. Dragonflies. Behaviour and Ecology of Odonata. Harley Books: Essex, UK; 1999. [Google Scholar]
- Cordero Rivera A, Lorenzo Carballa MO, Utzeri C, Vieira V. Parthenogenetic Ischnura hastata (Say, 1839), widespread in the Azores (Zygoptera: Coenagrionidae) Odonatologica. 2005;34:1–9. [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1997;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar O. Biogeography of parthenogenetic animals. Biogeographica. 1994;70:1–13. [Google Scholar]
- De Block M, Geenen S, Jordaens K, Backeljau T, Stoks R. Spatiotemporal allozyme variation in the damselfly, Lestes viridis (Odonata:Zygoptera): gene flow among permanent and temporary ponds. Genetica. 2005;124:137–144. doi: 10.1007/s10709-005-0623-4. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeland JR, Noble LR, Okamura B. Genetic consequences of the metapopulation biology of a facultatively sexual freshwater invertebrate. J Evol Biol. 2000;13:383–395. [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons LK, Michael Reed J, Chew FS. Habitat requirements and local persistence of three damselfly species (Odonata: Coenagrionidae) J Insect Conserv. 2002;6:47–55. [Google Scholar]
- Glesener RR, Tilman D. Sexuality and the components of environmental uncertainty: clues for Geographic Parthenogenesis in terrestrial animals. Am Nat. 1978;112:659–673. [Google Scholar]
- Haag CR, Ebert D. A new hypothesis to explain geographic parthenogenesis. Ann Zool Fenn. 2004;41:539–544. [Google Scholar]
- Hamilton WD, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proc Natl Acad Sci USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Blackmer F, Garrigan D, Nachman MW, Wilder JA. Human population structure and its effects on sampling Y chromosome sequence variation. Genetics. 2003;164:1495–1509. doi: 10.1093/genetics/164.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JL, Bohonak A, Kelley ST. Isolation by distance, web service. BMC Genet. 2005;6:13. doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SG. Geographic ranges, population structure and ages of sexual and parthenogenetic snail lineages. Evolution. 2006;60:1417–1426. [PubMed] [Google Scholar]
- Judson OP, Normark BB. Ancient asexual scandals. Trends Ecol Evol. 1996;11:41–46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- Kimmel M, Chakraborty R, King JP, Bamshad M, Watkins WS, Jorde LB. Signatures of population expansion in microsatellite repeat data. Genetics. 1998;148:1921–1930. doi: 10.1093/genetics/148.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JP, Kimmel M, Chakraborty R. A power analysis of microsatellite-based statistics for inferring past population growth. Mol Biol Evol. 2000;17 (12:1859–1868. doi: 10.1093/oxfordjournals.molbev.a026287. [DOI] [PubMed] [Google Scholar]
- Knowles LL. The burgeoning field of statistical phylogeography. J Evol Biol. 2004;17:1–10. doi: 10.1046/j.1420-9101.2003.00644.x. [DOI] [PubMed] [Google Scholar]
- Knowles LL, Maddison WP. Statistical phylogeography. Mol Ecol. 2002;11 (12:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–441. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D.2001Genetic Data Analysis: computer program for the analysis of allelic data. Version 1.0 (d16c)Free program distributed by the authors over the Internet fromhttp://lewis.eeb.uconn.edu/lewishome/software.html .
- Lively CM. A review of the Red Queen models for the persistence of obligate sexual reproduction. J Hered. 2010;101 (Suppl 1:S13–S20. doi: 10.1093/jhered/esq010. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Carballa MO, Cordero-Rivera A. Thelytokous parthenogenesis in the damselfly Ischnura hastata (Odonata, Coenagrionidae): genetic mechanisms and the lack of bacterial infection. Heredity. 2009;103:377–384. doi: 10.1038/hdy.2009.65. [DOI] [PubMed] [Google Scholar]
- Lorenzo Carballa MO, Cordero Rivera A, Giere S, Hadrys H. Isolation and characterization of microsatellite loci to study parthenogenesis in the citrine forktail, Ischnura hastata. Mol Ecol Notes. 2007;7:839–841. [Google Scholar]
- Luikart G, Sherwin WB, Steele BM, Allendorf FW. Usefulness of molecular markers for detecting population bottlenecks via monitoring genetic change. Mol Ecol. 1998;7:963–974. doi: 10.1046/j.1365-294x.1998.00414.x. [DOI] [PubMed] [Google Scholar]
- Lynch Destabilizing hybridization, general-purpose genotypes and geographic parthenogenesis. Q Rev Biol. 1984;59:257–290. [Google Scholar]
- Maynard-Smith J. The Evolution of Sex. Cambridge University Press: Cambridge; 1978. [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl. 2003;13:S146–S158. [Google Scholar]
- Panchal M, Beaumont MA. The automation and evaluation of nested clade phylogeographic analysis. Evolution. 2007;61:1466–1480. doi: 10.1111/j.1558-5646.2007.00124.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ. The coup de grâce for the nested clade phylogeographic analysis. Mol Ecol. 2008a;17:516–518. doi: 10.1111/j.1365-294X.2007.03589.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ. On the falsifiability of the nested clade phylogeographic analysis method. Mol Ecol. 2008b;17:1404. doi: 10.1111/j.1365-294X.2007.03589.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Grivet D. Optimal randomization strategies when testing the existence of a phylogeographic structure. Genetics. 2002;161:469–471. doi: 10.1093/genetics/161.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson GH, Taggart CT, Mesa KA, Boutilier RG. Isolation by distance in the Atlantic cod, Gadus morhua, at large and small geographic scales. Evolution. 2001;55:131–146. doi: 10.1111/j.0014-3820.2001.tb01279.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Templeton AR. GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol Ecol. 2000;9:487–488. doi: 10.1046/j.1365-294x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donelly P. Inference of population structure from multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak SE, Przeworski M. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 2002;18:559–563. doi: 10.1016/s0168-9525(02)02781-6. [DOI] [PubMed] [Google Scholar]
- Purse BV, Hopkins GW, Day KJ, Thompson DJ. Dispersal characteristics and management of a rare damselfly. J Appl Ecol. 2003;40:716–728. [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rouquette JR, Thompson DJ. Patterns of movement and dispersal in an endangered damselfly and the consequences for its management. J Appl Ecol. 2007;44:692–701. [Google Scholar]
- Rozas J, Sánchez del Barrio JC, Messeger X, Rozas R. DNAsp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Rannala B. The sampling distribution of disease-associated alleles. Genetics. 1997;147:1855–1861. doi: 10.1093/genetics/147.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf1995Biometry.3rd edn.W.H. Freeman: New York [Google Scholar]
- Stephens M, Smith NJ, Donelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Routman E, Phillips CA. Separating population structure from population history-a cladistic analysis if the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics. 1995;140:762–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek RC.1985Animal population genetics and disturbance: the effects of local extinctions and recolonizations on heterozygosity and fitnessIn: Pickett STA & White P (eds).The Ecology of Natural Disturbance and Patch Dynamics Academic Press: New York; 265–285. [Google Scholar]
- Wade MJ, McCauley DE. The effects of extinction and colonization on the genetic differentiation of populations. Evolution. 1988;42:995–1005. doi: 10.1111/j.1558-5646.1988.tb02518.x. [DOI] [PubMed] [Google Scholar]
- Zaykin DV, Pudovkin A, Weir BS. Correlation based inference for linkage disequilibrium with multiple alleles. Genetics. 2008;180:533–545. doi: 10.1534/genetics.108.089409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.