Abstract
Prion diseases are fatal neurodegenerative diseases of humans and animals caused by the misfolding and aggregation of prion protein (PrP). Mammalian prion diseases are under strong genetic control but few risk factors are known aside from the PrP gene locus (PRNP). No genome-wide association study (GWAS) has been done aside from a small sample of variant Creutzfeldt–Jakob disease (CJD). We conducted GWAS of sporadic CJD (sCJD), variant CJD (vCJD), iatrogenic CJD, inherited prion disease, kuru and resistance to kuru despite attendance at mortuary feasts. After quality control, we analysed 2000 samples and 6015 control individuals (provided by the Wellcome Trust Case Control Consortium and KORA-gen) for 491032-511862 SNPs in the European study. Association studies were done in each geographical and aetiological group followed by several combined analyses. The PRNP locus was highly associated with risk in all geographical and aetiological groups. This association was driven by the known coding variation at rs1799990 (PRNP codon 129). No non-PRNP loci achieved genome-wide significance in the meta-analysis of all human prion disease. SNPs at the ZBTB38–RASA2 locus were associated with CJD in the UK (rs295301, P = 3.13 × 10−8; OR, 0.70) but these SNPs showed no replication evidence of association in German sCJD or in Papua New Guinea-based tests. A SNP in the CHN2 gene was associated with vCJD [P = 1.5 × 10−7; odds ratio (OR), 2.36], but not in UK sCJD (P = 0.049; OR, 1.24), in German sCJD or in PNG groups. In the overall meta-analysis of CJD, 14 SNPs were associated (P < 10−5; two at PRNP, three at ZBTB38–RASA2, nine at nine other independent non-PRNP loci), more than would be expected by chance. None of the loci recently identified as genome-wide significant in studies of other neurodegenerative diseases showed any clear evidence of association in prion diseases. Concerning common genetic variation, it is likely that the PRNP locus contains the only strong risk factors that act universally across human prion diseases. Our data are most consistent with several other risk loci of modest overall effects which will require further genetic association studies to provide definitive evidence.
INTRODUCTION
Prion diseases are progressive neurodegenerative conditions of humans and animals caused by the misfolding and aggregation of the prion protein (PrP) (1). The most common human prion disease is sporadic or classical Creutzfeldt–Jakob disease (sCJD) which like other sporadic neurodegenerative disorders occurs with increasing incidence in older adults. Despite decades of investigation, no consistent risk factors for sCJD have been identified aside from age and common genetic variation at the human PrP gene (PRNP). Human prion diseases comprise three aetiologies: acquired, inherited and sporadic. While less common, acquired prion diseases are important because of public health concerns, such as those following the transmission of the cattle prion disease, bovine spongiform encephalopathy, to predominantly young British adults as variant Creutzfeldt–Jakob disease (vCJD) (2–4).
A common amino-acid polymorphism at codon 129 of PRNP, encoding either methionine or valine, is a strong genetic risk factor or modifier of the clinicopathological phenotype in all types of prion diseases (5–8). Codons 127 and 219 also harbour amino-acid polymorphisms which confer resistance to either kuru or sCJD (9,10). Although these are powerful effects, the transmissibility of prion diseases in laboratory rodents allows for genetic mapping studies which have demonstrated several non-Prnp modifier loci (11–13). Parallel human studies have also begun to suggest prion disease risk genes based on candidates derived from close functional links to PrP, screening human genes orthologous to mouse candidates, or genome-wide association studies (GWAS) in vCJD; examples include the RARB–THRB locus (14) and SPRN in vCJD (15); and HECTD2 (16) and STMN2 (14) in several human prion diseases. These human studies were underpowered by the necessarily small sample size of vCJD and the rarity of prion diseases in general.
Many of the neurodegenerative diseases share fundamental mechanisms involving protein misfolding and prion-like spreading of pathology associated with abnormally aggregated proteins in brain tissue (17,18). Such shared mechanisms might implicate joint genetic risk factors. As several GWAS have identified causal loci in Alzheimer's disease, frontotemporal dementia, Parkinson's disease and amyotrophic lateral sclerosis (19–22), testing these in prion diseases may provide insights into disease mechanisms more broadly.
Here, we present the first large GWAS in human prion diseases based on 2000 samples from three populations and relevant publicly available control series. In a single-stage design, we genotyped 579 sCJD, 133 vCJD, 137 inherited prion disease (IPD) and 32 iatrogenic Creutzfeldt–Jakob disease (iCJD) from the UK; 680 sCJD from Germany; and 568 samples from Papua New Guinea (PNG) including kuru and elderly female survivors of the kuru epidemic. The WTCCC (UK) or KORA (German) provided 6507 controls (23). Association analyses confirm the dominance of PRNP as a risk factor relative to all other genes. We are able to provide evidence for several additional genetic risk factors although none of these achieved genome-wide significance in meta-analyses between regions or aetiologies.
RESULTS
After quality control (QC), 8015 samples were analysed (see Materials and methods). Association analysis was first done in individual aetiological groups and geographical regions; these were then combined in meta-analyses. The predetermined primary study was a meta-analysis of human prion disease (allelic tests, case–control design, sCJD, vCJD, resistance to kuru) from all geographical regions. Other combinations and individual aetiological and geographical tests were secondary outcomes, for example, all sCJD, sCJD and vCJD in UK, sCJD (UK or German alone), vCJD, kuru (age of death), resistance to kuru, IPD (age of onset), and sCJD (age of onset). As a large proportion of the Fore population were affected by kuru including those with apparent genetic resistance at PRNP, we hypothesized that case–control study would not be the best strategy and instead kuru age of death was used as a quantitative trait. Age of death in kuru is significantly older in PRNP codon 129 heterozygous individuals compared with homozygous individuals (24). A similar effect is seen for several IPDs, and in a similar way, age was used as a quantitative trait (25–27).
In the overall meta-analysis of allelic tests, the top-ranked association was the known amino-acid polymorphism at PRNP codon 129 rs1799990 [overall P = 6.58 × 10−7, odds ratio (OR) = 0.77; in CJD groups, P = 1.24 × 10−8;Table 1]; however, there was considerable heterogeneity between the UK and Germany probably because of case ascertainment (UK, OR = 0.84; Germany, OR = 0.60, highly significant difference shown with the use of a Cochran–Mantel–Haenszel test for a 2 × 2 × 2 table and Breslow-Day test of homogeneity implemented using PLINK) (Fig. 1). Notably, in several prion diseases, rs1799990 confers resistance in the heterozygous state; therefore, an allelic model does not optimally capture the signal at this locus. Three other SNPs were associated with the PRNP locus in sCJD at P < 10−5: rs2756271, rs6107516 and rs6116477. Each of these SNPs had linkage disequilibrium (LD) with rs1799990 r2> 0.3, and conditional or stratified analyses suggested that the entire association signal was conferred by rs1799990. Although the primary analysis used an allelic model, a genotypic model showed a much stronger signal, as expected; for example, at rs1799990 in UK sCJD, Pgeno= 3.85 × 10−25 and in German sCJD, Pgeno= 2.51 × 10−31, with a deficit of heterozygous genotypes consistent with the known protective effect at this locus (6). Contrary to other studies, this GWAS provided no evidence of additional risk at the locus in sCJD (14,28–30). SNP rs2756271 was genotyped in the current study and showed very high LD with SNP rs1029273 (r2= 0.94) genotyped in previous studies in UK sCJD cases (14,28–30). Conditional analysis based on rs2756271–rs1799990 haplotype frequencies in cases and controls showed no evidence for additional risk conferred (rs2756271–rs1799990 haplotype OR, AA = 1.419, GA = 1.384, P = 0.65, implemented with PLINK).
Table 1.
Top ranked associations ordered by P-value based on meta-analysis of vCJD, sCJD and kuru resistance
| CHR | SNP | BP | Minorallele | Majorallele | P (overall meta) | q-value | OR | vCJD | OR | L95 | U95 | UK sCJD | OR | L95 | U95 | GermansCJD | OR | L95 | U95 | PGC corrected | iCJD P-value | OR | L95 | U95 | sCJD age atonset QT | Kuru age atdeath QT | Kururesistance | OR | L95 | U95 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | rs1799990 | 4628251 | G | A | 6.583E − 07 | 0.3323 | 0.7664 | 6.01E − 30 | 0.007496 | 0.001 | 0.053 | 0.015 | 0.844 | 0.74 | 0.97 | 1.543E − 09 | 0.601 | 0.509 | 0.709 | 9.778E − 09 | 0.0006295 | 2.578 | 1.47 | 4.53 | 0.0001313 | 0.2986 | 0.2579 | 0.8416 | 0.62 | 1.14 |
| 20 | rs2756271 | 4613262 | A | G | 1.005E − 06 | 0.3323 | 1.2614 | 2.26E − 08 | 2.04 | 1.58 | 2.63 | 0.4876 | 1.047 | 0.92 | 1.19 | 0.00003751 | 1.364 | 1.177 | 1.581 | 0.0000908 | 0.008182 | 0.4288 | 0.22 | 0.82 | 0.5318 | 0.2223 | 0.3211 | 1.537 | 0.65 | 3.62 |
| 3 | rs9857275 | 142560878 | A | C | 1.531E − 06 | 0.3323 | 0.7817 | 0.0008636 | 0.6053 | 0.45 | 0.82 | 6.08E − 05 | 0.747 | 0.65 | 0.86 | 0.1631 | 0.892 | 0.759 | 1.048 | 0.1855 | 0.145 | 1.503 | 0.87 | 2.61 | 0.2239 | – | – | – | – | |
| 7 | rs488333 | 83749207 | G | A | 2.683E − 06 | 0.3323 | 1.2681 | 0.05605 | 1.295 | 0.99 | 1.69 | 0.001928 | 1.24 | 1.08 | 1.42 | 0.001972 | 1.298 | 1.100 | 1.531 | 0.003303 | 0.6202 | 0.8531 | 0.45 | 1.6 | 0.1584 | – | 0.538 | 1.341 | 0.53 | 3.42 |
| 3 | rs13095453 | 142557652 | A | G | 3.297E − 06 | 0.3323 | 0.7892 | 0.0005654 | 0.595 | 0.44 | 0.8 | 0.0001092 | 0.756 | 0.66 | 0.87 | 0.2166 | 0.904 | 0.770 | 1.061 | 0.2408 | 0.09119 | 1.598 | 0.92 | 2.77 | 0.001122 | – | – | – | – | |
| 10 | rs12570947 | 109559840 | G | A | 4.287E − 06 | 0.3323 | 0.6877 | 0.001679 | 0.4778 | 0.3 | 0.77 | 8.63E − 05 | 0.663 | 0.54 | 0.82 | – | – | – | – | – | 0.2936 | 0.6126 | 0.24 | 1.54 | 0.04276 | 0.4233 | 0.3161 | 0.8587 | 0.64 | 1.16 |
| 4 | rs2903698 | 76551999 | G | A | 5.792E − 06 | 0.3323 | 1.2528 | 0.0001428 | 1.652 | 1.27 | 2.15 | 0.04848 | 1.153 | 1 | 1.33 | 0.004737 | 1.271 | 1.076 | 1.501 | 0.007326 | 0.6077 | 1.17 | 0.64 | 2.14 | 0.4199 | 0.8633 | 0.3116 | 1.194 | 0.85 | 1.69 |
| 10 | rs12761224 | 72197523 | A | C | 6.129E − 06 | 0.3323 | 1.3861 | 0.002949 | 1.685 | 1.19 | 2.39 | 0.007039 | 1.305 | 1.08 | 1.58 | 0.00881 | 1.385 | 1.085 | 1.768 | 0.01288 | 0.9479 | 0.9697 | 0.38 | 2.44 | 0.3808 | – | – | – | – | |
| 5 | rs17763373 | 173291937 | G | A | 6.807E − 06 | 0.3323 | 1.5106 | 0.09972 | 1.461 | 0.93 | 2.3 | 0.002878 | 1.427 | 1.13 | 1.81 | 0.001012 | 1.711 | 1.238 | 2.364 | 0.001801 | 0.2236 | 0.3124 | 0.04 | 2.27 | 0.3435 | – | – | – | – | |
| 3 | rs730566 | 48462052 | A | C | 7.374E − 06 | 0.3323 | 0.7848 | 0.5272 | 0.9102 | 0.68 | 1.22 | 0.0009991 | 0.773 | 0.66 | 0.9 | 0.000827 | 0.762 | 0.649 | 0.894 | 0.0015 | 0.2554 | 0.6714 | 0.34 | 1.34 | 0.2918 | 0.977 | 0.8383 | 0.8817 | 0.26 | 2.96 |
| 8 | rs10108954 | 8759788 | A | G | 7.498E − 06 | 0.3323 | 1.7359 | 0.3834 | 1.349 | 0.69 | 2.65 | 0.0001211 | 1.814 | 1.33 | 2.47 | 0.01196 | 1.767 | 1.127 | 2.770 | 0.01702 | 0.6089 | 1.445 | 0.35 | 5.97 | 0.07429 | – | – | – | – | |
| 19 | rs8105815 | 46365688 | A | C | 7.625E − 06 | 0.3323 | 1.3918 | 0.1643 | 1.31 | 0.89 | 1.92 | 0.001828 | 1.357 | 1.12 | 1.65 | 0.001617 | 1.490 | 1.161 | 1.911 | 0.002758 | 0.665 | 1.207 | 0.51 | 2.83 | 0.06505 | – | – | – | – | |
| 3 | rs6785073 | 142622020 | A | G | 8.223E − 06 | 0.3323 | 0.8 | 0.08123 | 0.7838 | 0.6 | 1.03 | 4.38E − 05 | 0.748 | 0.65 | 0.86 | 0.105 | 0.878 | 0.750 | 1.028 | 0.1238 | 0.0229 | 1.864 | 1.08 | 3.22 | 0.4644 | – | – | – | – | |
| 8 | rs2071598 | 99198683 | A | C | 8.966E − 06 | 0.3349 | 1.2646 | 0.5071 | 1.107 | 0.82 | 1.5 | 0.002574 | 1.256 | 1.08 | 1.46 | 0.001223 | 1.339 | 1.121 | 1.598 | 0.002141 | 0.7969 | 0.9133 | 0.46 | 1.82 | 0.1366 | 0.5076 | 0.1635 | 1.259 | 0.91 | 1.74 |
Additional association studies are shown with a small sample of iatrogenic CJD, sCJD age of onset (as a quantitative trait) and kuru age at death (as a quantitative trait). BP, base pair position build 36; OR, odds ratio of allelic test (with reference to the minor allele); L95/U95, 95% confidence intervals of OR; QT, quantitative trait.
Figure 1.
Manhattan plots showing P-values from allelic (1 d.f.) and genotypic (2 d.f.) chi-squared tests for five GWAS studies involving (A) vCJD versus UK controls (WTCCC2), (B) UK sCJD versus UK controls, (C) German sCJD versus German controls (KORA), (D) kuru using age of death as a quantitative trait and (E) elderly women survivors of the kuru epidemic (elderly women) versus healthy young (unexposed) Fore.
Aside from the PRNP locus, there were no genome-wide significant associations in the predetermined meta-analysis of all human prion disease; however, other loci did provide evidence of association in individual aetiological categories or regions. The top non-PRNP locus was at ZBTB38–RASA2 on chromosome 3 (Fig. 2) with rs9857275 (P = 1.53 × 10−6, OR = 0.7817), rs13095453 and rs6785073 (P < 10−5). In the UK, all CJD analysis, the top-ranked association aside from PRNP, was at the ZBTB38–RASA2 locus, rs295301 (P = 3.13 × 10−8, OR = 0.70; sCJD: P = 3.73 × 10−6, OR = 0.72; vCJD: P = 0.0014, OR = 0.6284); however, this SNP showed no evidence of association in German sCJD or in PNG. These SNPs have pairwise LD (r2 > 0.3) over 300 kb on chromosome 3 (Fig. 2). Rs295301 also showed weak evidence of a modifying effect on age of onset of sCJD (as a quantitative trait) with the risk allele being associated with an earlier age of onset (P = 0.04).
Figure 2.
Illustration of association signals from the top two associated genetic loci in this study (UK and German sCJD meta-analysis; PRNP locus, top; ZBTB38 locus, bottom). −Log10 P is shown from an allelic test using both genotyped and imputed SNPs (see Materials and methods). Blue line corresponds to recombination rate in cM/Mb (right-hand scale, Broad Institute SNAP Regional Association Plot, http://www.broadinstitute.org/mpg/snap/ldplot.php). Gene locations shown referencing build 37.
Other loci (P < 10−5) in the primary study meta-analysis are shown in Table 1. In total, 14 SNPs were associated, P < 10−5 (2 at PRNP and 12 SNPs at 10 independent non-PRNP loci). We examined whether we found an excess of P < 10−5 in three ways. First, by permuting the case–control status of all subgroups contributing to the meta-analysis (see Materials and methods), we found >13 SNPs on 8/500 permutations (γ-Poisson fitted P = 0.014) and >11 SNPs on 19/500 permutations (γ-Poisson P = 0.038). Second, the binomial probability of >13 SNPs (P < 10−5) (n = 511862 tests) was P = 0.001 and >11 SNPs was P = 0.007; we also accounted for LD by estimating the effective number of independent tests (see Materials and methods), >9 independent loci (n = 374943) P = 0.005 and >10 independent loci P = 0.002. Finally, the Q-value was 0.33 for all 14 SNPs, P < 10−5 (see Materials and methods). None of these loci showed any evidence of association in kuru, kuru resistance, or IPD (data not shown). Although several SNPs were not polymorphic in the Fore population, there were no association signals (P < 0.01) detected at these loci ±50 kb.
In the individual disease categories, there was only one non-PRNP finding at around levels of genome-wide significance in the CHN2 gene in vCJD, rs1016726, P = 1.5 × 10−7 (OR, 2.36), and marginally in UK sCJD P = 0.049 (OR, 1.24), but not German sCJD or in PNG groups. Associations previously described at the RARB–THRB locus and upstream of STMN2 were unsurprisingly similar but not stronger than those already published as there was an almost complete overlap in vCJD samples with the current study (14). All other disease categories showed no SNPs, P < 10−6.
Unlike vCJD, sCJD is a heterogeneous condition caused by multiple prion strain types (31). We therefore sought to explore evidence of association in defined subgroups of sCJD. The codon 129 polymorphism (rs1799990) is a powerful determinant of strain selection (32); however, we found no evidence of an interaction between these 10 loci and rs1799990. Stratification at rs1799990 did not strengthen any of the top associations or reveal any new loci (P < 10−6). Prion strains are known to determine key clinical features such as age of onset but no genome-wide significant loci were identified when testing only sCJD presenting prior to age 60.
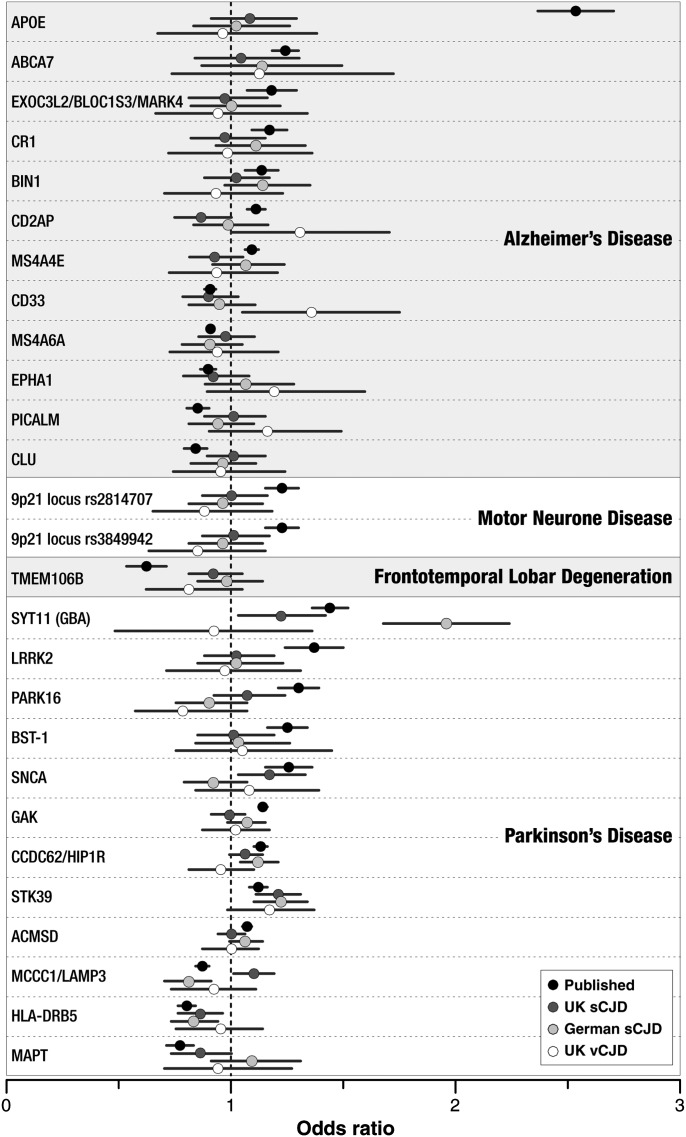
Known genome-wide significant associations in related neurodegenerative conditions showed no clear-cut evidence of association in a similar direction in prion disease (Fig. 3). Rs2102808 at STK39 showed an association with stronger effect than that described for Parkinson's disease in vCJD, UK sCJD and German sCJD with similar ORs in all groups (Fig. 3, Supplementary Material, Table S1).
Figure 3.
Strength of association at loci known to be involved in other neurodegenerative diseases. Dots refer to estimates of OR from the top-ranked SNPs in published association studies of alternative neurodegenerative diseases (see Supplementary Material, Table S1), and the same SNP and direction in prion diseases (the current study), with bars showing 95% confidence intervals of the OR. For further details of associations, SNP names and primary study citations, see Supplementary Material, Table S1.
DISCUSSION
We report the first GWAS of sCJD and kuru and the first study powered to detect moderately strong risk loci in human prion disease (OR > 1.5). We considered several different hypotheses of genetic risk, including whether effects were disease subtype or region specific, and whether possible risk alleles modify clinical phenotype. In 2009, we published findings from a GWAS in vCJD, suggesting SNPs upstream of the RARB and STMN2 loci were candidates for vCJD association; however, this study was only powered to detect very strong effects discovered in vCJD (14). Unsurprisingly, in the current study, PRNP genotypes were strongly associated with risk in all disease categories and regions. No other locus showed a similarly strong or universal association. In the meta-analysis, however, 10 other loci were found to approach the statistical threshold for suggestive GWAS significance (P < 10−5), beyond the number expected by chance, consistent with several modest genetic effects. Further genetic analyses are required to provide convincing evidence for association at these loci. We also demonstrate that it is feasible to conduct GWAS on old degraded and amplified DNA in the kuru archive collection.
A wealth of evidence places PrP centrally in the pathogenesis of all prion diseases. The genetic association at PRNP was very strong and present across regions, disease subtypes and clinical phenotype albeit with different genetic models in aetiological subgroups. For example, in vCJD, all but one sample was homozygous A (encoding methionine) at rs1799990 (codon 129), whereas both homozygous genotypes were at increased risk relative to heterozygous in all other disease groups.
Several studies have reported additional weak associations with sCJD or vCJD at the PRNP locus beyond that conferred by rs1799990 or other missense polymorphisms (14,28–30). In this study, our conditional analyses did not support these findings with no evidence for additional factors; possible explanations include different SNPs genotyped (although we have shown the genotyped SNP was an excellent surrogate for a previously reported risk allele), the failure of imputation to accurately genotype other SNPs, and false positive or false negative associations. This study suggests that additional association at the PRNP locus, if present, is likely to be of modest effect overall.
Several other loci approached genome-wide significance in overall meta-analysis. Some of these loci are in the vicinity of genes that might be plausible biological candidates for conferring risk of prion disease, others are in gene-dense regions or gene deserts. For example, rs9857275, top ranked in the meta-analysis, is intronic to ZBTB38, a zinc finger transcriptional activator that binds methylated DNA, is expressed in brain and several SNPs in the gene are associated with adult height in multiple populations (33). Second ranked in the overall meta-analysis was rs488333, upstream of SEMA3A, a secreted protein with chemoattractive or repulsive functions such as the inhibition of axonal outgrowth or stimulation of the growth of apical dendrites. Other loci (P < 10−5) in gene-dense regions or deserts make it difficult to generate hypotheses of a possible mechanism. The priority for future work is to generate robust genome-wide statistical significance by replication study in sCJD from other populations.
Several genes have already been proposed as prion disease risk factors, including those at the RARB–THRB locus, upstream of STMN2 and in the HECTD2 and SPRN genes. The meta-analysis across all prion diseases provided no additional support for these proposals; however, this is not surprising as all of these reports were heterogenous across different aetiological groups. We also considered whether associations in other related neurodegenerative diseases might be found in prion disease, for example, APOE, CLU, PICALM, BIN1 and the BLOC1S3 locus in Alzheimer's disease; MAPT, SNCA, LRRK2 and GBA in Parkinson's disease; and the Chr 9p21 locus in ALS; however, we found no evidence of even modest associations at these SNPs aside from very modest effects at STK39. These data suggest that these genetic pathways implicated in other neurodegenerative diseases are not generally shared or important in prion disease.
Overall our results are open to several different interpretations. Aside from PRNP, other genetic factors may be weak in prion diseases. Prion diseases are rare, and as a result, there is limited evidence regarding heritability. Inference of genetic susceptibility may be made from sibling concurrence (34), the heritability of age of onset in different IPDs (27), and by extrapolation from the clear evidence for genetic modifiers in mouse studies (12). Epidemiological studies have identified no modifiable risk factors, aside from inconsistent reports of an association with past surgery (35). The aetiology of sCJD therefore remains obscure to human studies, as mouse studies model acquired rather than sporadic prion disease.
Although the only genetic risk factor identified in this study was codon 129 of PRNP, it would be premature to conclude that no other proteins are involved in modifying the risk of disease. In comparison with successful GWAS in common diseases, the sample size we have analysed remains modest and weak effects would not be detected. We have not considered copy number variation or rare SNPs which may confer strong effects. Other explanations of the lack of associations in sCJD include the heterogeneity of this disease defined by a clinical syndrome. Several aetiologies are possible in the group including a proportion of zoonotic or iatrogenic disease. Each of these causes could have specific genetic risk factors that would be diluted in a study unable to correctly ascertain aetiology. Finally, it is possible that somatic mutation or stochastic (protein-based) events are central to aetiology and genetic modifiers play only a very small role in the downstream risk of clinical disease.
Future work should prioritize further GWAS in different populations and studies designed to ascertain rarer SNPs with possibly larger functional effects, analysis of copy number variation, pathway analysis and further study of the modification of clinical phenotype by genetic polymorphism.
MATERIALS AND METHODS
Human samples
The clinical and laboratory studies were approved by the local research ethics committee of University College London Institute of Neurology and National Hospital for Neurology and Neurosurgery and by the Medical Research Advisory Committee of the Government of PNG. Full participation of the PNG communities involved was established and maintained through discussions with village leaders, communities, families and individuals. Access to CJD patient genotype data is available through the European Genome-Phenome Archive at the European Bioinformatics Archive (accession number EGAS00000000097).
Variant Creutzfeldt–Jakob disease
Probable or definite vCJD patients, diagnosed according to established criteria (http://www.advisorybodies.doh.gov.uk/acdp/tseguidance/tseguidance_annexb.pdf), were recruited by the National Prion Clinic (NPC), London or the National CJD Research and Surveillance Unit, Edinburgh, from 1995 to 2010. Genomic DNA was usually extracted from peripheral blood; brain tissue was used as a source for some patients. Amplified DNA, using either multiple displacement amplification (Geneservice, Cambridge, UK) or fragmentation-PCR methods (Genomeplex, Sigma), was used for a small number (<10%) of samples. Samples were checked for degradation on 1% agarose gel and stored at 50 ng/µl in Tris-EDTA buffer. All patients were thought to have acquired the disease in the UK. Mean age of disease onset was 30 years, 56% were male.
Sporadic Creutzfeldt–Jakob disease
Probable or definite UK sCJD patients, according to WHO criteria, were recruited by the NPC, London, the NCJDRSU, Edinburgh, or numerous other referrers in the UK. DNA was sourced and amplified as for vCJD. All patients were of UK or northern European origin. Although the vast majority of patients were of white-British ethnicity, and all patients of known non-white ethnicity were excluded, this information was based on name and geography for some samples. DNA preparation and storage was similar to vCJD. In the UK 61% had pathologically confirmed sCJD, the remainder had a diagnosis of probable sCJD according to the published WHO criteria with a high specificity. Median age of onset of disease was 65 years (15–87); 272 participants were female. In Germany, all cases were pathologically confirmed, median age of onset was 66 (range 19–90); 306 participants were female.
Kuru/elderly women resistant to kuru
Prior to 1987, kuru surveillance was conducted by many different investigators (Gajdusek, Zigas, Baker, Alpers, Hornabrook, Moir and others) and from 1987 to 1995 solely by the Kuru Surveillance Team of the PNG Institute of Medical Research. From 1996 onwards, kuru surveillance was strengthened and a field base and basic laboratory for sample processing and storage was established in the village of Waisa in the South Fore. The kuru collection comprised young children, adolescents and adults from around the peak of the epidemic and elderly recent kuru cases with long incubation times.
Elderly exposed women were defined as aged over 50 years in 2000 from a kuru-exposed region. These women were unaffected at the time of sampling but were thought to have been exposed to kuru prions in childhood. Although these women may not be truly ‘resistant’ to kuru prions they would have incubation times in excess of 40 years. Additional controls were obtained from the young modern day healthy population that has not been exposed to kuru but came from villages in the exposed region by matching each elderly woman (‘resistant’) to at least two current residents of the same village aged <50 in 2000. These largely came from the South Fore, but with a significant number from the North Fore and a small number of individuals from Gimi, Keiagana and Yagaria linguistic groups. Further controls were obtained from young unexposed people from areas of PNG where no kuru has been recorded. Where identified by either genealogical data or microsatellite analysis, first degree relatives were excluded from these groups. DNA from degraded archival kuru sera, obtained from the NIH collection, was isolated by QIAGEN QIAamp Blood DNA minikit followed by whole genome amplification either through using a Φ29 protocol (Geneservice) or GenomePlex Complete Whole Genome Amplification Kit (Sigma).
Quality control
Totally, 840 KORA controls were genotyped on 550K Illumina arrays and 5667 WTCCC2 controls on the Illumina 1.2M Custom Duo array; cases were genotyped on the Illumina 660K. Cases included 680 German sCJD samples, 579 UK sCJD, 133 UK vCJD (and 5 non-UK vCJD), 165 kuru samples, 125 kuru resistant women and 286 geographically matched control individuals. There was no discrepancy between reported and genetic gender in the cases.
Prior to analysis, samples were removed from the WTCCC2 data set by their prescribed criteria. The WTCCC2 chose criteria to be similar to those often applied as standard in GWA studies. Individuals were excluded if they displayed a disproportionate number of heterozygous or missing calls. Related individuals were excluded according to identity by descent (IBD). Individuals were excluded on the basis of ancestry if they differed from the majority of the collection according to a principle component analysis of HapMap individuals. Gender discrepancy between the supplier and the inferred gender also led to sample removal. Individuals were excluded if the mean of their A and B allele intensities were outliers when compared with the sample at large. In total, 467 WTCCC2 individuals were removed leaving 5200 samples comprised of 2630 males and 2570 females.
We used a 98% sample genotyping call threshold for the main association tests; however, an 80% sample call rate was applied to the kuru samples. This allowed us to retain 90% of this small and unique case series. Observation of concordance between known duplicate pairs (n = 8) demonstrated that the overall genotype error was <0.00085 with a sample call rate above 80%. Analyses using kuru samples were restricted to this aetiological group alone using age at death as a quantitative trait to minimize the possibility of introducing bias by comparing cohorts with different QC. Cryptic duplicates were identified using the statistical genetics measure of relatedness, pi-hat. The threshold for exclusion was pi-hat > 0.8. A subset of 200K SNPs common to all three arrays and selected for the highest minor allele frequency (MAF) were used to derive pi-hat estimates. Only cryptic duplicates were discovered and no highly related individuals. Steps were taken to identify ethnic outliers within the sample set. As for the pi-hat estimates, the same 200K SNPs were used to derive identity by state estimates which were plotted on the first two axes of a multi-dimensional scaling plot. Samples outside the main cluster were excluded. Whole genome amplified samples did not appear as outliers and so were retained. After sample QC, 815 KORA, 634 German sCJD, 522 UKsCJD and 125 UKvCJD samples were retained for analysis. Five previously known non-UK vCJD samples appeared outside the main cluster.
QQ plots and genomic control indicated modest or no inflation of the association test statistic in all except the German study (lambda, UK vCJD = 1; UK sCJD = 1.02; German sCJD = 1.12; Fore = 0.99) (see Supplementary Material, Figure S1). We did not find evidence of technical artefacts related to the use of different genotyping arrays (through examination of QQ plots of SNPs with different allele frequencies, differential missingness and Hardy–Weinberg equilibrium, or associations with genotyping batches). We concluded that population stratification was the most likely explanation. Correction using principle components (Eigenstrat) reduced lambda in the German study from 1.11 to 1.037. For meta-analysis, German P-values and OR standard errors were corrected by the genomic control (GC) method (see Supplementary Material, Figure S1) (36). Allele frequencies were markedly different in the PNG population as expected, and so a different set of 175K SNPs was applied to the PNG samples. No PNG samples were removed as ethnic outliers.
SNP QC was applied to the control data together with the cases. SNPs with a missingness > 1%; MAF < 1%, or showing departure from Hardy–Weinberg equilibrium in controls (P < 1 × 10−3) were excluded. SNPs to be excluded varied depending upon the comparison being made. As expected the majority of SNPs excluded in the PNG comparisons were due to low minor allele frequencies in the PNG population. SNP rs1799990 was missing from the 550K KORA control set and the 660K cases but was present on the 1.2M Custom Duo WTCCC2 chips. SNP rs1799990 was genotyped for the missing samples using allelic discrimination PCR probes and the data added to the data set. This SNP passed all other QC measures and so was retained in the study to allow study of the PRNP associations and stratification.
The 1000 Genomes Pilot (June 2010) + HapMap3 (February 2009) CEU panel was used to impute SNPs 2.5 Mb upstream and downstream of our loci of interest (mentioned above). SNPs were strand aligned and phased, prior to imputation using the program Impute_v2 which employs a Markov chain Monte Carlo framework (37). Imputed SNPs with a missingness > 1%; MAF < 1%, or showing departure from Hardy–Weinberg equilibrium in controls (<1 × 10−3) were excluded. Subsequent tests of association were performed using SNPtestv2 which takes genotype improbability into account when comparing imputed genotypes (38).
Statistical analysis
Data manipulation and statistics were performed using PLINK (39). Cohorts of vCJD, UK sCJD, German sCJD, iatrogenic CJD and kuru resistant women were compared with relevant controls using allelic and genotypic models. Age at death (or sampling) was used as a quantitative trait in analysis of kuru patients. Age at clinical onset expressed as deviation from the mean expected onset for each IPD mutation was used as a quantitative trait in the IPD cohort. Meta-analyses were done using PLINK for all UK CJD, all sCJD, all CJD and all CJD including resistance to kuru. Imputation was done using IMPUTE2 using data provided by the WTCCC and 1000 genomes project. Conditional analyses were done at the PRNP locus using rs1799990 in additive and genotypic models. Stratified analyses were done using all three genotypes at rs1799990. Age of clinical onset in UK sCJD was used either as a quantitative trait or as a threshold for stratification (include only sCJD age at onset <60 years). We performed a permutation analysis to evaluate the significance of observing n SNPs with P < 10−5, when approximately five such SNPs were expected under the null hypothesis. Case–control labels were permuted within each of the four cohorts, and a genome-wide meta-analysis was performed on the permuted data set using the same analysis as for the observed data set. This was repeated 500 times generating crude estimates for the significance of this statistic (see Results). To improve on this estimate, we fitted a gamma-Poisson distribution (also known as the negative binomial) to the 500 counts of significant SNPs by matching the empirical mean and variance to their theoretical values (method of moments estimation). Although the choice of 10−5 as a significance threshold for this analysis is arbitrary, it is a natural choice and the only one we considered so we have not implemented any formal correction for multiple testing. We estimated the effective number of independent tests using simpleM (40).
SUPPLEMENTARY MATERIAL
FUNDING
Funding for the project was provided by the Wellcome Trust and Medical Research Council. The kuru studies were initially funded by a Wellcome Trust Principal Research Fellowship in the Clinical Sciences to J.C., and since 2001, all other aspects of the work by the Medical Research Council. Some of this work was undertaken at UCLH/UCL who received a proportion of funding from the Department of Health's NIHR Biomedical Research Centres funding scheme.
Supplementary Material
ACKNOWLEDGEMENTS
This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. We would like to thank patients, their families and carers, UK neurologists and other referring physicians, co-workers at the National Prion Clinic, our colleagues at the National Creutzfeldt-Jakob Disease Research and Surveillance Unit, Edinburgh, and the Fore communities in PNG. We thank our team of local kuru reporters, including Tuli Anua, Auyana Winagaiya, the late Anua Senavaiyo, Igana Aresagu, Kabina Yaraki, Anderson Puwa, David Pako, Pibi Auyana, Jolam Ove, Jack Kosinto, Dasta Hutu and James Kisava. We are grateful to Anthony Jackson and Peter Siba, John Reeder, Charles Mgone and other staff of the PNG Institute of Medical Research for their support. We gratefully acknowledge the help of the late Carleton Gajdusek, the late Joseph Gibbs and their associates from the former Laboratory of Central Nervous System Studies of the National Institutes of Health, Bethesda, USA, for archiving and sharing old kuru samples. Genotype data will be made available through the European Genome-phenome Archive.
Conflicts of Interest statement. None declared.
REFERENCES
- 1.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Collinge J., Sidle K.C., Meads J., Ironside J., Hill A.F. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 3.Bruce M.E., Will R.G., Ironside J.W., McConnell I., Drummond D., Suttie A., McCardle L., Chree A., Hope J., Birkett C., et al. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 4.Hill A.F., Desbruslais M., Joiner S., Sidle K.C.L., Gowland I., Collinge J. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 5.Collinge J., Palmer M.S., Dryden A.J. Genetic predisposition to iatrogenic Creutzfeldt–Jakob disease. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-v. [DOI] [PubMed] [Google Scholar]
- 6.Palmer M.S., Dryden A.J., Hughes J.T., Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt–Jakob disease. Nature. 1991;352:340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 7.Poulter M., Baker H.F., Frith C.D., Leach M., Lofthouse R., Ridley R.M., Shah T., Owen F., Collinge J., Brown J., et al. Inherited prion disease with 144 base pair gene insertion: I: genealogical and molecular studies. Brain. 1992;115:675–685. doi: 10.1093/brain/115.3.675. [DOI] [PubMed] [Google Scholar]
- 8.Collinge J. Molecular neurology of prion disease. Journal of Neurology Neurosurgery and Psychiatry. 2005;76:906–919. doi: 10.1136/jnnp.2004.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mead S., Whitfield J., Poulter M., Shah P., Uphill J., Campbell T., Al Dujaily H., Hummerich H., Beck J., Mein C.A., et al. A novel protective prion protein variant that colocalizes with kuru exposure. N. Engl. J. Med. 2009;361:2056–2065. doi: 10.1056/NEJMoa0809716. [DOI] [PubMed] [Google Scholar]
- 10.Shibuya S., Higuchi J., Shin R.W., Tateishi J., Kitamoto T. Protective prion protein polymorphisms against sporadic Creutzfeldt–Jakob disease. Lancet. 1998;351:419. doi: 10.1016/S0140-6736(05)78358-6. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson D.A., Chiotti K., Ebeling C., Groth D., DeArmond S.J., Prusiner S.B., Carlson G.A. Quantitative trait loci affecting prion incubation time in mice. Genomics. 2000;69:47–53. doi: 10.1006/geno.2000.6320. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd S., Onwuazor O.N., Beck J., Mallinson G., Farrall M., Targonski P., Collinge J., Fisher E. Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc. Natl Acad. Sci. USA. 2001;98:6279–6283. doi: 10.1073/pnas.101130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd S., Uphill J.B., Targonski P.V., Fisher E., Collinge J. Identification of genetic loci affecting mouse-adapted bovine spongiform encephalopathy incubation time in mice. Neurogenetics. 2002;4:77–81. doi: 10.1007/s10048-002-0133-9. [DOI] [PubMed] [Google Scholar]
- 14.Mead S., Poulter M., Uphill J., Beck J., Whitfield J., Webb T.E., Campbell T., Adamson G., Deriziotis P., Tabrizi S.J., et al. Genetic risk factors for variant Creutzfeldt–Jakob disease: a genome-wide association study. Lancet Neurol. 2009;8:57–66. doi: 10.1016/S1474-4422(08)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck J.A., Campbell T., Adamson G., Poulter M., Uphill J., Molou E., Collinge J., Mead S. Association of a null allele of SPRN with variant Creutzfeldt–Jakob disease. J. Med. Genet. 2008;45:813–817. doi: 10.1136/jmg.2008.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd S.E., Maytham E.G., Pota H., Grizenkova J., Molou E., Uphill J., Hummerich H., Whitfield J., Alpers M.P., Mead S., Collinge J. HECTD2 is associated with susceptibility to mouse and human prion disease. PLoS Genet. 2009;5:e1000383. doi: 10.1371/journal.pgen.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller G. Neurodegeneration. Could they all be prion diseases? Science. 2009;326:1337–1339. doi: 10.1126/science.326.5958.1337. [DOI] [PubMed] [Google Scholar]
- 18.Meyer-Luehmann M., Coomaraswamy J., Bolmont T., Kaeser S., Schaefer C., Kilger E., Neuenschwander A., Abramowski D., Frey P., Jaton A.L., et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 19.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Deerlin V.M., Sleiman P.M.A., Martinez-Lage M., Chen-Plotkin A., Wang L.S., Graff-Radford N.R., Dickson D.W., Rademakers R., Boeve B.F., Grossman M., et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP43 inclusions. Nat. Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon-Sanchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shatunov A., Mok K., Newhouse S., Weale M.E., Smith B., Vance C., Johnson L., Veldink J.H., van Es M.A., van den Berg L.H., et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurology. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mead S., Whitfield J., Poulter M., Shah P., Uphill J., Beck J., Campbell T., Al Dujaily H., Hummerich H., Alpers M.P., Collinge J. Genetic susceptibility, evolution and the kuru epidemic. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:3741–3746. doi: 10.1098/rstb.2008.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mead S., Poulter M., Beck J., Webb T., Campbell T., Linehan J., Desbruslais M., Joiner S., Wadsworth J.D., King A., et al. Inherited prion disease with six octapeptide repeat insertional mutation–molecular analysis of phenotypic heterogeneity. Brain. 2006;129:2297–2317. doi: 10.1093/brain/awl226. [DOI] [PubMed] [Google Scholar]
- 26.Webb T.E., Poulter M., Beck J., Uphill J., Adamson G., Campbell T., Linehan J., Powell C., Brandner S., Pal S., et al. Phenotypic heterogeneity and genetic modification of P102L inherited prion disease in an international series. Brain. 2008;131:2632–2646. doi: 10.1093/brain/awn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb T.E.F., Whittaker J., Collinge J., Mead S. Age of onset and death in inherited prion disease are heritable. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:496–501. doi: 10.1002/ajmg.b.30844. [DOI] [PubMed] [Google Scholar]
- 28.Mead S., Mahal S.P., Beck J., Campbell T., Farrall M., Fisher E., Collinge J. Sporadic—but not variant—Creutzfeldt–Jakob disease is associated with polymorphisms upstream of PRNP Exon 1. Am. J. Hum. Genet. 2001;69:1225–1235. doi: 10.1086/324710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormack J.E., Baybutt H.N., Everington D., Will R.G., Ironside J.W., Manson J.C. PRNP contains both intronic and upstream regulatory regions that may influence susceptibility to Creutzfeldt–Jakob disease. Gene. 2002;288:139–146. doi: 10.1016/s0378-1119(02)00466-3. [DOI] [PubMed] [Google Scholar]
- 30.Vollmert C., Windl O., Xiang W., Rosenberger A., Zerr I., Wichmann H.E., Bickeboller H., Illig T., Kretzschmar H.A. Significant association of a M129V independent polymorphism in the 5′ UTR of the PRNP gene with sporadic Creutzfeldt–Jakob disease in a large German case-control study. J. Med. Genet. 2006;43:e53. doi: 10.1136/jmg.2006.040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill A.F., Joiner S., Wadsworth J.D., Sidle K.C., Bell J.E., Budka H., Ironside J.W., Collinge J. Molecular classification of sporadic Creutzfeldt–Jakob disease. Brain. 2003;126:1333–1346. doi: 10.1093/brain/awg125. [DOI] [PubMed] [Google Scholar]
- 32.Collinge J., Clarke A. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 33.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 34.Webb T.E., Pal S., Siddique D., Heaney D.C., Linehan J.M., Wadsworth J.D., Joiner S., Beck J., Wroe S.J., Stevenson V., et al. First report of Creutzfeldt–Jakob disease occurring in 2 siblings unexplained by PRNP mutation. J. Neuropathol. Exp. Neurol. 2008;67:838–841. doi: 10.1097/NEN.0b013e318182f36e. [DOI] [PubMed] [Google Scholar]
- 35.Ward H.J., Everington D., Cousens S.N., Smith-Bathgate B., Gillies M., Murray K., Knight R.S., Smith P.G., Will R.G. Risk factors for sporadic Creutzfeldt–Jakob disease. Ann. Neurol. 2008;63:347–354. doi: 10.1002/ana.21294. [DOI] [PubMed] [Google Scholar]
- 36.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 37.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X.Y., Becker L.C., Becker D.M., Starmer J.D., Province M.A. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet. Epidemiol. 2010;34:100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.