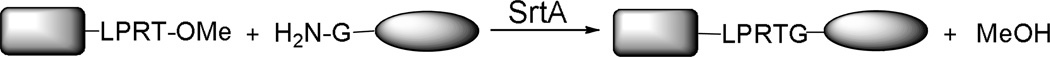Abstract
Sortases are a family of transpeptidases found in Gram–positive bacteria responsible for covalent anchoring of cell surface proteins to bacterial cell walls. It has been discovered that sortase A (SrtA) of Staphylococcus aureus origin is rather promiscuous and can accept various molecules as substrates. As a result, SrtA has been widely used to ligate peptides and proteins with a variety of nucleophiles, and the ligation products are useful for research in chemical biology, proteomics, biomedicine, etc. This review summarizes the recent applications of SrtA with special emphasis on SrtA–catalyzed ligation of carbohydrates with peptides and proteins.
Keywords: sortase, enzymatic transpeptidation, peptide, protein, glycopeptide, GPI anchor
1. Introduction
Sortases are a family of membrane–anchored transpeptidases found in Gram–positive bacteria. They are responsible for the so-called bacterial “cell wall sorting” process, which anchors surface proteins to the cell wall.1 Cell wall sorting is believed to be important for bacterial virulence.2 Therefore, since their discovery,3 sortases have become the focus of numerous studies aiming at understanding their structure, function, and reaction mechanism,4–14 as well as discovering inhibitors for these enzymes.15–17 The transpeptidation mechanisms of sortases have been well established.9 They recognize and react with a short peptide sequence, known as the “sorting signal”, near the target protein C–terminus to generate reactive C–terminal thioesters, and then transfer the acyl group to the N–terminus of the oligoglycine side chain of proteoglycans on the cell surface to form an amide bond between the target protein and proteoglycan.
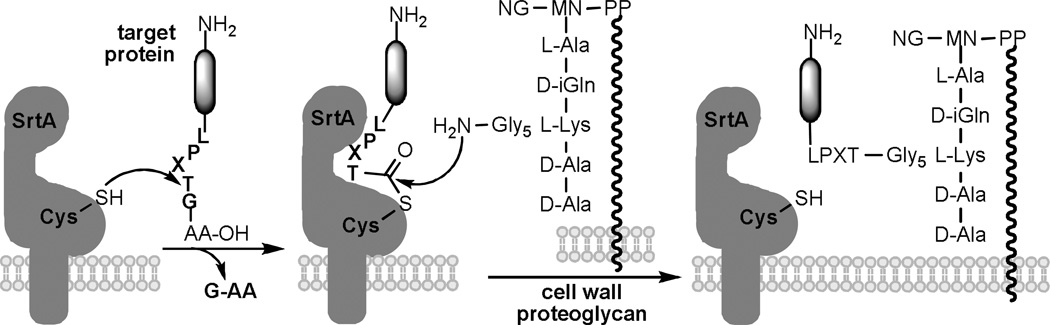
Sortase A (SrtA) of Staphylococcus aureus origin is one of the typical sortases. Its prototype is a 206 amino acid protein having an N–terminal hydrophobic domain and a positively charged tail.1,2,18,19 SrtA was found to recognize and react with a pentapeptide signal, LPXTG, where X is a variable amino acid, break the peptide bond between T and G to form a thioester with the Cys184 thiol group, and finally transfer the acyl group of T to the N–terminus of the oligoglycine side chain of cell wall proteoglycans (Figure 1).9, 1
Figure 1.
SrtA–mediated cell surface protein sorting in S. aureus.
In the past six years, the application of SrtA as a tool to site–specific modification of proteins or to site–specific ligation of peptides, proteins, and various other molecules has become a very hot topic. This application is made possible largely because of the easy access to bioactive and water–soluble recombinant SrtA.12,20 For example, the recombinant extracellular part of SrtA without N–terminal hydrophobic domain and the charged tail retained the same transpeptidase activity as native SrtA. This application is also made possible because it is relatively easy to engineer proteins to carry a sorting signal and to engineer proteins or other peptide receptors to contain an oligoglycine moiety so as to satisfy the transpeptidation requirements of SrtA. More importantly, it has been demonstrated that SrtA is rather substrate promiscuous; therefore, it can be utilized to ligate proteins with a variety of nucleophiles and be useful for the study of chemical biology, proteomics, biomedicine, and many other areas.21–23 There have been several extensive reviews about SrtA and its application.21–24 Consequently, while focusing on the synthetic applications of SrtA, this review will put special emphasis on recent progress in utilizing SrtA to catalyze the ligation of carbohydrates with peptides and proteins for the synthesis of glycoconjugates.
2. SrtA–mediated peptide/protein–peptide/protein, nucleic acid, and lipid ligation
2.1 SrtA–mediated peptide/protein–peptide/protein ligation
In 2004, Pollok and co-workers first demonstrated that SrtA could be used as a ligation tool for site–specific peptide to peptide, peptide to protein, and protein to protein fusions.25 Using a series of short peptides, they first determined the number of glycine residues required at the N–terminus of peptide acceptors for efficient SrtA–catalyzed ligation with peptides containing the sorting signal, LPXTG. It was concluded that one glycine residue at the peptide N–terminus would be sufficient for effective ligation with peptide donors in the presence of SrtA, although substrates with two or more glycine residues had slightly faster reaction rates. Furthermore, the reaction efficiency of branched peptides was similar to that of linear ones, and proteins were also successfully stitched together.
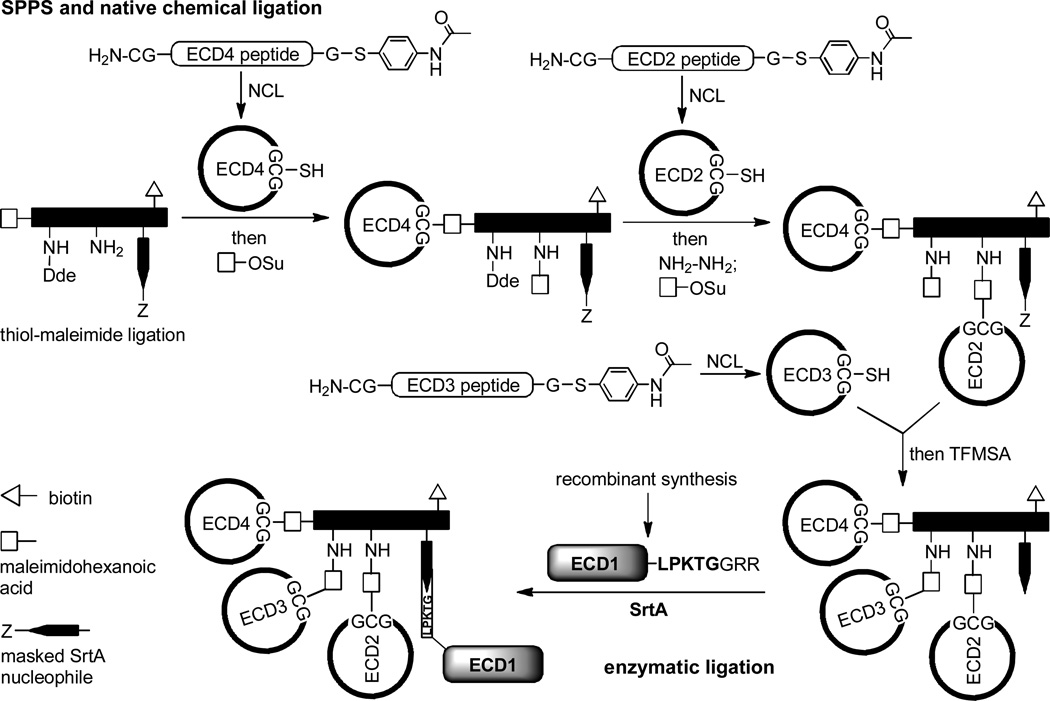
Since then, SrtA has been employed to ligate many full size proteins. For example, Boder and co-workers described the efficient ligation of eGFP carrying a sorting signal with eGFP carrying a triglycine moiety at the N–terminus to obtain a dimeric eGFP conjugate.26 Tanaka and co-workers used SrtA to prepare streptavidin–modified eGFP and glucose oxidase GOD conjugates,27 which showed tight and specific binding to biotin, as well as elevated glucose oxidase activity in the case of GOD conjugate as compared to the streptavidin–GOD conjugates prepared by other methods. In order to modify antibodies, Tanaka and co-workers introduced a LPETG motif to the C–terminus of ZZ domain that has a high affinity to the Fc region of all kinds of antibodies.28 In the presence of SrtA, ZZ domain was successfully ligated with proteins such as pentaglycine–appended alkaline phosphatase (Gly5–AP) and luciferase (Gly5–Luc) and triglycine–appended GOD (Gly3–GOD). The resultant ZZ domain–protein conjugates were then used to prepare functionalized antibody–protein complexes via simply mixing these conjugates with antibodies. In another application, Beyermann and co-workers combined SrtA–mediated ligation strategy with technologies such as recombinant protein expression and enzymatic/chemical synthesis to create very complex protein structures containing unnatural, multiply branched and multicyclic backbone topology (Scheme 1). In this study, three cyclic ECD domains, including ECD2, ECD3 and ECD4 which were prepared by solid phase peptide synthesis (SPPS) and then cyclization via intramolecular native chemical ligation (NCL), were linked to a peptide template containing a masked oligoglycine motif by a series of regioselective thiol–maleimide ligation. In the meantime, a recombinant ECD1 domain containing the sorting signal LPKTG at its C–terminus was expressed in E. coli. Finally, ECD1 was coupled to the peptide conjugate carrying multiple ECD domains by SrtA–mediated ligation to afford the GRF1 receptor mimic (Scheme 1), which showed high binding affinity to Sauvagine and Urcortin 1.
Scheme 129.
SrtA was also used to achieve intramolecular ligation reactions to generate cyclic peptides and proteins. For example, Boder and co-workers26 observed that in the presence of SrtA an eGFP derivative carrying N–terminal glycine and C–terminal LPETG–His6 motifs formed cyclic eGFP as a side product, in addition to a mixture of eGFP oligomers up to pentamer. Ploegh,30,31 Bolscher,32 and co-workers studied SrtA–catalyzed protein cyclization reactions in greater detail and found that the reactions gave moderate to excellent yields of the desired cyclic protein products. It was also reported that cyclic proteins had improved thermal stability compared to the linear counterparts.31 Roy and co-workers found that SrtA could catalyze an “isopeptide” ligation, namely, transferring peptide substrates with a LPXTG motif to the ε–amino group of Lys residue to form cyclic and/or branched oligomers.33 Recently, Guo and co-workers34 applied SrtA to cyclic peptide and glycopeptide synthesis and identified the minimal size required for peptide head to tail cyclization. It was concluded that this method is applicable to the preparation of macrocyclic peptides and glycopeptides containing 15 or more amino acids. Clearly, SrtA–catalyzed protein and peptide cyclization can be developed into a powerful complement to current technologies, such as NCL, EPL and PTS,35 for the synthesis of cyclic proteins and peptides of biological and pharmacological interest.
Typically, SrtA–mediated reactions are reversible, which can reach equilibrium in a short period, thus they give only moderate yields of the ligation products. The use of a large excess of the enzyme or one of the substrates may improve ligation yields. However, increasing the reaction time may result in decreased ligation efficiency due to substrate and product hydrolysis. Nagamune and co-workers recently demonstrated that the ligation efficiency could be improved by introducing a rigid secondary structure, such as β–hairpin, around the ligation site in the newly formed product,36 because the product became unrecognizable by SrtA so as to drive the reaction equilibrium toward completion (Scheme 2). For example, they found that the efficiency of a reaction between peptide MBP2 and protein Trx–2, which generated the stable β–hairpin structure Wzip2 in the product, was significantly higher (>70%) than that of reactions between MBP–1 and Trx–1 and between MBP–c and Trx–C (40–50%), which generated weak or no β–hairpin structure.
Scheme 236.
Another strategy developed to improve SrtA–catalyzed ligation efficiency was to use C–terminal methyl esters, that is, LPRT–OMe, to replace the LPRTG motif in the peptide donors. The reaction gave MeOH (Scheme 3), instead of a short peptide with N–terminal glycine, as the major side product during the cleavage of the sorting signals of the substrate proteins, to avoid the competitive reactions caused by the side product. This method led to the formation of almost quantitative yields of protein conjugates.37
Scheme 3.
2.2 SrtA–mediated peptide–nucleic acid ligation
SrtA–mediated transpeptidation has also found applications in the synthesis of peptide–nucleic acid (PNA) conjugates, which, as DNA analogs, are useful in the development of new therapeutics and diagnostics, even though solid–phase synthesis is proved to be a straight forward and flexible approach for these molecules.38 For example, Pritz and co-workers synthesized a cell–penetrating PNA conjugate, which showed improved delivery into mammalian cells and biological activity.39 For this purpose, they designed and synthesized an 18–mer nucleic acid containing the sorting signal, LPKTG, and a well–known amphipathic peptide carrying three additional glycine residues at the N–terminus, and coupled these two fragments together in a 38% yield using SrtA. However, when the reaction was performed in dialysis with a molecular mass cutoff of 2000 Da to restrain the reverse reaction, a 94% conversion was achieved.39 One of the advantages of this synthetic strategy over total chemical synthesis is the simplified product purification.
2.3 SrtA–mediated ligation of peptides/proteins with lipids and other molecular tags
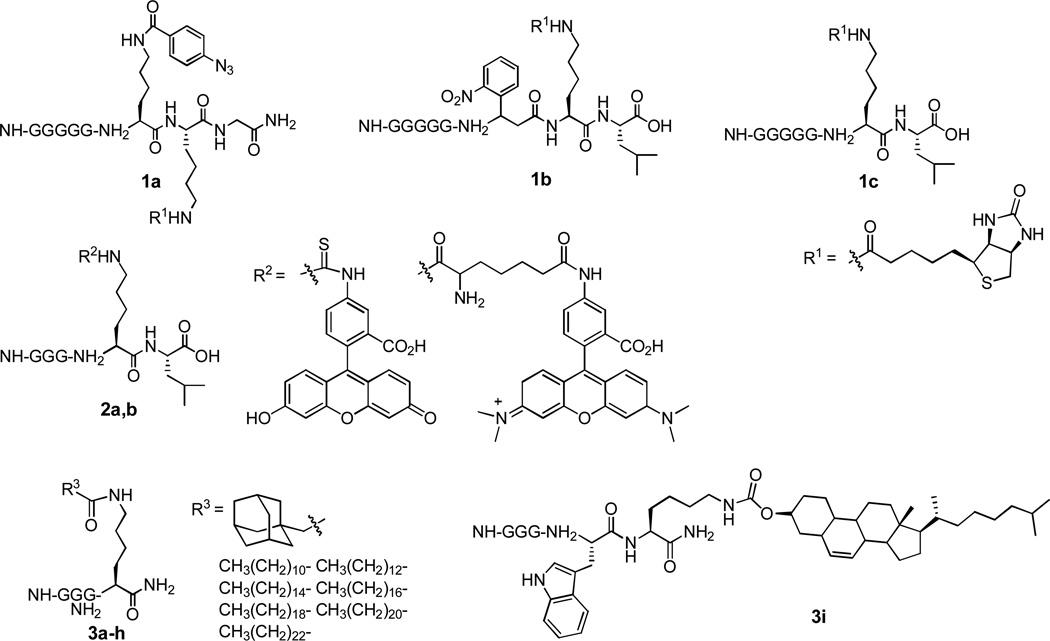
Ploegh and co-workers40 employed the SrtA–mediated ligation technology to couple proteins to a diverse set of lipids and molecular tags (Figure 2), such as biotin and fluorescent probes. The resultant protein conjugates are useful for the study of protein interactions and protein trafficking. For example, several lipid tails were attached to GFP to give good to excellent yields (60–90%) of lipoproteins, which were demonstrated to associate with mammalian cells in a lipid tail–dependent fashion and localize in the plasma membrane and endosomes.41
Figure 2.
Biotin (1a–c), fluorescent tag (2a,b), and lipid (3a–i) modified peptides that were used to label proteins via SrtA–catalyzed ligation.
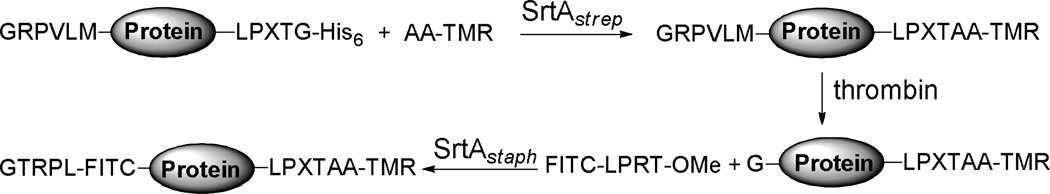
Taking advantage of the different substrate specificities of sortases, Ploegh and co-workers37 have recently developed a strategy for dual site–specific labeling of proteins as outlined in Scheme 4. They utilized SrtAstrep, which recognizes the same sorting signal LPXTG as SrtAStaph but only accepts alanine as peptide acceptor, for protein C–terminal labeling. The LPXTA sequence in the resultant conjugates was unrecognized by SrtAStaph, allowing subsequently selective modification of the N–terminus by SrtAStaph with LPRT methyl esters as the peptide donor.
Scheme 4.
SrtA–mediated ligation technology was also employed to modify proteins for improving their solubility and NMR spectroscopic resolution for protein structural studies. For example, Inagati and co-workers42 attached GB1, a solubility enhancement motif, to isotope–labeled proteins through SrtA–mediated ligation to obtain soluble samples for NMR study. Tsang and co-workers43 used a similar technology to address the NMR resolution challenge as the protein size increases.
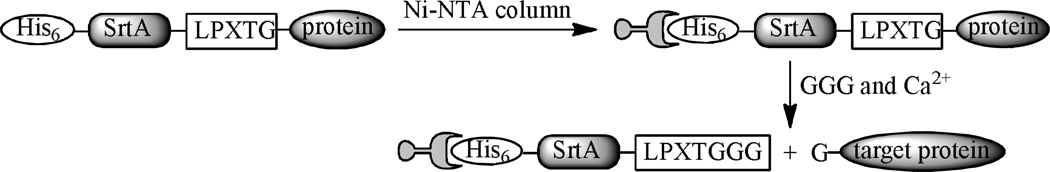
Mao44 designed a strategy to facilitate protein purification by using separation tags that can be subsequently removed by SrtA–mediated reactions. First, proteins with a His6–SrtA–linker–LPXTG tag, which can be captured by an affinity column, were expressed in E. coli. Proteins attached to the column were then released with triglycine through a SrtA–catalyzed transpeptidation reaction while the other part of the molecule remained on the column (Scheme 5).
Scheme 5.
3. SrtA–mediated attachment of peptides and proteins to solid materials and living cells
3.1 SrtA–mediated protein attachment to resins, gels, and other solid materials
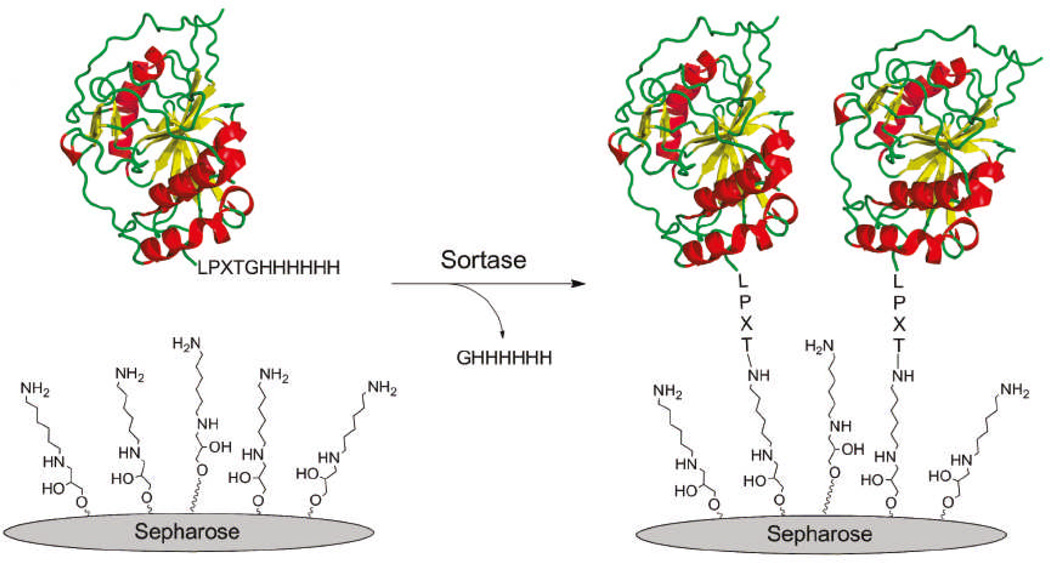
Protein immobilization to solid surfaces has a wide range of applications in the development of recyclable catalysts, affinity matrices, micro devices, protein assays, and so on. The current protein immobilization methods include simple absorption, trapping protein with a gel matrix, and covalent linkage, among which covalent linkage is the most robust. A number of selective ligation methods, such as Staudinger ligation, NCL, and expressed protein ligation, have been developed for covalent attachment of proteins to solid supports.45 Recently, SrtA–mediated ligation has been shown to be a powerful and convenient alternative to exiting methods. For instance, Boder and co-workers26 stapled eGFP–LEPG to triglycine–modified polystyrene beads via SrtA–mediated ligation and to other amine–terminated beads but with considerably reduced efficiency. Neylon and co-workers46 have also demonstrated that a range of proteins could be covalently linked to solid supports. In their study, eGFP–LPETGG–His6, DsRed–LPETGG–His6, and fragile Tus–LPETGG–His6 were efficiently linked to oligoglycine–modified glycidyl methacrylate (GMA) beads, glass slips, and Affi–Gel 102 resin, respectively. Proft and co-workers47 successfully immobilized rFba–LPETG onto the surface of biacore sensor chips in a site–specific manner via SrtA–mediated reactions, which allowed close examination of protein binding specificity, kinetics, and affinity. More recently, Nishimura and co-workers48 immobilized recombinant enzymes (Scheme 6), such as glycosyltransferases, and other proteins onto commercial Sepharose gel through SrtA–mediated ligation. The immobilized enzymes displayed improved stability, desirable glycosyl transfer activity, and practical reusability, which should be very valuable for the construction of carbohydrate and glycoconjugate libraries. Therefore, SrtA–catalyzed ligation has been demonstrated to be a practical method for protein immobilization under mild conditions that does not affect protein functions.
Scheme 648.
3.2 SrtA–mediated labeling of living cells
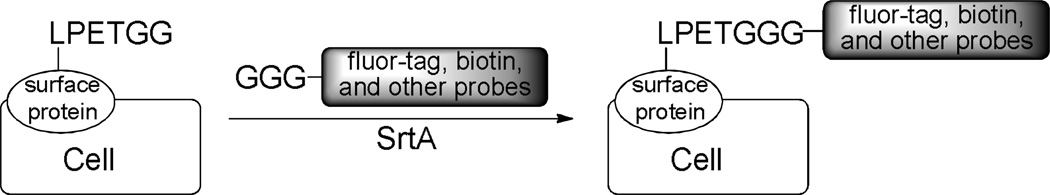
Site–specific modification of proteins on cell surface is a powerful technique for the elucidation of protein functions, thus, many efforts have been focused on the incorporation of synthetic probes into surface proteins in living cells, among which enzymatic modification of proteins is particularly attractive owing to the site and substrate specificity of enzymes.49 SrtA–mediated ligation has also been explored in the labeling of proteins on living cell surfaces. Ploegh and co-workers40 proved that surface protein CD40L carrying C–terminal LPETG motif on living HEK 293T cell could be successfully labeled upon incubation with SrtA and synthetic probes carrying an oligoglycine tag. Nagamune and co-workers 50 have installed both small molecule probes, such as biotin and Alexa, and full size proteins, such as eGFP, onto living cell surfaces by the same strategy (Scheme 7). However, this strategy can only be used to label surface proteins with extracellular C–terminus.
Scheme 7.
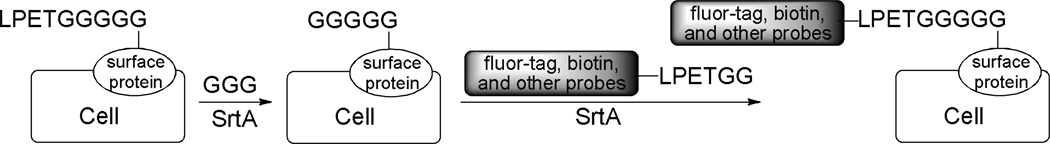
Nagamune and co-workers have also developed a strategy for site–specific N–terminal labeling of surface proteins on living cells (Scheme 8).51 First, a LPETGGGGG tag was introduced to the target protein close to its N–terminus. Thereafter, the tag was cleaved by SrtA with triglycine as a substrate to expose pentaglycine epitope as a peptide acceptor. Finally, the target protein reacted with synthetic probes carrying the sorting signal in the presence of SrtA to achieve cell surface protein labeling. In conclusion, SrtA–catalyzed ligation has been demonstrated to be a useful tool for the modification of membrane proteins on the surface of living cells.
Scheme 8.
5. SrtA–mediated ligation for the synthesis of carbohydrate–peptide, glycopeptide and protein conjugates
Glycopeptides, glycoproteins, and other glycoconjugates play a pivotal role in many biological processes, such as cell adhesion, communication, growth, and differentiation.52 To understand their functions at the molecular level, it is essential to have sufficient and homogeneous samples. Despite the recent progress in the chemical synthesis of glycopeptides and glycoproteins, this area remains an important challenge. Thus, chemoenzymatic synthesis, which combines the advantages of chemical and enzymatic syntheses, has gained great attention.53 In this context, SrtA–mediated ligation has been explored for the synthesis of various glycoconjugates.
4.1 SrtA–mediated peptide–carbohydrate ligation for neoglycopeptide/protein synthesis
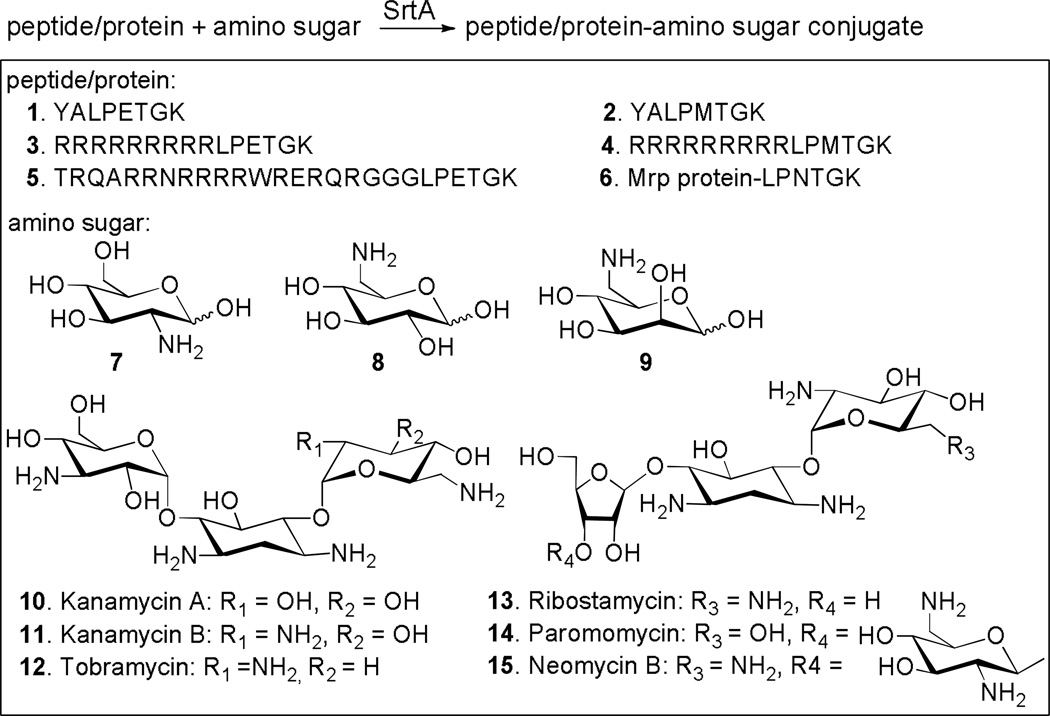
Roy and co-workers54 reported the first application of SrtA–mediated ligation of peptides with sugars for neoglycoconjugate synthesis (Scheme 9). In this research, 6–aminohexoses, including 6–deoxy–6–aminoglucose 8 and 6–deoxy–6–aminomannose 9, were successfully coupled with short peptides YALPETGK 1 and YALPMTGK 2 in the presence of SrtA. In contrast, when glucosamine 7 was used as the nucleophile, only peptide hydrolysis was observed. Consequently, the –CH2–NH2 motif in 8 and 9 was believed to be essential for the enzymatic ligation to occur. Sugar substrates were subsequently expanded to aminoglycoside antibiotics, such as 10–15, containing 6–amino and 2,6–diamino functionality. Biologically relevant peptides, such as the arginine–rich peptides 3, 5 and 4 derived from Tat and Rev proteins, and full size proteins, such as MrP protein 6, containing the SrtA–recognition motif at the C–terminus were ligated with the above–mentioned aminosugars to afford modest to excellent yields (18%–70%) of the corresponding glycoconjugates. Electronic spray mass spectrometry analysis of the products proved that the ligation was at the sugar 6–amino position. Furthermore, preliminary binding studies have demonstrated that these conjugates had improved biological properties.
Scheme 9.
4.2 SrtA–mediated GPI anchor–peptide/glycopeptide ligation
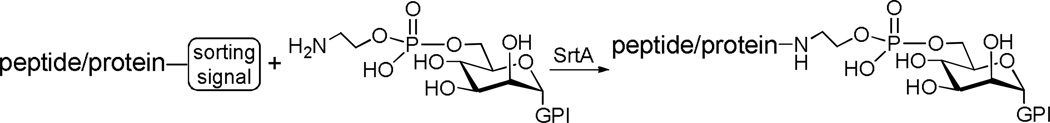
Glycosylphosphatidylinositols (GPIs) are a class of glycophospholipids ubiquitously expressed by eukaryotic cells, which anchor surface proteins and glycoproteins to the cell membrane. GPIs and GPI–anchored proteins and glycoproteins play a pivotal role in a range of biological events.55 To study the biological functions of these molecules, it is essential to have access to abundant and homogeneous samples, which remains a great challenge despite the recent progress in the chemical synthesis of GPI–anchored peptides/glycopeptides56 and proteins.57,58
To address this issue, Guo and co-workers59–61 have recently exploited SrtA–mediated ligation of GPI anchors with peptides, glycopeptides, and proteins for GPI–anchored peptide, glycopeptide, and protein synthesis (Scheme 10). SrtA should be particularly suitable to this application, because all of the GPI–anchored proteins and glycoproteins have their polypeptide C–terminus linked to the phosphoetanolamine group at the non–reducing end of the GPI core glycan. Moreover, peptides and proteins carrying the sorting signal at their C–termini can be easily obtained by solid–phase peptide synthesis and by recombinant protein technology, respectively. Therefore, the synthetic strategy can be widely applicable.
Scheme 10.
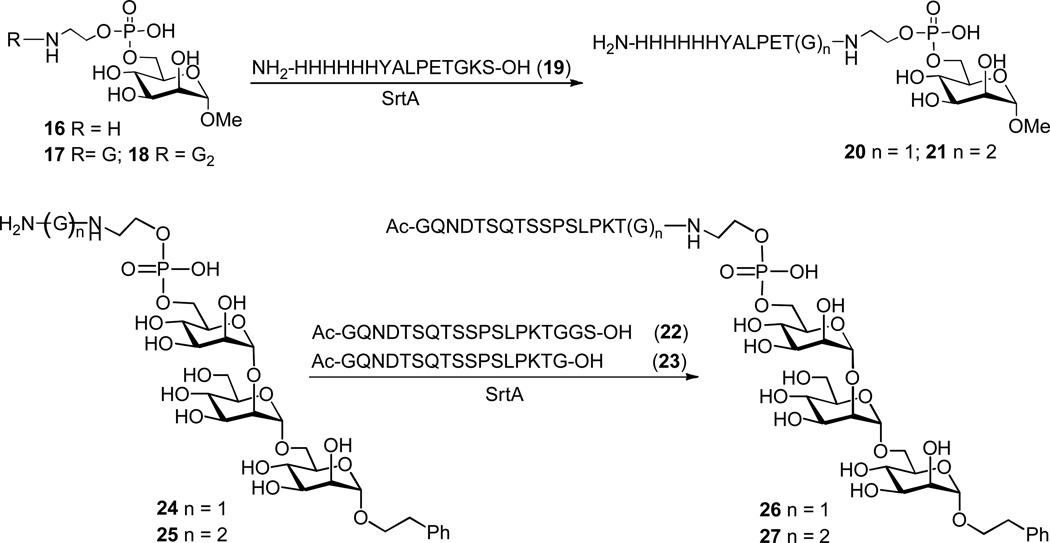
To probe the concept outlined in Scheme 10, Guo and co-workers examined the ligation of GPI analogs with a short peptide 19 catalyzed by SrtA (Scheme 11). They found that 16 was not an ideal SrtA substrate but, after introduction of one or two glycine residues to the phosphoethanolamine moiety, the resultant GPI analogs 17 and 18 were coupled to 19 to give the desired GPI conjugates 20 and 21 in excellent yields (>95%). Moreover, 17 and 18 were found to have similar reactivity. The results indicate that, for GPIs to be efficiently accepted by SrtA for transpeptidation reactions, they need to have one or two glycine residues linked to the phosphoethanolamine moiety.59
Scheme 11.
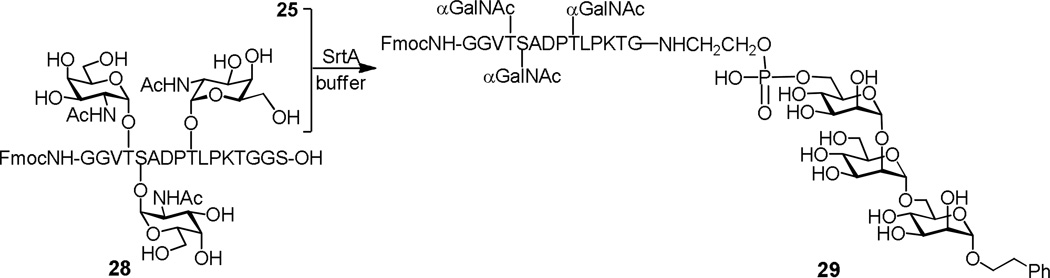
SrtA–catalyzed ligation of GPI analogs 24 and 25 with CD52 peptide 22, as well as some other peptides, proceeded smoothly under the conditions described above to afford 26 and 27 in 78–81% yields (Scheme 11). Interestingly, these reactions were more effective than that of the corresponding monosaccharide analog 17, suggesting that SrtA may accept more complex GPI derivatives for ligating with peptides. On the other hand, the reactions of 24 with 23 gave only very low yield (10%) of the desired GPI conjugates, suggesting that for peptide substrates to be efficiently recognized and accepted by SrtA, the sorting signal should not be directly exposed at the peptide C–terminus. It was further revealed that SrtA effected the ligation of a small synthetic protein with 24 to obtain an analog of the human CD24 antigen in 73% yield.60 Guo and co-workers also studied the SrtA–catalyzed ligation of GPIs with glycopeptides.61 They demonstrated that glycopeptide 28, a partial sequence of MUC1 carrying three O–linked T antigens, was efficiently coupled to GPI analog 25 in the presence of SrtA (Scheme 12). These studies have therefore shown that SrtA may be generally applicable to GPI ligation with peptides, glycopeptides, and proteins for the synthesis of GPI–anchored peptides, glycopeptides and proteins.
Scheme 12.
4.3 SrtA–mediated glycopeptide–glycopeptide and glycopeptides–protein ligation
Protein glycosylation is a common posttranslational modification, most frequently with glycans linked to an asparagine residue within the Asn–X–Ser/Thr sequence and to a Ser/Thr residue, which are known as N– and O–glycosylations, respectively. Glycoproteins are biosynthesized as mixtures of numerous glycoforms. To obtain homogeneous glycoproteins for functional studies, a variety of synthetic methods have been developed.62,63 Similarly, SrtA–catalyzed peptide ligation may be used to couple synthetic glycopeptides with peptides or proteins for the preparation of structurally well–defined glycopeptides and glycoproteins. For example, Guo and co-workers found that, after removal of the N–terminal protecting group in the GPI conjugate 29, its glycopeptide chain could be further elongated with glycopeptide 28 to afford more complex GPI–anchored glycopeptide 30 in a 59% yield (Scheme 13).61
Scheme 13.
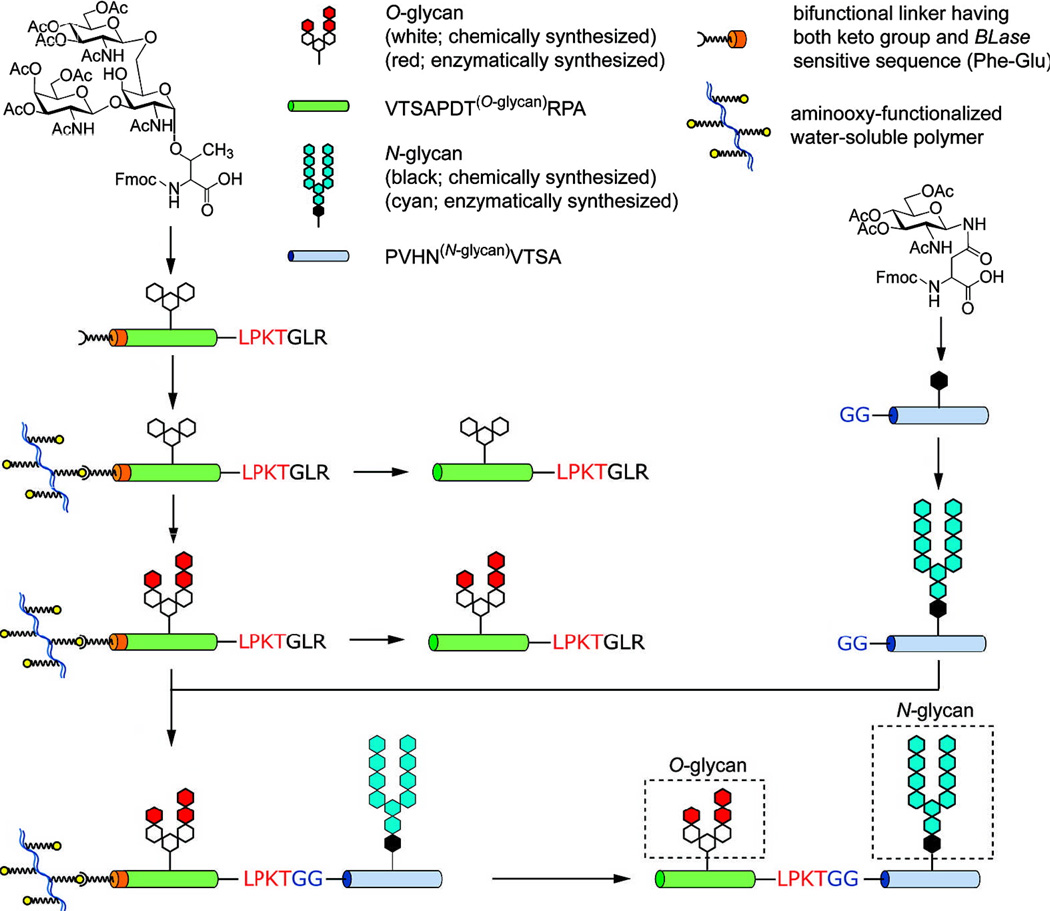
Nishimura and co-workers have reported an elegant chemoenzymatic synthesis of a structurally complex and well–defined glycopeptide containing N– and O–glycans by combining SrtA–mediated glycopeptide ligation with polymer–supported glycopeptide synthesis and enzymatic elongation of glycans.64,65 As shown in Scheme 14, after glycopeptides carrying simple glycans were prepared by SPPS with glycosylated amino acids as key building blocks, the glycans were elongated through a series of enzymatic glycosylations in solution or on water–soluble polymer support. Thereafter, the two glycopeptide segments were coupled together via SrtA–mediated peptide ligation, which was followed by detachment from the water–soluble polymer support to eventually afford glycopeptide in a 77% yield.
Scheme 1464.
5. Closing Remarks
SrtA–mediated ligation has gained great attention since its first report six years ago. The simple and easily achievable requirements for this unique ligation technology are: (1) donor substrate being equipped with a LPXTG motif and (2) acceptor substrate containing an oligoglycine motif. Both substrates can be readily prepared via chemical synthesis or genetic engineering/recombinant protein expression. In the meanwhile, the promiscuous character of SrtA allowed its application to a diversity of substrates. As a result, this new ligation technology can be broadly useful. Furthermore, the mild reaction conditions for SrtA–mediated ligation, which can be achieved in aqueous and physiologically tolerable organic solvents, such as 20% aq. DMSO or polyethylene glycol, made this technology even more attractive for application to biological systems.39 It is thus easily imaginable that SrtA–mediated ligation will find more and more applications in the future.
Acknowledgement
The authors gratefully acknowledge National Science Foundation (CHE-0320878, 0715275, and 1053848), National Institutes of Health (R01 GM090270 and CA95142), and Wayne State University for their generous financial supports of this work.
References
- 1.Marraffini LA, DeDent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of Gram–positive bacteria. Microbiol. Mol. Biol. Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cossart P, Jonquieres R. Sortase, a universal target for therapeutic agents against gram–positive bacteria? Proc Natl Acad Sci U S A. 2000;97:5013–5015. doi: 10.1073/pnas.97.10.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneewind O, Mazmanian SK, Liu G, Hung TT. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 4.Ton–That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate–imidazolium ion pair for catalysis. J Biol Chem. 2002;277:7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- 5.Connolly KM, Smith BT, Pilpa R, Ilangovan U, Jung ME, Clubb RT. Sortase from Staphylococcus aureus does not contain a thiolate–imidazolium ion pair in its active site. J Biol Chem. 2003;278:34061–34065. doi: 10.1074/jbc.M305245200. [DOI] [PubMed] [Google Scholar]
- 6.Kruger RG, Dostal P, McCafferty DG. Development of a high–performance liquid chromatography assay and revision of kinetic parameters for the Staphylococcus aureus sortase transpeptidase SrtA. Anal Biochem. 2004;326:42–48. doi: 10.1016/j.ab.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini LA, Ton–That H, Zong Y, Narayana SV, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. A conserved arginine residue is required for efficient catalysis of sortase A. J Biol Chem. 2004;279:37763–377670. doi: 10.1074/jbc.M405282200. [DOI] [PubMed] [Google Scholar]
- 8.Aulabaugh A, Ding W, Kapoor B, Tabei K, Alksne L, Dushin R, Zatz T, Ellestad G, Huang X. Development of an HPLC assay for Staphylococcus aureus sortase: evidence for the formation of the kinetically competent acyl enzyme intermediate. Anal Biochem. 2007;360:14–22. doi: 10.1016/j.ab.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Huang X, Aulabaugh A, Ding W, Kapoor B, Alksne L, Tabei K, Ellestad G. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry. 2003;42:11307–11315. doi: 10.1021/bi034391g. [DOI] [PubMed] [Google Scholar]
- 10.Schneewind O, Ton–That H, Mazmanian SK, Alksne L. Anchoring of surface proteins to the cell wall of Staphylococcus aureus – Cysteine 184 and histidine 120 of sortase form a thiolate–imidazolium ion pair for catalysis. J Biol Chem. 2002;277:7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- 11.Ilangovan U, Iwahara J, Ton–That H, Schneewind O, Clubb RT. Assignment of the 1H: 13C and 15N signals of Sortase. J Biomol NMR. 2001;19:379–380. doi: 10.1023/a:1011299500628. [DOI] [PubMed] [Google Scholar]
- 12.Ilangovan U, Ton–That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zong Y, Bice TW, Ton–That H, Schneewind O, Narayana SV. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 14.Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Sessions RB, Kehoe MA, McCafferty DG, Banfield MJ. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J Biol Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung ME, Suree N, Clubb RT. Recent advances towards new anti–infective agents that inhibit cell surface protein anchoring in Staphylococcus aureus and other Gram–Positive pathogens. Mini Rev Med Chem. 2007;7:991–1000. doi: 10.2174/138955707782110097. [DOI] [PubMed] [Google Scholar]
- 16.Maresso AW, Schneewind O. Sortase as a target of anti–infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- 17.Suree N, Yi SW, Thieu W, Marohn M, Damoiseaux R, Chan A, Jung ME, Clubb RT. Discovery and structure–activity relationship analysis of Staphylococcus aureus sortase A inhibitors. Bioorg Med Chem. 2009;17:7174–7185. doi: 10.1016/j.bmc.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev. 2008;32:307–320. doi: 10.1111/j.1574-6976.2008.00102.x. [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Ton–That H, Schneewind O. Sortase–catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 20.Ton–That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritz S. Enzymes in protein ligation: The coupling of peptides, peptide nucleic acids and proteins by sortase A. Mini–Rev. Org.Chem. 2008;5:47–52. [Google Scholar]
- 22.Tsukiji S, Nagamune T. Sortase–mediated ligation: a gift from Gram–positive bacteria to protein engineering. Chembiochem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 23.Proft T. Sortase–mediated protein ligation: an emerging biotechnology tool for protein modification and immobilisation. Biotechnol. Lett. 2010;32:1–10. doi: 10.1007/s10529-009-0116-0. [DOI] [PubMed] [Google Scholar]
- 24.Popp MW–L, Ploegh HL. Making and Breaking Peptide Bonds: Protein Engineering Using Sortase. Angew. Chem. Int. Ed. 2011;50:5024–5032. doi: 10.1002/anie.201008267. [DOI] [PubMed] [Google Scholar]
- 25.Mao HY, Hart SA, Schink A, Pollok BA. Sortase–mediated protein ligation: A new method for protein engineering. J. Am. Chem. Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 26.Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular "stapler" for sequence–specific protein conjugation. Bioconjugate Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto T, Sawamoto S, Sakamoto T, Tanaka T, Fukuda H, Kondo A. Site–specific tetrameric streptavidin–protein conjugation using sortase A. J. Biotechol. 2011;152:37–42. doi: 10.1016/j.jbiotec.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto T, Sawamoto S, Tanaka T, Fukuda H, Kondo A. Enzyme–Mediated Site–Specific Antibody–Protein Modification Using a ZZ Domain as a Linker. Bioconjugate Chem. 2010;21:2227–2233. doi: 10.1021/bc100206z. [DOI] [PubMed] [Google Scholar]
- 29.Pritz S, Kraetke O, Klose A, Klose J, Rothemund S, Fechner K, Bienert M, Beyermann M. Synthesis of Protein Mimics with Nonlinear Backbone Topology by a Combined Recombinant, Enzymatic, and Chemical Synthesis Strategy. Angew. Chem. Int. Ed. 2008;47:3642–3645. doi: 10.1002/anie.200705718. [DOI] [PubMed] [Google Scholar]
- 30.Antos JM, Popp MWL, Ernst R, Chew GL, Spooner E, Ploegh HL. A Straight Path to Circular Proteins. J. Biol. Chem. 2009;284:16028–16036. doi: 10.1074/jbc.M901752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poppa MW, Dougana SK, Chuanga T–Y, Spoonera E, Ploegha HL. Sortase–catalyzed transformations that improve the properties of cytokines. Proc. Natl. Acad. Sci. USA. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolscher JGM, Oudhoff MJ, Nazmi K, Antos JM, Guimaraes CP, Spooner E, Haney EF, Vallejo JJG, Vogel HJ, van't Hof W, Ploegh HL, Veerman ECI. Sortase A as a tool for high–yield histatin cyclization. FASEB Journal. 2011;25:2650–2658. doi: 10.1096/fj.11-182212. [DOI] [PubMed] [Google Scholar]
- 33.Roy RP, Dasgupta S, Samantaray S, Sahal D. Isopeptide Ligation Catalyzed by Quintessential Sortase. J. Biol. Chem. 2011;286:23996–24006. doi: 10.1074/jbc.M111.247650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z, Guo X, Guo Z. Sortase A–catalyzed peptide cyclization for the synthesis of macrocyclic peptides and glycopeptides. Chem. Commun. 2011;47:9218–9220. doi: 10.1039/c1cc13322e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.avassoli AN, Todd A, Benkovic, Stephen J. Production of cyclic proteins and peptides. Nuc. Aci. and Mol. Bio. 2005;16:293–305. [Google Scholar]
- 36.Yamamura Y, Hirakawa H, Yamaguchi S, Nagamune T. Enhancement of sortase A–mediated protein ligation by inducing a beta–hairpin structure around the ligation site. Chem. Commun. 2011;47:4742–4744. doi: 10.1039/c0cc05334a. [DOI] [PubMed] [Google Scholar]
- 37.Antos JM, Chew GL, Guimaraes CP, Yoder NC, Grotenbreg GM, Popp MW, Ploegh HL. Site–specific N– and C–terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Koning MC, van der Marel GA, Overhand M. Synthetic developments towards PNA–peptide conjugates. Curr.Opin.Chem.Biol. 2003;7:734–740. doi: 10.1016/j.cbpa.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Pritz S, Wolf Y, Kraetke O, Klose J, Bienert M, Beyermann M. Synthesis of Biologically Active Peptide Nucleic Acid–Peptide Conjugates by Sortase–Mediated Ligation. J. Org. Chem. 2007;72:3909–3912. doi: 10.1021/jo062331l. [DOI] [PubMed] [Google Scholar]
- 40.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat. Chem. Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 41.Antos JM, Miller GM, Grotenbreg GM, Ploegh HL. Lipid Modification of Proteins through Sortase–Catalyzed Transpeptidation. J. Am. Chem. Soc. 2008;130:16338–16343. doi: 10.1021/ja806779e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobashigawa Y, Kumeta H, Ogura K, Inagaki F. Attachment of an NMR–invisible solubility enhancement tag using a sortase–mediated protein ligation method. J. Biomol.NMR. 2009;43:145–150. doi: 10.1007/s10858-008-9296-5. [DOI] [PubMed] [Google Scholar]
- 43.Refaei MA, Combs A, Kojetin DJ, Cavanagh J, Caperelli C, Rance M, Sapitro J, Tsang P. Observing selected domains in multi–domain proteins via sortase–mediated ligation and NMR spectroscopy. J. Biomol.NMR. 2010;49:3–7. doi: 10.1007/s10858-010-9464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao H. A self–cleavable sortase fusion for one–step purification of free recombinant proteins. Protein. Expres. Purif. 2004;37:253–263. doi: 10.1016/j.pep.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Camarero JA. Recent developments in the site–specific immobilization of proteins onto solid supports. Biopolymers. 2008;90:450–458. doi: 10.1002/bip.20803. [DOI] [PubMed] [Google Scholar]
- 46.Chan LY, Cross HF, She JK, Cavalli G, Martins HFP, Neylon C. Covalent Attachment of Proteins to Solid Supports and Surfaces via Sortase–Mediated Ligation. PLoS One. 2007;2:e1164. doi: 10.1371/journal.pone.0001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clow F, Fraser JD, Proft T. Immobilization of proteins to biacore sensor chips using Staphylococcus aureus sortase A. Biotechnol. Lett. 2008;30:1603–1607. doi: 10.1007/s10529-008-9718-1. [DOI] [PubMed] [Google Scholar]
- 48.Ito T, Sadamoto R, Naruchi K, Togame H, Takemoto H, Kondo H, Nishimura SI. Highly Oriented Recombinant Glycosyltransferases: Site–Specific Immobilization of Unstable Membrane Proteins by Using Staphylococcus aureus Sortase A. Biochemistry. 2010;49:2604–2614. doi: 10.1021/bi100094g. [DOI] [PubMed] [Google Scholar]
- 49.Johnsson N, George N, Johnsson K. Protein chemistry on the surface of living cells. Chembiochem. 2005;6:47–52. doi: 10.1002/cbic.200400290. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. Site–Specific Protein Modification on Living Cells Catalyzed by Sortase. ChemBioChem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto T, Nagamune T. Expansion of the sortase–mediated labeling method for site–specific N–terminal labeling of cell surface proteins on living cells. Chem. Commun. 2009:1022–1024. doi: 10.1039/b818792d. [DOI] [PubMed] [Google Scholar]
- 52.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 53.Bennett CS, Wong CH. Chemoenzymatic approaches to glycoprotein synthesis. Chem. Soc. Rev. 2007;36:1227–1238. doi: 10.1039/b617709c. [DOI] [PubMed] [Google Scholar]
- 54.Samantaray S, Marathe U, Dasgupta S, Nandicoori VK, Roy RP. Peptide–sugar ligation catalyzed by transpeptidase sortase: A facile approach to neoglycoconjugate synthesis. J. Am. Chem. Soc. 2008;130:2132–2133. doi: 10.1021/ja077358g. [DOI] [PubMed] [Google Scholar]
- 55.Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: a complex membrane–anchoring structure for proteins. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao N, Xue B, Guo ZW. Chemical synthesis of a skeleton structure of sperm CD52 – A GPI–anchored glycopeptide. Angew. Chem. Int. Ed. 2004;43:1569–1573. doi: 10.1002/anie.200353251. [DOI] [PubMed] [Google Scholar]
- 57.Paulick MG, Wise AR, Forstner MB, Groves JT, Bertozzi CR. Synthetic analogues of glycosylphosphatidylinositol–anchored proteins and their behavior in supported lipid bilayers. J. Am. Chem. Soc. 2007;129:11543–11550. doi: 10.1021/ja073271j. [DOI] [PubMed] [Google Scholar]
- 58.Becker CFW, Liu XY, Olschewski D, Castelli R, Seidel R, Seeberger PH. Semisynthesis of a Glycosylphosphatidylinositol–Anchored Prion Protein. Angew. Chem. Int. Ed. 2008;47:8215–8219. doi: 10.1002/anie.200802161. [DOI] [PubMed] [Google Scholar]
- 59.Guo X, Wang Q, Swarts BM, Guo Z. Sortase–catalyzed peptide–glycosylphosphatidylinositol analog ligation. J. Am. Chem. Soc. 2009;131:9878–9879. doi: 10.1021/ja903231v. [DOI] [PubMed] [Google Scholar]
- 60.Wu ZM, Guo XQ, Wang QL, Swarts BM, Guo ZW. Sortase A–Catalyzed Transpeptidation of Glycosylphosphatidylinositol Derivatives for Chemoenzymatic Synthesis of GPI–Anchored Proteins. J. Am. Chem. Soc. 2010;132:1567–1571. doi: 10.1021/ja906611x. [DOI] [PubMed] [Google Scholar]
- 61.Wu ZM, Guo XQ, Guo ZW. Chemoenzymatic synthesis of glycosylphosphatidylinositol–anchored glycopeptides. Chem. Commun. 2010;46:5773–5774. doi: 10.1039/c0cc00828a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis BG. Synthesis of glycoproteins. Chem. Rev. 2002;102:579–601. doi: 10.1021/cr0004310. [DOI] [PubMed] [Google Scholar]
- 63.Gamblin DP, Scanlan EM, Davis BG. Glycoprotein Synthesis: An Update. Chem. Rev. 2009;109:131–163. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 64.Matsushita T, Sadamoto R, Ohyabu N, Nakata H, Fumoto M, Fujitani N, Takegawa Y, Sakamoto T, Kurogochi M, Hinou H, Shimizu H, Ito T, Naruchi K, Togame H, Takemoto H, Kondo H, Nishimura SI. Functional Neoglycopeptides: Synthesis and Characterization of a New Class of MUC1 Glycoprotein Models Having Core 2–Based O–Glycan and Complex–Type N–Glycan Chains. Biochemistry. 2009;48:11117–11133. doi: 10.1021/bi901557a. [DOI] [PubMed] [Google Scholar]
- 65.Matsushita T, Nishimura SI. Novel Synthesis of Functional Mucin Glycopeptides Containing Both N– and O– Glycans. Methods Enzymol. 2010;478:485–502. doi: 10.1016/S0076-6879(10)78023-X. [DOI] [PubMed] [Google Scholar]