Abstract
Herpesviruses are a complex family of dsDNA viruses that are a major cause of human disease. All family members share highly related viral replication proteins, such as DNA polymerase, ssDNA-binding proteins and processivity factors. Consequently, it is generally thought that lytic replication occurs through a common and conserved mechanism. However, considerable evidence indicates that proteins controlling initiation of DNA replication vary greatly among the herepesvirus subfamilies. In this article, we focus on some of the known mechanisms that regulate Epstein-Barr virus lytic-cycle replication, and compare this to other herpesvirus family members. Our reading of the literature leads us to conclude that diverse viral mechanisms generate a common nucleoprotein prereplication structure that can be recognized by a highly conserved family of viral replication enzymes.
Keywords: BZLF1, EBV, Epstein–Barr, OriLyt, recombination, repair, replication, Zta
Epstein–Barr virus (EBV) is the prototypical γ-herpesvirus because of its early discovery as a human tumor virus [1]. EBV is now known to contribute to a variety of human disorders, including infectious mononucleosis, oral hairy leukoplakia, nasopharyngeal carcinoma, Burkitt’s lymphoma and lymphoproliferative diseases occurring in immunocompromised individuals. A second human γ-herpesvirus has been identified as the causative agent of Kaposi’s sarcoma, and has been designated Kaposi’s sarcoma-associated herpes virus (KSHV) [2]. KSHV, a close relative of EBV, is also found to be associated with lymphoid disorders, including pleural effusion lymphomas and Castlemen’s disease. Both EBV and KSHV preferentially establish latent infection in B lymphocytes and contribute to malignant transformations in both lymphoid and epithelial tissues.
Although latent viral infection is typically thought to contribute to lymphomagenesis, it is becoming increasingly apparent that lytic replication of the virus is a strong contributing factor to cancer cell evolution. Lytic EBV has been implicated in nasopharyngeal carcinoma [3] and methotrexate-induced lymphomas, arising in patients treated for rheumatoid arthritis and polymyositis [4]. Chronic lytic EBV infection caused by co-infection of malaria is suspected of promoting endemic Burkitt’s lymphoma [5,6]. KSHV lytic infection in endothelial cells is strongly correlated with progression of Kaposi’s sarcoma. Pharmacological inhibitors of herpesvirus lytic replication can ameliorate disease progression, but do not prevent recurrence due to drug resistance [7]. Considering the importance of lytic-cycle replication to pathogenesis, a further understanding of the early events that control initiation of viral DNA replication will improve our ability to develop therapeutics of viral-associated disease.
Initiation of γ-herpesvirus lytic cycle
Like all herpesviruses, lytic replication can initiate at two points in the viral lifecycle: during primary infection or upon reactivation of latent infection. It is unclear whether these mechanisms are significantly different, but it is likely that different chromosome configurations and cell-response factors must be involved in these different pathways to productive infection. For EBV and KSHV, the immediate-early (IE) proteins must be expressed and functional for productive infection to progress. While EBV and KSHV share partial conservation of these IE proteins, they have remarkably different biological and biochemical properties. The most conserved IE protein is referred to as Rta, and is essential for transcription activation and lytic replication in both viruses [8,9]. In EBV, a second IE protein, referred to as Zta (encoded by the BZLF1 gene and also known as, Z, ZEBRA and EB1) plays a primary role in lytic activation and lytic replication [10–13]. The KSHV ortholog, K8, does not appear to activate transcription [8,14] and overexpression of BRLF1, the gene that encodes Rta, can overcome the block to lytic replication in a K8-null virus [15]. This divergence in Zta/K8 requirement emphasizes the variation in mechanisms of initiation of lytic replication, even among two highly related γ-herpesviruses.
The requirement for Zta and Rta in EBV lytic replication have been demonstrated by numerous genetic and biochemical studies. Genetic disruption of either gene prevents lytic replication [9]. Although viruses lacking Zta can still immortalize primary B lymphocytes in culture, these cells fail to form tumors in severe-combined immunodeficient mice [16,17]. Since Zta is a potent transcription activator, it is likely that Zta expression promotes tumor formation through activation of viral and cellular factors, including viral cytokines that promote tumor formation [18–20]. Rta and Zta can be coexpressed from a single bicistronic transcript in EBV [21], and a similar gene organization exists for KSHV ORF50, the gene that encodes Rta, and the K8 gene [22]. Signaling pathways that activate Rta or Zta transcription are known to initiate lytic-cycle gene expression. Numerous cellular factors can bind the transcriptional regulatory regions of these IE genes and are subject to complex regulation. In addition, both Rta and Zta can interact with numerous cellular factors and are subject to post-translational modifications that can affect their function in lytic replication and transcription activation. Thus, regulation of these IE genes represents an important level of control for initiation of lytic-cycle replication. Many of these controls have been reviewed else-where [23–25]. In this article, we focus on the role of these and other proteins in the establishment of an active origin of lytic replication, and consider their function in this later stage process where they have essential and direct functions at the origins of DNA replication.
Identification of the conserved Herpesviridae core replication machinery
In 1986, a landmark complementation assay was developed to identify six core herpes simplex virus (HSV)1 genes required to support lytic replication of a plasmid containing the HSV1 repeated short-region origin of replication (OriS) [26]. Subsequent studies revealed that orthologs of these six replication proteins were required for lytic replication of other herpesviruses, including EBV [27–30]. In the case of EBV, these include a viral polymerase (BALF5), a polymerase processivity factor (BMRF1/EA-D), a helicase–primase complex (BBLF4, BSLF1 and BBLF1/2) and a ssDNA-binding protein (BALF2) [27]. The BALF5 polymerase is highly processive, able to efficiently add more than 7200 nucleotides to an RNA primer on a DNA template at a rate of 12 nucleotides per second before falling off and its affinity for RNA–DNA hybrids is greater than its affinity for ssDNA [31]. BALF5 protein has 3′–5′ exonuclease activity and preferentially excises a terminal mismatched nucleotide [32]. Affinity analysis has shown that BALF5 polymerase is able to interact with the helicase/primase proteins BSLF1, BBLF4 and BBLF2/3 [33], which themselves are able to form an enzymatic complex independently [34]. The processivity power of BALF5 is enhanced by its interaction with the BMRF1 gene product, EA-D [35,36]. EA-D, a sliding clamp protein resembling the cellular PCNA protein, is required for lytic replication [37] and has a unique role as a transcription factor [38,39]. These herpesvirus DNA replication enzymes bear high sequence homology across family members, are capable of both leading and lagging strand synthesis in vitro [40] and can synthesize DNA at a preformed replication fork in vitro [41]. Furthermore, many of these core replication proteins can be interchanged between different herpresviruses; HSV1 core replication proteins replicate the varicella zoster virus genome [42]; the EBV core proteins replicate human cytomegalovirus (CMV) [43] and herpesvirus papio DNA [44]; and the KSHV core proteins replicate the EBV genome [30]. The herpesvirus core replication machinery can even replicate DNA viruses from other viral families, such as adeno-associated virus [45,46] and simian virus 40 [47]. However, each viral lytic origin requires specific recognition conferred by its own origin-binding protein. Without the correct origin-binding protein, the core enzymatic complex cannot initiate replication.
Diverse group of origin-binding proteins
A herpes lytic origin-binding activity was first described for HSV1 [48] and later identified as the product of the UL9 gene [49]. UL9 encodes an ATP-dependent helicase [50,51], which, in cooperation with the ssDNA-binding protein ICP8, is able to bind, loop, distort and unwind OriS DNA [52–56]. Human CMV uses a UTPase encoded by the UL84 gene as its origin-binding protein [43,57,58]. UL84 protein binds an RNA stem-loop structure within the CMV origin of lytic replication (OriLyt) [59] where it functions as a transcription factor along with the IE2 protein at the origin’s bidirectional promoters [60]. In the case of EBV, the IE transcription factor Zta is the best candidate for a viral-encoded origin-binding protein required for lytic replication [27,61–63].
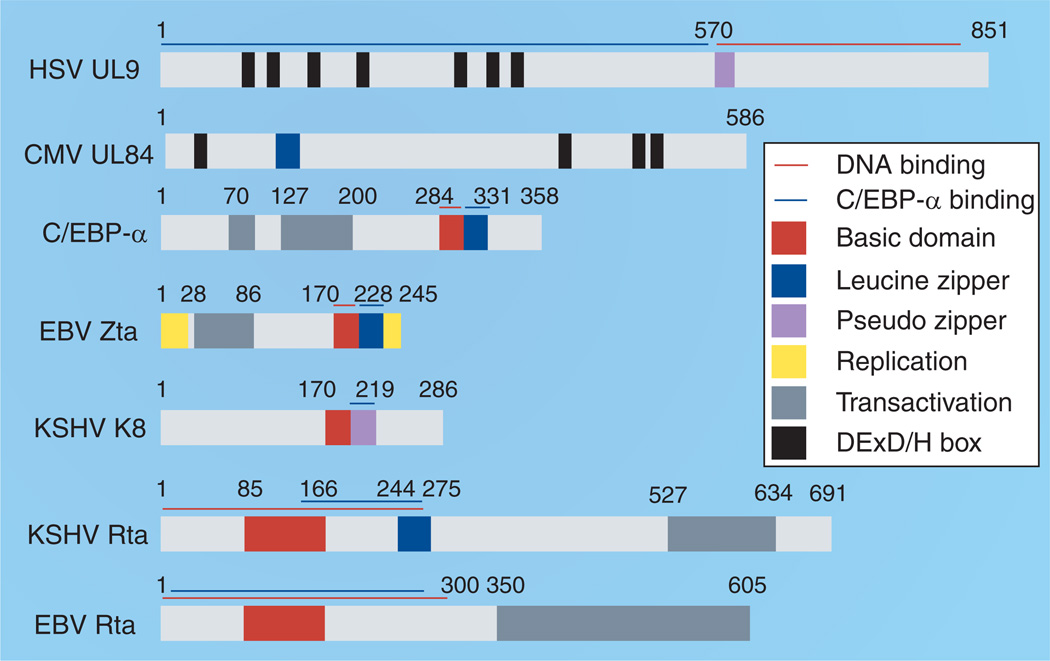
Zta binds directly to multiple sites in OriLyt and recruits components of the viral core replication machinery to OriLyt [64–67]. In addition to OriLyt binding, Zta activates transcription from multiple viral promoters, including the two divergent promoters within the approximately 1 kb OriLyt. Zta consists of an amino-terminal transcription activation domain and carboxy-terminal basic-zipper (bZip) domain (Figure 1). The bZip domain has closest homology to the cellular transcription factors C/EBP-α, c-Fos and c-Jun [68,69]. The basic region (residues 170–198) permits binding to origin and promoter DNA via pseudo-palindromic heptad Zta response elements (ZREs), which include AP1/TRE sites [70–72] and others conforming to the loose consensus sequence 5′-T(G>T>C) (A>G>T)(C/G)(T>C>A)(C>A>G)A-3′ found in the promoters of both cellular [72–79] and viral [69,72,80–83] genes. The crystal structure of the bZip domain has revealed a unique fold-back structure at the C-terminus that distinguishes it from all other known bZip structures [84–86]. Recent studies reveal that Zta binds preferentially to methylated cytosines, and this has been proposed to regulate the tendency of EBV to establish latent infection and to efficiently reactivate latent genomes that have been subject to cytosine methylation [83,87–89].
Figure 1. Domain structure of the herpesvirus lytic origin-binding proteins.
Although herpesvirus lytic origin-binding proteins are quite diverse, there are a few limited similarities. HSV UL9 and CMV UL84 share DExD/H box similarities, and most of these proteins contain leucine zipper or pseudo zipper domains (blue and purple boxes). Many of these regions bind to the cellular protein C/EBP-α (also shown).
CMV: Cytomegalovirus; EBV: Epstein–Barr virus; HSV: Herpes simplex virus; KSHV: Kaposi’s sarcoma-associated herpesvirus.
Since Zta lacks any known enzymatic activity, it is thought that cellular-interacting proteins contribute essential activities to replication initiation. Among the many interacting proteins, the interaction with C/EBP-α is most notable because of its potential common role at other herpesvirus lytic origins [90,91] and because it shares the most extensive homology with Zta (Figure 2A). C/EBP-α has been implicated in both Zta-mediated replication and cell-cycle arrest [92–95]. Zta physically interacts with C/EBP-α through contacts in the conserved zipper domain, where the two proteins are thought to form tetramers [96]. The Zta–C/EBP-α interaction is responsible for the induction of CDK inhibitors (p21WAF-1/CIP-1 and p27KIP-1) [95], and in the absence of C/EPB-α, Zta is unable to activate the p21 or p27 promoters or cause cell-cycle arrest [92]. The precise role of cell-cycle arrest prior to viral DNA replication is not completely understood, but is thought to be important for preventing competition of virus with the host cell demands during DNA replication. Cell-cycle arrest prior to DNA replication appears to be a common event for all of the herpesviruses [97].
Figure 2. Zta basic-zipper alignments.
(A) Zta most closely resembles the C/EBP-α protein, the greatest homology is in Zta’s C-terminal tail that includes its critical basic-zipper region. (B) The critical basic region of Zta shares limited homology with the region of HSV1 UL9 protein involved in ssDNA binding.
Like EBV, the KSHV genome encodes a protein, K8 (K-bZip, RAP), which resembles and is syngenic to Zta. Like Zta, K8 binds to C/EPB-α and causes C/EBP-α-dependent G0/G1 cell-cycle arrest [98]. However, the requirement for K8 in DNA replication is somewhat controversial. Unlike Zta, ectopic expression of K8 cannot reactivate virus from latency [14]. Furthermore, K8 does not possess any intrinsic DNA-binding activity. Initially known as KSHV RAP, K8 associates with KSHV OriLyt and recruits some viral and cellular proteins to the site of DNA replication [99,100], but its DNA binding is indirect, mediated by interaction with C/EBP-α [101,102], Rta [103] or the viral latency-associated nuclear antigen (LANA) [104]. Initial studies using OriLyt-containing plasmids indicated that K8 is required for lytic replication [105,106], and a K8-knockout (BAC36ΔK8) virus is compromised for DNA replication [8]. However, this defect can be rescued by overexpression of Rta [15], suggesting that, to some extent, K8 is dispensable. By contrast, the overexpression of EBV Rta is unable to rescue a BZLF1-knockout virus [107]. K8 also differs from Zta in that its ‘zipper’ region (residues 190–237), while required for multimerization [108], folds into a β-sheet rather than as an α-helix [109]. K8 can bind to Rta via its zipper motif and this interaction may attenuate Rta transcription activity and, consequently, activate its replication function [103,110]. Thus, K8 has diverged significantly from its EBV ortholog Zta, while KSHV Rta takes on a more prominent role in replication initiation and viral reactivation from latency [8,111,112].
Identification of lytic origins
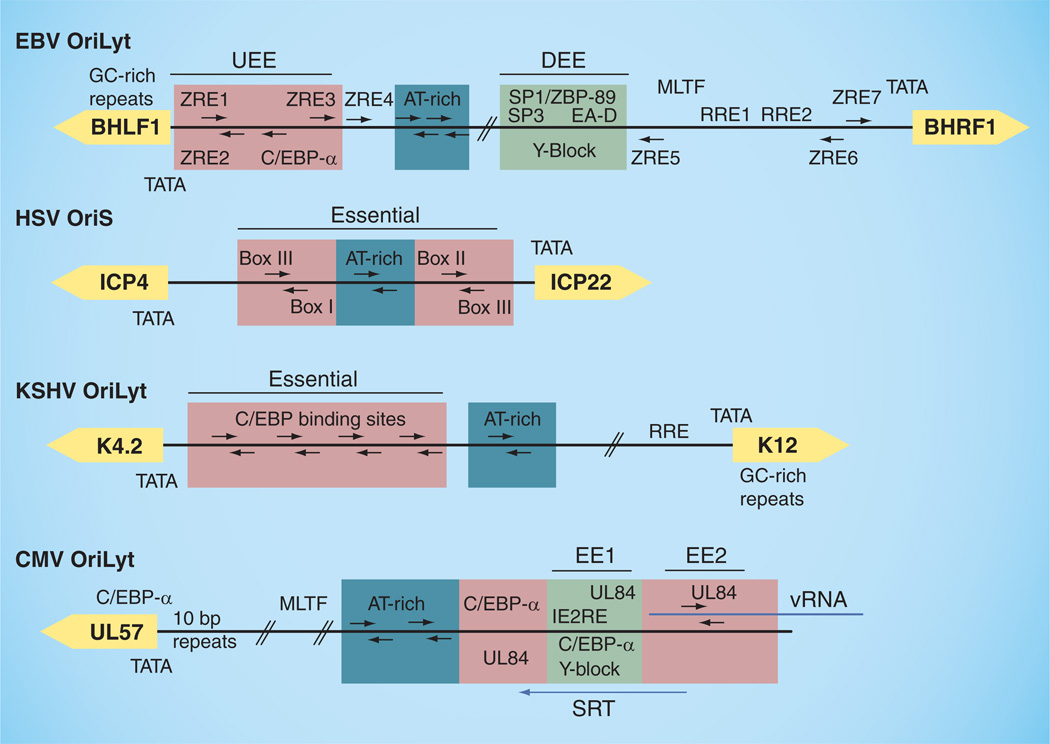
Early studies with HSV revealed that DNA replication could initiate at three homologous cis-acting elements (two copies of OriS and one copy of the unique long origin of lytic replication [OriL]), each capable of functioning as an origin (Figure 3) [113–116]. Each α-herpesvirus origin (and those of roseolavirus β-herpesviruses) includes a necessary palindrome containing two initiator protein-binding sites (box I and II) flanking an AT-rich sequence [117,118] and a third (box III) adjacent to box I also required for replication in vivo [115,119,120]. EBV typically encodes two homologous copies of OriLyt [121], although there are functional strains that only encode one copy, which includes binding sites for Zta [72,121]. Unlike the origins of α-herpes- and roseolaviruses, the minimal EBV OriLyt sequence is comprised of two essential elements flanking dispensable AT-rich palindromes [122]. These elements are located between two divergent promoters. The upstream essential element (UEE) contains the TATA box for the BHLF1 (or L3) gene, two ZREs (ZRE1/2) [62,122,123], which form an inverted repeat that is similar to the UL9 box III–I binding sites in HSV1 OriS, and a CCAAT box. The downstream essential element (DEE) contains binding sites for the Sp1, Sp3 and ZBP-89 proteins, which interact with the core viral-replication proteins [62,65,124,125], including the EA-D processivity factor that is able to activate the BHRF1 promoter [67] via the downstream element [38]. The DEE also contains a homopurine–homopyrimidine ‘Y-box’ sequence capable of forming a triple helix in vitro, and mutations that impair the triple-helix formation in vitro also disrupt DNA replication in vivo [126]. The BHRL1 promoter also contains two binding sites for Rta, although these are found outside of the minimal OriLyt sequence [127]. The critical regions identified in EBV OriLyt have also been shown to be evolutionarily conserved in the related herpesvirus papio [44].
Figure 3. Human herpesvirus lytic origins of replication.
Inverted repeats containing binding sites for known origin-binding proteins are noted with red boxes. AT-rich regions are boxed in blue and Y-block elements are boxed in green. Flanking divergent genes are shown in yellow. Essential elements have also been noted.
CMV: Cytomegalovirus; DEE: Downstream essential element; EBV: Epstein–Barr virus; EE1: Essential element 1; EE2: Essential element 2; KSHV: Kaposi’s sarcoma-associated herpesvirus; HSV: Herpes simplex virus; IE2RE: Immediate-early gene 2 response element; OriLyt: Origin of lytic replication; OriS: Repeated short-region origin of replication; REE: Rta response element; SRT: Cytomegalovirus OriLyt transcript; UEE: Upstream essential element; ZRE: Zta response element.
Like EBV, KSHV encodes two nearly identical OriLyt sequences found between divergent promoters that are close proximity to GC-rich repeats [111,112,128,129]. These include a region similar to the EBV DEE, containing an Rta responsive element in association with a TATA box, an AT-rich palindrome sequence, and eight CCAAT boxes organized as four head–tail pairs [130] reminiscent of the UL9 (pointed out in [129]) and Zta UEE binding sites on their respective genomes. The closely related murine herpesvirus (MHV)-68 has a set-up similar to KSHV, albeit with two pairs of inverted CCAAT boxes required for efficient replication rather than four [131,132]. Mutation of an OriLyt sequence within MHV-68 not only resulted in the impairment of lytic but also latent replication [133], an interesting observation in light of the recent connection drawn between KSHV LANA and OriLyt [104].
Unlike the other herpesviruses, human CMV encodes only one known OriLyt occupying more than 2.5 kb of the genome [134–139]. This large and complex element, although obviously different from α- and γ-herpesvirus origins, does retain some similarities with the others, including a bidirectional promoter (OriLytPM), many GC-rich inverted repeats, potential transcription-factor binding sites and several AT-rich segments, all of which have currently poorly defined functions [140]. Like EBV, there are two regions of CMV OriLyt known to be essential for replication [60,137]. Essential element 1 (EE1) contains a necessary ‘Y-block’ promoter element [141], similar to that of the EBV OriLyt DEE, which contains binding sites for CMV IE2, UL84 and C/EBP-α [91]. The second essential element (EE2) overlaps a long RNA transcript (CMV OriLyt transcript [SRT]) and contains a base-sensitive RNA–DNA hybrid that can form an RNA stem-loop, to which UL84 also binds [59,142].
Origin strand unwinding & ssDNA binding proteins
Little is known about the early initiation events leading to strand unwinding of γ-herpesvirus lytic origins. The best-characterized lytic origin-binding protein of the family is the HSV1 UL9 helicase, which appears to work in concert with the single-stranded binding protein ICP8 to accomplish strand separation. The functional domains of UL9 have been mapped, including those required for DNA and ICP8 binding [143–146]. In vitro, the UL9 protein binds cooperatively to the OriS box I–III sequence in an ATP-dependent manner [52] and, together with ICP8, is able to unwind several OriS-containing DNA substrates [147,148]. Binding of UL9 and ICP8 causes a conformational change in OriS that is detectable by nuclease probing [149] and electromobility shift assays [150,151]. This activated form of OriS, termed OriS*, has been shown to contain a DNA hairpin formed by complementary intrastrand base pairing of box I and III [152,153]. Nuclear magnetic resonance and DNA melting experiments have been used to demonstrate that a large number of α-herpesviruse have the capacity to form hairpins in OriS at origin-binding protein-recognition sites [154]. It is still unclear as to the role that DNA secondary structure plays in the origins of other herpesviruses, although the presence of so many inverted repeats within their sequences raises the possibility that intrastrand binding may be part of a conserved mechanism of replication initiators.
Although EBV Zta and HSV1 UL9 are structurally two very different proteins, they both possess ssDNA-binding activity [Rennekamp AJ et al., Unpublished Data] [155] and have some sequence homology in the regions known to bind ssDNA (Figure 2B). In addition, the ssDNA-binding proteins encoded by EBV BALF2 and UL29 (ICP8) have approximately 30% sequence homology and high structural equivalency [156–159]. The ICP8 protein is a multifunctional zinc metalloprotein [160], which preferentially binds ssDNA in a nonsequence-specific manner [161]. ICP8 also binds to the C-terminus of the UL9 protein, stimulating helicase activity [55,162–164]. Both ICP8 and EBV BALF2 proteins have the properties of a ssDNA strand annealing protein, similar to the λ RED β recombination protein [165]. ICP8 can displace short DNA strands from their complementary sequences [166], promote DNA strand transfer [167,168] and strand invasion [169–171], and can renature complementary strands of DNA [172]. There is evidence suggesting that the BALF2 protein can perform some, if not all, of these functions as well [173].
θ, rolling circle & recombination?
It has been proposed that herpesviruses copy their genomes via a ‘rolling-circle’ method of lytic replication [174–177]. Several lines of evidence have been used to support this model. First, herpesvirus genomes, including EBV, adopt a circular conformation within the cell quickly following infection and upon lytic induction [178–184]. Circular DNA would provide the template necessary for a rolling-circle mechanism. Second, concatemeric forms of intracellular DNA have been observed as replicative intermediates during lytic replication [121,181,185,186], as well as a reduction in the copy number of genomic termini [187,188]. These intermediates also sediment rapidly in sucrose gradients, demonstrating the presence of viral DNA with molecular weights beyond that of single genomes [189,190]. In addition, some encapsidated defective genomes have been identified as head–tail repeats [191]. Finally, as a proof of principle, it has been demonstrated that the core herpesvirus enzymes and extracts of infected human cells are able to replicate certain templates via a rolling-circle mechanism in vitro [192,193].
However, a simple rolling-circle mechanism does not adequately explain every observation. Further examination of lytic replicative intermediates, using pulse-field gels and electron microscopy, reveals a highly branched network of DNA, containing multiple forks on single molecules [186,194]. In addition, analyses of the kinetics of lytic replication reveal that viral DNA accumulates exponentially and is amplified several hundred-fold in just a few hours [195], while a rolling-circle mechanism would produce a linear amplification [196]. Finally, replicative concatamers of HSV1 genomes contain genomic inversions suggestive of strand-transfer events [181,197]. While these observations do not exclude a rolling-circle mechanism, they suggest that, at the very least, other modes of genome replication are at work. Indeed, EBV OriLyt is able to direct semiconservative replication and production of monomeric progeny soon after lytic induction when incorporated into a plasmid [196]. These observations have led to a dual-mechanism model, similar to that observed in λ-phage replication, where lytic replication is initiated via a plasmid or 'θ’ mode, where copy number is enriched, followed by a switch to rolling circle.
It is also likely that herpesvirus lytic replication involves a recombination mechanism. Herpesvirus genomes are highly recombinogenic, containing frequent genomic inversions anchored by repeat regions [191,198–203]. Homologous recombination at these regions (e.g., the terminal repeats in the EBV genome) occurs frequently and is dependent on lytic replication [168,204–206], and specifically on the core herpes replication machinery and the viral origin (in these experiments, OriS), which together are also sufficient to induce recombination [207,208]. Surprisingly, however, even herpesviruses that naturally lack invertible repeat elements have replication machinery able to support efficient segment inversion, suggesting that the recombinatory function of the conserved core herpes replication machinery plays some additional role, perhaps in replication [209]. The core single-stranded binding protein (e.g., ICP8) is known to promote ssDNA strand invasion, homologous pairing and D-loop formation [168,170,171], not unlike the λRED β recombination protein [165]. In the case of EBV, this protein, encoded by the BALF2 gene, is known to associate with the viral alkaline nuclease (BGLF5) [210], which structurally resembles the λ RED α exonuclease [211]. BGLF5 has 5′–3′ exonuclease activity, as well as endonuclease activity on linear ssDNA, linear dsDNA, nicked dsDNA circles and super-coiled plasmid DNA, in addition to an RNase activity [211–214]. This nuclease is known to contribute to, although not absolutely required for, genome replication [215].
Role for cellular recombination & DNA damage response proteins
In addition to encoding their own proteins capable of promoting recombination, herpesviruses also interface with host cell replication machinery. Many of these proteins are involved in cellular DNA damage repair–recombination pathways [216–218].
Herpes simplex virus 1/2 infection induces phosphorylation of p53, ATM and Mre11–Rad50–Nbs1 (MRN) complex members, as well as several other DNA damage proteins (e.g., RPA, Chk2, Rad50 and 53BP) [219–221]. At the same time, several of these proteins are recruited to viral-replication compartments (e.g., p53, ATM, the MRN complex, DNA–PKCs, Rad50, Ku80/86 and WRN) [219,220,222] and interact with ICP8 [222] and/or OriS [223]. This damage response is not present in latency or latency-like situations, and not inhibited by the viral DNA polymerase inhibitor phosphonoacetic acid [219], suggesting that this is an early replication event. Indeed, RPA, Rad51 and Nbs1 are recruited to prereplicative compartments containing only UL9, ICP8 and the helicase–primase complex in the absence of polymerase.
Generally speaking, activation of the DNA damage response is beneficial for viral replication as Mre11 or ATM (and perhaps WRN) mutant cells have reduced capacity to support lytic replication [219,222]. By contrast, viral replication in Ku70-deficient murine embryonic fibroblasts is increased by almost 50-fold [222], a second subunit of DNA–PK, Ku80/Ku86, is excluded from replication compartments [221], and the DNA–PK core subunit (DNA–PKCS) is degraded in a proteosome-dependent manner upon expression of HSV1 ICP0 [224]. These observations suggest that the cellular homologous recombination (HR) pathway is important for lytic replication while the nonhomologous end-joining (NHEJ) pathway is inhibitory [221,222]. ICP0 is also capable of inducing the phosphorylation/activation of Chk2 via ATM [225], and knocking out the DNA damage pathway induced by ssDNA and mediated by ATR/ATRIP/RPA [226].
In terms of the DNA damage response, CMV infection appears to be very different from HSV1/2. p53 is increased, phosphorylated and relocated to viral replication compartments [227], and p53-null fibroblasts, while permissive for CMV infection, show a decrease in viral DNA and particle production [228]. However, ATM is not activated and the MRN complex is excluded from replication compartments. Although the quantity and phosphorylation of Nbs1 does increase, both ATM and Mre11 are dispensable for CMV replication [227].
Epstein–Barr virus lytic replication resembles HSV1/2 in this regard; EBV elicits ATM signal transduction (with minimal activation of ATR) and recruits phosphorylated p53, ATM and the MRN complex to replication compartments. However, ATM activation was not required, as caffeine treatment, which inhibits ATM activation, did not affect lytic replication [229]. As in the case of HSV1/2, proteins involved in HR, including RPA, Rad51, Rad52 and the MRN complex, are recruited and loaded onto the EBV genome in replication compartments. Furthermore, EBV replication compartments contained dsDNA breaks and Rad51 and RPA32 were required for viral DNA synthesis [230]. Zta binds to RPA subunits [231] and forms a functional interaction with 53BP1 [232], which is also involved in the detection and repair of dsDNA breaks. The finding that a DNA ligsase IV syndrome patient, lacking the important ligase required for NHEJ repair, developed EBV-positive B-cell lymphoma is also intriguing if one assumes that the virus thrives in a situation where the balance between NHEJ and HR is dramatically shifted exclusively toward homologous repair [233]. In addition to containing HR proteins, EBV replication compartments have been shown to contain mismatch repair (MMR) proteins, including PCNA, RC-F, MSH2, MSH6, MLH1 and hPSM2, which are loaded on to the viral genome and copurified with the BMLR1 protein [234] and the RecQL helicase, which are associated with Zta [235]. The BGLF4 viral kinase interacts with the XPC protein, a member of a third DNA repair pathway, nucleotide excision repair. BGLF4 or XPC knockdown results in decreased viral replication [236,237], and BGLF4 expression enhances cellular XPC-mediated DNA repair in vivo [236].
The BGLF4 homolog in γ-MHV-68, encoded by ORF36, also plays a role in the induction of the DNA damage response by directly phosphorylating γ-H2AX, the dsDNA break sensor. This activation of γ-H2AX is further enhanced by ATM, and all three proteins are required for efficient replication of the virus in primary mouse macrophage cells [238]. However, the requirement for induction of a DNA damage response may be cell-type specific as fibroblast infection results in the inhibition of NHEJ/HR. This inhibition is mediated by the unique viral M2 protein, which, although sufficient to induce expression of ATM, also binds to ATM causing inhibition of the downstream effectors γ-H2AX, Nbs1 and 53BP. M2 is also capable of inhibiting nucleotide excision repair through interaction with the DDB1–COP9–cullin repair complex [239]. KSHV also encodes four interferon response factor-like proteins capable of blocking DNA repair pathways by the inhibition and degradation of p53 [240,241]. Consequently, neither p53 nor active ATM accumulates in KSHV-infected fibroblasts. By contrast, in the KSHV-infected lymphocyte cell line BCBL1, several DNA repair proteins were found bound to OriLyt and localized to replication compartments. These include RecQL helicase [242], the MMR proteins MSH2 and MSH6, NHEJ proteins DNA–PKCS, Ku86 and Ku70, and poly-ADP ribose polymerase 1 (PARP1). PARP1 inhibitors were shown to diminish replication whereas hydroxyurea, which raises PARP1 activity, caused an increase in the DNA replication [242]. These observations provide compelling arguments that DNA recombination and repair activities play a critical role in the early stages of herpesvirus lytic replication.
Transcriptional requirements
There is mounting evidence in support of a role for RNA and transcription in the initiation of lytic herpesvirus replication. First, the lytic origin-binding proteins of both EBV and KSHV (Zta and Rta) are both transcription activators. It has also been known for quite some time that RNA polymerase II, TATA binding protein and TATA binding protein-associated factors [243] are recruited to HSV1 replication compartments [244,245]. What has become clearer recently is that transcription plays a major role in the selection of origins in mammalian cells [246]. All herepsvirus lytic origins consist of promoters containing transcription-factor binding sites important for replication. For example, varicella zoster virus OriS-dependent DNA replication and origin promoter transcription both require binding of the cellular factors Sp1 and Sp3 [247]. EBV replication is dependent on transcription of the BHRF1 promoter and, possibly, the BHLF1 promoter [122,248], independent of the gene product or even promoter sequence [121]. In the same way, KSHV and CMV OriLyt-mediated replication are also dependent on activation of their bidirectional promoters [60,90]. Several RNA species transversing herpesvirus origins have also been identified. These include OriS-RNA2, which overlaps HSV1/2 OriS [249,250], and a family of small bottom strand RNAs, which overlap the EBV OriLyt UEE [251]. The best-characterized lytic origin RNA, identified in CMV OriLyt, forms a persistent RNA–DNA hybrid structure [142] containing an RNA stem-loop sequence that is bound by the UL84 origin-binding protein [59]. Interestingly, an RNA–DNA hybrid has also been found at the human mitochondrial heavy-strand origin [252]. RNA may also play a regulatory role in latent replication of EBV at OriP [253,254]. These observations suggest a role for transcription and possible RNA itself in the initiation of lytic replication.
Additional factors involved in EBV lytic replication
Several additional viral proteins have also been shown to play important, although not always necessary, roles in enhancing lytic replication. These include the BGLF5 alkaline exo-nuclease [215] and BGLF4 kinase [237] described earlier. In addition to its role in promoting DNA recombination, the BGLF4 protein also localizes to replication compartments where it phosphorylates Zta [255], EA-D [256] and the MCM4–MCM6–MCM7 complex, which it also activates [257]. BGLF4 also phosphorylates EBV nuclear antigen 1, which remains expressed and bound to OriP during lytic replication [258], resulting in disruption of latent genome maintenance [259].
Uracil DNA glycosylases (UDGs) are also important since the inhibition of both viral and cellular UDGs significantly impairs lytic replication [260,261]. Studies involving the CMV homolog of EBV BKRF3 (UL114) suggest that UDGs are an integral part of the early-to-late replication switch mechanism of the virus, whereby incorporation of uracils into the viral genome followed by UDG and exonuclease activity may lead to strand breaks creating substrates for recombination-dependent replication [262]. This is an intriguing possibility given that uracil incorporation into HSV1 OriS would abrogate UL9 binding [263], perhaps mediating a switch from UL9-dependent to UL9-independent replication. Further proof of principle has been provided by demonstration of the ability of HSV1 UDG (UL2), processivity factor (UL42) and DNA polymerase (UL30) to cooperate with human AP endonuclease to create a DNA single-strand break in vitro [264]. Additional lines of evidence suggest that viral UDGs are not merely occasional repair proteins. These include the observation that:
-
▪
Recombinant CMV genome lacking the viral UDG gene (UL114) did not accumulate more uracil compared with the wild-type virus [265];
-
▪
The catalytic activity of viral UDG was very inefficient as compared with human UDG;
-
▪
The viral processivity factor is required for the viral UDG to be loaded onto DNA [266].
Conclusion & future perspective
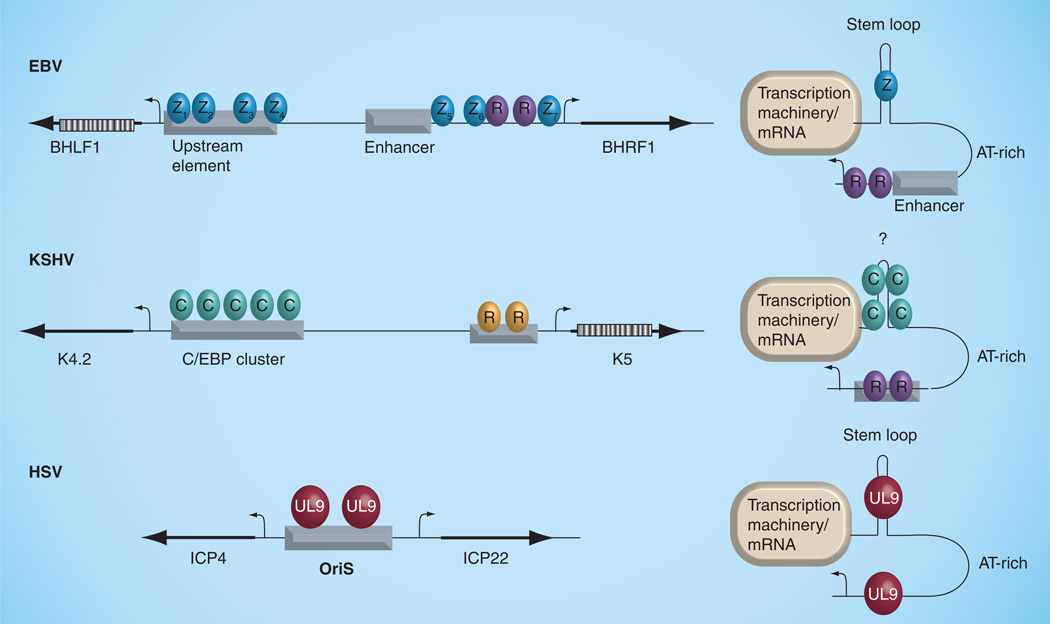
The most successful therapeutic interventions currently used against herpesvirus infection and associated diseases target the lytic replication of viral DNA. A better understanding of the enzymes and mechanisms involved in this process will likely yield additional drug targets and improved treatment options in the future. Our survey of the literature suggests that a large diversity of mechanisms are employed to generate the initiating nucleoprotein structures and host cell environment conducive to lytic replication. Nevertheless, these various mechanisms converge on common pathways that include highly conserved viral-replication enzymes and cellular factors involved in host cell DNA recombination, repair and replication. Other common requirements include the host cell-cycle arrest, a nuclear reorganization into replication compartments and a near-universal requirement for RNA transcription. We suggest that common elements, such as DNA hairpin structures bound to origin-binding proteins, transcription initiation factors and RNA transcripts, contribute to the formation of a higher-order structure that is recognized by the core viral DNA replication machinery (Figure 4). A major focus of future research will be investigating the mechanistic contributions of DNA repair proteins (e.g., viral and cellular endonucleases and recombinases) to the initiation and progression of lytic DNA replication. Certainly, the role of virus proteins on cell-cycle control have been investigated extensively, but the precise mechanism of cell-cycle rerouting used by herpesviruses deserves further attention. For γ-herpesviruses, the common use of C/EBP-α factors for lytic-cycle replication and cell-cycle arrest raises some important questions, including whether other viruses use this family of proteins to control their origins of replication, and whether this is coordinated with cell metabolic state and differentiation status, where C/EBP-α is known to play an important role. Finally, it will be necessary to determine whether RNA transcription contributes to lytic replication initiation. Active transcription may facilitate formation of an active prereplication complex by stimulating strand unwinding and torsional strain. Alternatively, transcription factors and RNA polymerase accessory factors may facilitate chromatin remodeling and replisome assembly. It will also be interesting to determine whether the transcribed RNAs (both coding and noncoding) contribute directly to origin protein assembly or function during lytic replication. These and other areas of future investigation suggest that exciting discoveries will soon emerge from further studies of herpesvirus DNA replication.
Figure 4. Hypothetical model of common structures formed by herpesvirus origins.
EBV, KSHV and HSV OriS are envisioned to form higher-order structures that include inverted repeat hairpins that bind viral (UL9, Zta) or cellular (C/EBP-α) proteins. Transcription complexes formed at both ends of OriLyts are envisioned to induce topological changes that promote DNA unwinding and templates suitable for viral DNA replisomes to assemble.
EBV: Epstein–Barr virus; HSV: Herpes simplex virus; KSHV: Kaposi’s sarcoma-associated herpesvirus; OriS: Repeated short-region origin of replication.
Executive summary.
Introduction
-
▪
All herpesviruses share a common requirement of lytic replication to produce progeny virus. Although core replication enzymes are conserved among the family members, significant variation is observed at the level of initiation and origin-binding factors.
-
▪
Lytic replication is a major source of all herpesvirus pathogenesis and contributes to cancer cell evolution during chronic infections of Epstein–Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus.
Herpesviridae core replication proteins
-
▪
All herpesviruses share a core set of highly conserved lytic-replication proteins, which can be substituted for one another in many cases.
-
▪
In the case of EBV, these include a viral polymerase (BALF5, currently the major target of herpesvirus antivirals), polymerase processivity factor (BMRF1/EA-D), helicase–primase complex (BBLF4, BSLF1 and BBLF1/2) and a ssDNA-binding protein (BALF2).
Diverse group of origin-binding proteins
-
▪
In contrast to the homologous core replication proteins, herpesviruses each encode very different lytic origin-binding proteins.
-
▪
The EBV lytic origin-binding proteins are Rta and Zta, which also function as immediate-early transcriptional activators. Although not very similar to the other origin-binding proteins, it appears to share some intriguing overlapping functions.
Herpesvirus origins of lytic replication
-
▪
Genetic experiments and plasmid replication assays have been used to identify important cis-acting viral elements required for lytic DNA replication.
-
▪
Although EBV OriLyt and the lytic origins of other herpesviruses vary greatly in sequence composition, they share many tantalizing similarities including the presence of bidirectional promoters, CCAAT boxes and inverted repeat sequences bound by replication initiator proteins.
θ, rolling circle & recombination?
-
▪
It has been proposed that herpesviruses copy their genomes via a ‘rolling-circle’ method of lytic replication. Several lines of evidence support this model. However, a simple rolling-circle mechanism does not adequately explain the highly branched DNA intermediates observed during replication.
-
▪
This has led to a dual-mechanism model, where lytic replication is initiated via a plasmid or ’θ’ mode where copy number is enriched followed by a switch to rolling circle.
Role for cellular recombination & DNA damage-response proteins
-
▪
It is likely that herpesvirus lytic replication involves a recombination mechanism. In addition to encoding their own proteins capable of promoting recombination, herpesviruses that interface with host cell proteins are involved in cellular DNA damage repair–recombination pathways.
Transcriptional requirements
-
▪
There is mounting evidence in support of a role for RNA and transcription in the initiation of lytic herpesvirus replication. Herepsvirus lytic origins consist of promoters containing transcription-factor binding sites important for replication and several RNA species transversing hepesvirus origins, which may play a role in replication.
Additional factors involved in EBV lytic replication
-
▪
Several additional proteins have also been shown to play important, although not always necessary, roles in enhancing lytic replication. These include endonucleases and uracil DNA glycosylases, which may provide insight into the replication mechanisms.
Conclusion & future perspective
-
▪
The most successful therapeutic interventions currently used against herpesvirus infection and associated diseases target the lytic replication of viral DNA. A better understanding of the enzymes and mechanisms involved in this process will likely yield additional drug targets and improved treatment options in the future.
-
▪
Three major focuses of future research will be investigation into the mechanistic contributions of DNA repair proteins (e.g., viral and cellular endonucleases and recombinases) to the initiation of DNA replication upon lytic reactivation, the use of the C/EBP family of proteins as origin controls and the role of RNA and RNA transcription in origin function.
Acknowledgments
The authors’ research was supported by grants from NIH (CA86678) and the Wistar Cancer Center (NCI) to Paul M Lieberman. Andrew J Rennekamp was also supported by a predoctoral fellowship on the Wistar Institute Cancer Biology Training Grant from NIH (1T32 CA09171).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Andrew J Rennekamp, The Wistar Institute, 3601 Spruce Street, Philadelphia, PA 19104, USA and The University of Pennsylvania, Biomedical Graduate Program in Cell & Molecular Biology, The School of Medicine, Philadelphia, PA 19104, USA, Tel.: +1 215 898 9523, Fax: +1 251 898 0663, andrewre@med.upenn.edu.
Paul M Lieberman, The Wistar Institute, 3601 Spruce Street, Philadelphia, PA 19104, USA, Tel.: +1 215 898 9491, Fax: +1 215 898 0663, lieberman@wistar.org.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 2.Schulz TF. The pleiotropic effects of Kaposi’s sarcoma herpesvirus. J. Pathol. 2006;208(2):187–198. doi: 10.1002/path.1904. [DOI] [PubMed] [Google Scholar]
- 3. Joab I, Nicolas JC, Schwaab G, et al. Detection of anti-Epstein–Barr-virus transactivator (ZEBRA) antibodies in sera from patients with nasopharyngeal carcinoma. Int. J. Cancer. 1991;48(5):647–649. doi: 10.1002/ijc.2910480503.. ▪ Implicates Epstein–Barr virus (EBV) lytic-cycle gene products as a causative agent in the formation of nasopharyngeal carcinoma.
- 4. Feng WH, Cohen JI, Fischer S, et al. Reactivation of latent Epstein–Barr virus by methotrexate: a potential contributor to methotrexate-associated lymphomas. J. Natl Cancer Inst. 2004;96(22):1691–1702. doi: 10.1093/jnci/djh313.. ▪ Implicates EBV lytic cycle as a causative agent in the formation of lymphomas in rheumatiod arthritis and poliomyositis patients treated with methotrexate.
- 5.Moormann AM, Chelimo K, Sumba OP, et al. Exposure to holoendemic malaria results in elevated Epstein–Barr virus loads in children. J. Infect. Dis. 2005;191(8):1233–1238. doi: 10.1086/428910. [DOI] [PubMed] [Google Scholar]
- 6. Chene A, Donati D, Guerreiro-Cacais AO, et al. A molecular link between malaria and Epstein–Barr virus reactivation. PLoS Pathog. 2007;3(6):e80. doi: 10.1371/journal.ppat.0030080.. ▪ The plasmodium falciparum 1 membrane protein is able to induce lytic EBV.
- 7.Billaud G, Thouvenot D, Morfin F. Drug targets in herpes simplex and Epstein–Barr virus infections. Infect. Disord. Drug Targets. 2009;9(2):117–125. doi: 10.2174/187152609787847703. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, AuCoin DP, Huete AR, Cei SA, Hanson LJ, Pari GS. A Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 2005;79(6):3479–3487. doi: 10.1128/JVI.79.6.3479-3487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feederle R, Kost M, Baumann M, et al. The Epstein–Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19(12):3080–3089. doi: 10.1093/emboj/19.12.3080.. ▪ Classical genetic study where the BRLF1 or BZLF1 genes were knocked out in the context of the full viral genome.
- 10.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein–Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5(12):3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Countryman J, Miller G. Activation of expression of latent Epstein–Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl Acad. Sci. USA. 1985;82(12):4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooney CM, Rowe DT, Ragot T, Farrell PJ. The spliced BZLF1 gene of Epstein–Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 1989;63(7):3109–3116. doi: 10.1128/jvi.63.7.3109-3116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada K, Shimizu N, Sakuma S, Ono Y. trans activation of the latent Epstein–Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 1986;57(3):1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polson AG, Huang L, Lukac DM, et al. Kaposi’s sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 2001;75(7):3175–3184. doi: 10.1128/JVI.75.7.3175-3184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato-Noah T, Xu Y, Rossetto CC, Colletti K, Papouskova I, Pari GS. Overexpression of the Kaposi’s sarcoma-associated herpesvirus transactivator K-Rta can complement a K-bZIP deletion BACmid and yields an enhanced growth phenotype. J. Virol. 2007;81(24):13519–13532. doi: 10.1128/JVI.00832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong GK, Gulley ML, Feng WH, Delecluse HJ, Holley-Guthrie E, Kenney SC. Epstein–Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 2005;79(22):13993–14003. doi: 10.1128/JVI.79.22.13993-14003.2005.. ▪▪ Demonstrates the importance of lytic replication and, specifically, the BZLF1 gene in EBV-induced tumorigenesis.
- 17.Hong GK, Kumar P, Wang L, et al. Epstein–Barr virus lytic infection is required for efficient production of the angiogenesis factor vascular endothelial growth factor in lymphoblastoid cell lines. J. Virol. 2005;79(22):13984–13992. doi: 10.1128/JVI.79.22.13984-13992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki I, Cheung RK, Dosch HM. Viral interleukin 10 is critical for the induction of B cell growth transformation by Epstein–Barr virus. J. Exp. Med. 1993;178(2):439–447. doi: 10.1084/jem.178.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart AD, Stewart JP, Arrand JR, Mackett M. The Epstein–Barr virus encoded cytokine viral interleukin-10 enhances transformation of human B lymphocytes. Oncogene. 1995;11(9):1711–1719. [PubMed] [Google Scholar]
- 20.Rousset F, Garcia E, Defrance T, et al. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl Acad. Sci. USA. 1992;89(5):1890–1893. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manet E, Gruffat H, Trescol-Biemont MC, et al. Epstein–Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 1989;8(6):1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seaman WT, Ye D, Wang RX, Hale EE, Weisse M, Quinlivan EB. Gene expression from the ORF50/K8 region of Kaposi’s sarcoma-associated herpesvirus. Virology. 1999;263(2):436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 23.Amon W, Farrell PJ. Reactivation of Epstein–Barr virus from latency. Rev. Med. Virol. 2005;15(3):149–156. doi: 10.1002/rmv.456. [DOI] [PubMed] [Google Scholar]
- 24.Speck SH, Chatila T, Flemington E. Reactivation of Epstein–Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5(10):399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 25.Kenney SC. Reactivation and lytic replication of EBV. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., editors. Human Herpesviruses. NY, USA: Cambridge University Press; 2007. pp. 403–433. [PubMed] [Google Scholar]
- 26. Challberg MD. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl Acad. Sci. USA. 1986;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094.. ▪▪ Landmark study that identified the herpesvirus core lytic replication proteins using complementation assays and a plasmid containing the lytic origin.
- 27. Fixman ED, Hayward GS, Hayward SD. trans-acting requirements for replication of Epstein–Barr virus Ori-Lyt. J. Virol. 1992;66(8):5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992.. ▪ Identification of the core replication proteins required for lytic EBV replication.
- 28.Pari GS, Kacica MA, Anders DG. Open reading frames UL44, IRS1/TRS1, and UL36–38 are required for transient complementation of human cytomegalovirus OriLyt-dependent DNA synthesis. J. Virol. 1993;67(5):2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pari GS, Anders DG. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus OriLyt-dependent DNA replication. J. Virol. 1993;67(12):6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu FY, Ahn JH, Alcendor DJ, et al. Origin-independent assembly of Kaposi’s sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 2001;75(3):1487–1506. doi: 10.1128/JVI.75.3.1487-1506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsurumi T. Primer terminus recognition and highly processive replication by Epstein–Barr virus DNA polymerase. Biochem. J. 1991;280(Pt 3):703–708. doi: 10.1042/bj2800703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsurumi T. Characterization of 3′-to 5′-exonuclease activity associated with Epstein–Barr virus DNA polymerase. Virology. 1991;182(1):376–381. doi: 10.1016/0042-6822(91)90685-5. [DOI] [PubMed] [Google Scholar]
- 33.Fujii K, Yokoyama N, Kiyono T, et al. The Epstein–Barr virus pol catalytic subunit physically interacts with the BBLF4–BSLF1–BBLF2/3 complex. J. Virol. 2000;74(6):2550–2557. doi: 10.1128/jvi.74.6.2550-2557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama N, Fujii K, Hirata M, et al. Assembly of the Epstein–Barr virus BBLF4, BSLF1 and BBLF2/3 proteins and their interactive properties. J. Gen. Virol. 1999;80(Pt 11):2879–2887. doi: 10.1099/0022-1317-80-11-2879. [DOI] [PubMed] [Google Scholar]
- 35.Kiehl A, Dorsky DI. Cooperation of EBV DNA polymerase and EA-D(BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;184(1):330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 36.Li JS, Zhou BS, Dutschman GE, Grill SP, Tan RS, Cheng YC. Association of Epstein–Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J. Virol. 1987;61(9):2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuhierl B, Delecluse HJ. The Epstein–Barr virus BMRF1 gene is essential for lytic virus replication. J. Virol. 2006;80(10):5078–5081. doi: 10.1128/JVI.80.10.5078-5081.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Holley-Guthrie E, Ge JQ, Dorsky D, Kenney S. The Epstein–Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV OriLyt. Virology. 1997;230(1):22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama S, Murata T, Murayama K, et al. Epstein–Barr virus polymerase processivity factor enhances BALF2 promoter transcription as a coactivator for the BZLF1 immediate-early protein. J. Biol. Chem. 2009;284:21557–21568. doi: 10.1074/jbc.M109.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falkenberg M, Lehman IR, Elias P. Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc. Natl Acad. Sci. USA. 2000;97(8):3896–3900. doi: 10.1073/pnas.97.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabkin SD, Hanlon B. Herpes simplex virus DNA synthesis at a preformed replication fork in vitro. J. Virol. 1990;64(10):4957–4967. doi: 10.1128/jvi.64.10.4957-4967.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stow ND, Davison AJ. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J. Gen. Virol. 1986;67(Pt 8):1613–1623. doi: 10.1099/0022-1317-67-8-1613. [DOI] [PubMed] [Google Scholar]
- 43.Sarisky RT, Hayward GS. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting OriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 1996;70(11):7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryon JJ, Fixman ED, Houchens C, et al. The lytic origin of herpesvirus papio is highly homologous to Epstein–Barr virus Ori-Lyt: evolutionary conservation of transcriptional activation and replication signals. J. Virol. 1993;67(7):4006–4016. doi: 10.1128/jvi.67.7.4006-4016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stracker TH, Cassell GD, Ward P, et al. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 2004;78(1):441–453. doi: 10.1128/JVI.78.1.441-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glauser DL, Strasser R, Laimbacher AS, et al. Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction. J. Virol. 2007;81(9):4732–4743. doi: 10.1128/JVI.02476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matz B. Herpes simplex virus infection generates large tandemly reiterated simian virus 40 DNA molecules in a transformed hamster cell line. J. Virol. 1987;61(5):1427–1434. doi: 10.1128/jvi.61.5.1427-1434.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elias P, O’Donnell ME, Mocarski ES, Lehman IR. A DNA binding protein specific for an origin of replication of herpes simplex virus type 1. Proc. Natl Acad. Sci. USA. 1986;83(17):6322–6326. doi: 10.1073/pnas.83.17.6322.. ▪ Describes a study that led to the identification of UL9 as the herpes simplex virus (HSV) origin-binding protein.
- 49. Olivo PD, Nelson NJ, Challberg MD. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc. Natl Acad. Sci. USA. 1988;85(15):5414–5418. doi: 10.1073/pnas.85.15.5414.. ▪ Describes a study that led to the identification of UL9 as the herpes simplex virus (HSV) origin-binding protein.
- 50.Bruckner RC, Crute JJ, Dodson MS, Lehman IR. The herpes simplex virus 1 origin binding protein: a DNA helicase. J. Biol. Chem. 1991;266(4):2669–2674. [PubMed] [Google Scholar]
- 51.Fierer DS, Challberg MD. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J. Virol. 1992;66(7):3986–3995. doi: 10.1128/jvi.66.7.3986-3995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gustafsson CM, Hammarsten O, Falkenberg M, Elias P. Herpes simplex virus DNA replication: a spacer sequence directs the ATP-dependent formation of a nucleoprotein complex at OriS. Proc. Natl Acad. Sci. USA. 1994;91(11):4629–4633. doi: 10.1073/pnas.91.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koff A, Schwedes JF, Tegtmeyer P. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J. Virol. 1991;65(6):3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. The herpes simplex virus type 1 origin-binding protein carries out origin specific DNA unwinding and forms stem-loop structures. EMBO J. 1996;15(7):1742–1750. [PMC free article] [PubMed] [Google Scholar]
- 55.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol. 1996;258(5):789–799. doi: 10.1006/jmbi.1996.0287. [DOI] [PubMed] [Google Scholar]
- 56.Makhov AM, Lee SS, Lehman IR, Griffith JD. Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: visualization of a specific preunwinding complex. Proc. Natl Acad. Sci. USA. 2003;100(3):898–903. doi: 10.1073/pnas.0237171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colletti KS, Xu Y, Cei SA, Tarrant M, Pari GS. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of OriLyt-dependent DNA replication: evidence that IE2–UL84 and UL84–UL84 interactions are required for lytic DNA replication. J. Virol. 2004;78(17):9203–9214. doi: 10.1128/JVI.78.17.9203-9214.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colletti KS, Xu Y, Yamboliev I, Pari GS. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 2005;280(12):11955–11960. doi: 10.1074/jbc.C400603200. [DOI] [PubMed] [Google Scholar]
- 59. Colletti KS, Smallenburg KE, Xu Y, Pari GS. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of OriLyt. J. Virol. 2007;81(13):7077–7085. doi: 10.1128/JVI.00058-07.. ▪ Identifcation of a stem-loop RNA structure, bound by UL84, within an RNA/DNA hybrid region of cytomegalovirus OriLyt.
- 60.Xu Y, Cei SA, Rodriguez Huete A, Colletti KS, Pari GS. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within OriLyt. J. Virol. 2004;78(21):11664–11677. doi: 10.1128/JVI.78.21.11664-11677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lieberman PM, Berk AJ. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein–Barr virus Zta protein. J. Virol. 1990;64(6):2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990.. ▪ Describes the identification of Zta as a candidate origin-binding protein for EBV.
- 62. Schepers A, Pich D, Hammerschmidt W. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein–Barr virus. EMBO J. 1993;12(10):3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x.. ▪ Describes the identification of Zta as a candidate origin-binding protein for EBV.
- 63. Fixman ED, Hayward GS, Hayward SD. Replication of Epstein–Barr virus OriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 1995;69(5):2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995.. ▪ Describes the identification of Zta as a candidate origin-binding protein for EBV.
- 64.Gao Z, Krithivas A, Finan JE, et al. The Epstein–Barr virus lytic transactivator Zta interacts with the helicase–primase replication proteins. J. Virol. 1998;72(11):8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao G, Huang J, Fixman ED, Hayward SD. The Epstein–Barr virus replication protein BBLF2/3 provides an origin-tethering function through interaction with the zinc finger DNA binding protein ZBRK1 and the KAP-1 corepressor. J. Virol. 2005;79(1):245–256. doi: 10.1128/JVI.79.1.245-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao G, Wu FY, Hayward SD. Interaction with the Epstein–Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 2001;75(18):8792–8802. doi: 10.1128/JVI.75.18.8792-8802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q, Hong Y, Dorsky D, et al. Functional and physical interactions between the Epstein–Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J. Virol. 1996;70(8):5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang YN, Dong DL, Hayward GS, Hayward SD. The Epstein–Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 1990;64(7):3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Packham G, Economou A, Rooney CM, Rowe DT, Farrell PJ. Structure and function of the Epstein–Barr virus BZLF1 protein. J. Virol. 1990;64(5):2110–2116. doi: 10.1128/jvi.64.5.2110-2116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. Epstein–Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8(1):127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein–Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8(5):1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein–Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 1990;64(3):1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990.. ▪ Characterization of Zta binding sites within EBV OriLyt.
- 73.Flemington E, Speck SH. Epstein–Barr virus BZLF1 trans activator induces the promoter of a cellular cognate gene c-fos. J. Virol. 1990;64(9):4549–4552. doi: 10.1128/jvi.64.9.4549-4552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato H, Takeshita H, Furukawa M, Seiki M. Epstein–Barr virus BZLF1 transactivator is a negative regulator of Jun. J. Virol. 1992;66(8):4732–4736. doi: 10.1128/jvi.66.8.4732-4736.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li D, Qian L, Chen C, et al. Down-regulation of MHC class II expression through inhibition of CIITA transcription by lytic transactivator Zta during Epstein–Barr virus reactivation. J. Immunol. 2009;182(4):1799–1809. doi: 10.4049/jimmunol.0802686. [DOI] [PubMed] [Google Scholar]
- 76.Jones RJ, Dickerson S, Bhende PM, Delecluse HJ, Kenney SC. Epstein–Barr virus lytic infection induces retinoic acid-responsive genes through induction of a retinol-metabolizing enzyme, DHRS9. J. Biol. Chem. 2007;282(11):8317–8324. doi: 10.1074/jbc.M608667200. [DOI] [PubMed] [Google Scholar]
- 77.Chang Y, Lee HH, Chen YT, et al. Induction of the early growth response 1 gene by Epstein–Barr virus lytic transactivator Zta. J. Virol. 2006;80(15):7748–7755. doi: 10.1128/JVI.02608-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsu M, Wu SY, Chang SS, et al. Epstein–Barr virus lytic transactivator Zta enhances chemotactic activity through induction of interleukin-8 in nasopharyngeal carcinoma cells. J. Virol. 2008;82(7):3679–3688. doi: 10.1128/JVI.02301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahot S, Sergeant A, Drouet E, Gruffat H. A novel function for the Epstein–Barr virus transcription factor EB1/Zta: induction of transcription of the hIL-10 gene. J. Gen. Virol. 2003;84(Pt 4):965–974. doi: 10.1099/vir.0.18845-0. [DOI] [PubMed] [Google Scholar]
- 80.Hung CH, Liu ST. Characterization of the Epstein–Barr virus BALF2 promoter. J. Gen. Virol. 1999;80(Pt 10):2747–2750. doi: 10.1099/0022-1317-80-10-2747. [DOI] [PubMed] [Google Scholar]
- 81.Granato M, Farina A, Gonnella R, et al. Regulation of the expression of the Epstein–Barr virus early gene BFRF1. Virology. 2006;347(1):109–116. doi: 10.1016/j.virol.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 82.Quinlivan EB, Holley-Guthrie EA, Norris M, Gutsch D, Bachenheimer SL, Kenney SC. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein–Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21(14):1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dickerson SJ, Xing Y, Robinson AR, Seaman WT, Gruffat H, Kenney SC. Methylation-dependent binding of the Epstein–Barr virus BZLF1 protein to viral promoters. PLoS Pathog. 2009;5(3):e1000356. doi: 10.1371/journal.ppat.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Petosa C, Morand P, Baudin F, Moulin M, Artero JB, Muller CW. Structural basis of lytic cycle activation by the Epstein–Barr virus ZEBRA protein. Mol. Cell. 2006;21(4):565–572. doi: 10.1016/j.molcel.2006.01.006.. ▪▪ Describes the acquisition of the first partial crystal structure of Zta.
- 85.Morand P, Budayova-Spano M, Perrissin M, Muller CW, Petosa C. Expression, purification, crystallization and preliminary x-ray analysis of a C-terminal fragment of the Epstein–Barr virus ZEBRA protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62(Pt 3):210–214. doi: 10.1107/S1744309106002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sinclair AJ. bZIP proteins of human γ-herpesviruses. J. Gen. Virol. 2003;84(Pt 8):1941–1949. doi: 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- 87. Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 2004;36(10):1099–1104. doi: 10.1038/ng1424.. ▪ First to describe the preferential binding of Zta to methylated DNA.
- 88.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue186. J. Virol. 2005;79(12):7338–7348. doi: 10.1128/JVI.79.12.7338-7348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karlsson QH, Schelcher C, Verrall E, Petosa C, Sinclair AJ. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein–Barr virus lytic switch protein. PLoS Pathog. 2008;4(3):e1000005. doi: 10.1371/journal.ppat.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y, Tang Q, Maul GG, Yuan Y. Kaposi’s sarcoma-associated herpesvirus Ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 2006;80(24):12171–12186. doi: 10.1128/JVI.00990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kagele D, Gao Y, Smallenburg K, Pari GS. Interaction of HCMV UL84 with C/EBPα transcription factor binding sites within OriLyt is essential for lytic DNA replication. Virology. 2009;392(1):16–23. doi: 10.1016/j.virol.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu FY, Chen H, Wang SE, et al. CCAAT/enhancer binding protein α interacts with ZTA and mediates ZTA-induced p21(CIP-1) accumulation and G(1) cell cycle arrest during the Epstein–Barr virus lytic cycle. J. Virol. 2003;77(2):1481–1500. doi: 10.1128/JVI.77.2.1481-1500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cayrol C, Flemington E. G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein–Barr virus transactivator Zta. J. Biol. Chem. 1996;271(50):31799–31802. doi: 10.1074/jbc.271.50.31799. [DOI] [PubMed] [Google Scholar]
- 94.Cayrol C, Flemington EK. The Epstein–Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15(11):2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 95.Rodriguez A, Armstrong M, Dwyer D, Flemington E. Genetic dissection of cell growth arrest functions mediated by the Epstein–Barr virus lytic gene product, Zta. J. Virol. 1999;73(11):9029–9038. doi: 10.1128/jvi.73.11.9029-9038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu FY, Wang SE, Chen H, Wang L, Hayward SD, Hayward GS. CCAAT/enhancer binding protein α binds to the Epstein–Barr virus (EBV) ZTA protein through oligomeric interactions and contributes to cooperative transcriptional activation of the ZTA promoter through direct binding to the ZII and ZIIIB motifs during induction of the EBV lytic cycle. J. Virol. 2004;78(9):4847–4865. doi: 10.1128/JVI.78.9.4847-4865.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flemington EK. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 2001;75(10):4475–4481. doi: 10.1128/JVI.75.10.4475-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wu FY, Tang QQ, Chen H, et al. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-α. Proc. Natl Acad. Sci. USA. 2002;99(16):10683–10688. doi: 10.1073/pnas.162352299.. ▪ First to describe a role for C/EBP-α in herpesvirus lytic replication.
- 99.Izumiya Y, Izumiya C, Van Geelen A, et al. Kaposi’s sarcoma-associated herpesvirus-encoded protein kinase and its interaction with K-bZIP. J. Virol. 2007;81(3):1072–1082. doi: 10.1128/JVI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katano H, Ogawa-Goto K, Hasegawa H, Kurata T, Sata T. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology. 2001;286(2):446–455. doi: 10.1006/viro.2001.1005. [DOI] [PubMed] [Google Scholar]
- 101.Wu FY, Wang SE, Tang QQ, et al. Cell cycle arrest by Kaposi’s sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein α and p21(CIP-1) J. Virol. 2003;77(16):8893–8914. doi: 10.1128/JVI.77.16.8893-8914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang SE, Wu FY, Yu Y, Hayward GS. CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 2003;77(17):9590–9612. doi: 10.1128/JVI.77.17.9590-9612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liao W, Tang Y, Lin SF, Kung HJ, Giam CZ. K-bZIP of Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 2003;77(6):3809–3815. doi: 10.1128/JVI.77.6.3809-3815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rossetto C, Yamboliev I, Pari GS. Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 K-bZIP modulates LANA mediated suppression of lytic origin-dependent DNA synthesis. J. Virol. 2009;83(17):8492–8501. doi: 10.1128/JVI.00922-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lefort S, Flamand L. Kaposi’s sarcoma-associated herpesvirus K-bZIP protein is necessary for lytic viral gene expression, DNA replication, and virion production in primary effusion lymphoma cell lines. J. Virol. 2009;83(11):5869–5880. doi: 10.1128/JVI.01821-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin CL, Li H, Wang Y, Zhu FX, Kudchodkar S, Yuan Y. Kaposi’s sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 2003;77(10):5578–5588. doi: 10.1128/JVI.77.10.5578-5588.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang P, Day L, Dheekollu J, Lieberman PM. A redox-sensitive cysteine in Zta is required for Epstein–Barr virus lytic cycle DNA replication. J. Virol. 2005;79(21):13298–13309. doi: 10.1128/JVI.79.21.13298-13309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hwang S, Gwack Y, Byun H, Lim C, Choe J. The Kaposi’s sarcoma-associated herpesvirus K8 protein interacts with CREB-binding protein (CBP) and represses CBP-mediated transcription. J. Virol. 2001;75(19):9509–9516. doi: 10.1128/JVI.75.19.9509-9516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Al Mehairi S, Cerasoli E, Sinclair AJ. Investigation of the multimerization region of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) protein K-bZIP: the proposed leucine zipper region encodes a multimerization domain with an unusual structure. J. Virol. 2005;79(12):7905–7910. doi: 10.1128/JVI.79.12.7905-7910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rossetto C, Gao Y, Yamboliev I, Papouskova I, Pari G. Transcriptional repression of K-Rta by Kaposi’s sarcoma-associated herpesvirus K-bZIP is not required for OriLyt-dependent DNA replication. Virology. 2007;369(2):340–350. doi: 10.1016/j.virol.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.AuCoin DP, Colletti KS, Cei SA, Papouskova I, Tarrant M, Pari GS. Amplification of the Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP) Virology. 2004;318(2):542–555. doi: 10.1016/j.virol.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 112.Wang Y, Li H, Chan MY, Zhu FX, Lukac DM, Yuan Y. Kaposi’s sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 2004;78(16):8615–8629. doi: 10.1128/JVI.78.16.8615-8629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Stow ND. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x.. ▪▪ Describes identification of the first herpesvirus lytic origins.
- 114. Stow ND, McMonagle EC. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1.. ▪▪ Describes identification of the first herpesvirus lytic origins.
- 115. Weller SK, Spadaro A, Schaffer JE, Murray AW, Maxam AM, Schaffer PA. Cloning, sequencing, and functional analysis of OriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol. Cell. Biol. 1985;5(5):930–942. doi: 10.1128/mcb.5.5.930.. ▪▪ Describes identification of the first herpesvirus lytic origins.
- 116.Balliet JW, Min JC, Cabatingan MS, Schaffer PA. Site-directed mutagenesis of large DNA palindromes: construction and in vitro characterization of herpes simplex virus type 1 mutants containing point mutations that eliminate the OriL or OriS initiation function. J. Virol. 2005;79(20):12783–12797. doi: 10.1128/JVI.79.20.12783-12797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deb S, Doelberg M. A 67-base-pair segment from the Ori-S region of herpes simplex virus type 1 encodes origin function. J. Virol. 1988;62(7):2516–2519. doi: 10.1128/jvi.62.7.2516-2519.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lockshon D, Galloway DA. Sequence and structural requirements of a herpes simplex viral DNA replication origin. Mol. Cell. Biol. 1988;8(10):4018–4027. doi: 10.1128/mcb.8.10.4018.. ▪ Identification of the critical elements within the HSV lytic origins.
- 119.Koff A, Tegtmeyer P. Characterization of major recognition sequences for a herpes simplex virus type 1 origin-binding protein. J. Virol. 1988;62(11):4096–4103. doi: 10.1128/jvi.62.11.4096-4103.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weir HM, Stow ND. Two binding sites for the herpes simplex virus type 1 UL9 protein are required for efficient activity of the oriS replication origin. J. Gen. Virol. 1990;71(Pt 6):1379–1385. doi: 10.1099/0022-1317-71-6-1379. [DOI] [PubMed] [Google Scholar]
- 121. Hammerschmidt W, Sugden B. Identification and characterization of OriLyt, a lytic origin of DNA replication of Epstein–Barr virus. Cell. 1988;55(3):427–433. doi: 10.1016/0092-8674(88)90028-1.. ▪ Describes the identification of the EBV OriLyt.
- 122. Schepers A, Pich D, Mankertz J, Hammerschmidt W. cis-acting elements in the lytic origin of DNA replication of Epstein–Barr virus. J. Virol. 1993;67(7):4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993.. ▪ Describes the identification of the EBV OriLyt.
- 123.Schepers A, Pich D, Hammerschmidt W. Activation of OriLyt, the lytic origin of DNA replication of Epstein–Barr virus, by BZLF1. Virology. 1996;220(2):367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 124.Baumann M, Feederle R, Kremmer E, Hammerschmidt W. Cellular transcription factors recruit viral replication proteins to activate the Epstein–Barr virus origin of lytic DNA replication, OriLyt. EMBO J. 1999;18(21):6095–6105. doi: 10.1093/emboj/18.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gruffat H, Renner O, Pich D, Hammerschmidt W. Cellular proteins bind to the downstream component of the lytic origin of DNA replication of Epstein–Barr virus. J. Virol. 1995;69(3):1878–1886. doi: 10.1128/jvi.69.3.1878-1886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Portes-Sentis S, Sergeant A, Gruffat H. A particular DNA structure is required for the function of a cis-acting component of the Epstein–Barr virus OriLyt origin of replication. Nucleic Acids Res. 1997;25(7):1347–1354. doi: 10.1093/nar/25.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein–Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 1990;18(23):6835–6843. doi: 10.1093/nar/18.23.6835.. ▪ Characterization of Rta as a lytic origin-binding protein.
- 128.AuCoin DP, Colletti KS, Xu Y, Cei SA, Pari GS. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 2002;76(15):7890–7896. doi: 10.1128/JVI.76.15.7890-7896.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lukac DM, Yuan Y. Reactivation and lytic replication of KSHV. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., editors. Human Herpesviruses. NY, USA: Cambridge University Press; 2007. pp. 434–460. [PubMed] [Google Scholar]
- 130.Xu Y, Rodriguez-Huete A, Pari GS. Evaluation of the lytic origins of replication of Kaposi’s sarcoma-associated virus/human herpesvirus 8 in the context of the viral genome. J. Virol. 2006;80(19):9905–9909. doi: 10.1128/JVI.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gong D, Qi J, Arumugaswami V, Sun R, Deng H. Identification and functional characterization of the left origin of lytic replication of murine γ-herpesvirus 68. Virology. 2009;387(2):285–295. doi: 10.1016/j.virol.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Deng H, Chu JT, Park NH, Sun R. Identification of cis sequences required for lytic DNA replication and packaging of murine γ-herpesvirus 68. J. Virol. 2004;78(17):9123–9131. doi: 10.1128/JVI.78.17.9123-9131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Flach B, Steer B, Thakur NN, Haas J, Adler H. The M10 locus of murine γ-herpesvirus 68 contributes to both the lytic and latent phase of infection. J. Virol. 2009 doi: 10.1128/JVI.00629-09. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hamzeh FM, Lietman PS, Gibson W, Hayward GS. Identification of the lytic origin of DNA replication in human cytomegalovirus by a novel approach utilizing ganciclovir-induced chain termination. J. Virol. 1990;64(12):6184–6195. doi: 10.1128/jvi.64.12.6184-6195.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Anders DG, Kacica MA, Pari G, Punturieri SM. Boundaries and structure of human cytomegalovirus OriLyt, a complex origin for lytic-phase DNA replication. J. Virol. 1992;66(6):3373–3384. doi: 10.1128/jvi.66.6.3373-3384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Borst EM, Messerle M. Analysis of human cytomegalovirus OriLyt sequence requirements in the context of the viral genome. J. Virol. 2005;79(6):3615–3626. doi: 10.1128/JVI.79.6.3615-3626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu Y, Huang L, Anders DG. Human cytomegalovirus OriLyt sequence requirements. J. Virol. 1998;72(6):4989–4996. doi: 10.1128/jvi.72.6.4989-4996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Anders DG, Punturieri SM. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J. Virol. 1991;65(2):931–937. doi: 10.1128/jvi.65.2.931-937.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Masse MJ, Karlin S, Schachtel GA, Mocarski ES. Human cytomegalovirus origin of DNA replication (OriLyt) resides within a highly complex repetitive region. Proc. Natl Acad. Sci. USA. 1992;89(12):5246–5250. doi: 10.1073/pnas.89.12.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Anders DG, Kerry JA, Pari GS. DNA synthesis and late viral gene expression. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., editors. Human Herpesviruses. NY, USA: Cambridge University Press; 2007. pp. 295–310. [PubMed] [Google Scholar]
- 141.Huang L, Zhu Y, Anders DG. The variable 3μ ends of a human cytomegalovirus OriLyt transcript (SRT) overlap an essential, conserved replicator element. J. Virol. 1996;70(8):5272–5281. doi: 10.1128/jvi.70.8.5272-5281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Prichard MN, Jairath S, Penfold ME, St Jeor S, Bohlman MC, Pari GS. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J. Virol. 1998;72(9):6997–7004. doi: 10.1128/jvi.72.9.6997-7004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Malik AK, Weller SK. Use of transdominant mutants of the origin-binding protein (UL9) of herpes simplex virus type 1 to define functional domains. J. Virol. 1996;70(11):7859–7866. doi: 10.1128/jvi.70.11.7859-7866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marintcheva B, Weller SK. Residues within the conserved helicase motifs of UL9, the origin-binding protein of herpes simplex virus-1, are essential for helicase activity but not for dimerization or origin binding activity. J. Biol. Chem. 2001;276(9):6605–6615. doi: 10.1074/jbc.M007743200. [DOI] [PubMed] [Google Scholar]
- 145.Marintcheva B, Weller SK. Existence of transdominant and potentiating mutants of UL9, the herpes simplex virus type 1 origin-binding protein, suggests that levels of UL9 protein may be regulated during infection. J. Virol. 2003;77(17):9639–9651. doi: 10.1128/JVI.77.17.9639-9651.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chattopadhyay S, Weller SK. DNA binding activity of the herpes simplex virus type 1 origin binding protein, UL9, can be modulated by sequences in the N terminus: correlation between transdominance and DNA binding. J. Virol. 2006;80(9):4491–4500. doi: 10.1128/JVI.80.9.4491-4500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee SS, Lehman IR. Unwinding of the box I element of a herpes simplex virus type 1 origin by a complex of the viral origin binding protein, single-strand DNA binding protein, and single-stranded DNA. Proc. Natl Acad. Sci. USA. 1997;94(7):2838–2842. doi: 10.1073/pnas.94.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.He X, Lehman IR. Unwinding of a herpes simplex virus type 1 origin of replication (Ori(S)) by a complex of the viral origin binding protein and the single-stranded DNA binding protein. J. Virol. 2000;74(12):5726–5728. doi: 10.1128/jvi.74.12.5726-5728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Elias P, Gustafsson CM, Hammarsten O. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication OriS. J. Biol. Chem. 1990;265(28):17167–17173. [PubMed] [Google Scholar]
- 150.Aslani A, Simonsson S, Elias P. A novel conformation of the herpes simplex virus origin of DNA replication recognized by the origin binding protein. J. Biol. Chem. 2000;275(8):5880–5887. doi: 10.1074/jbc.275.8.5880. [DOI] [PubMed] [Google Scholar]
- 151.Macao B, Olsson M, Elias P. Functional properties of the herpes simplex virus type I origin-binding protein are controlled by precise interactions with the activated form of the origin of DNA replication. J. Biol. Chem. 2004;279(28):29211–29217. doi: 10.1074/jbc.M400371200. [DOI] [PubMed] [Google Scholar]
- 152. Aslani A, Macao B, Simonsson S, Elias P. Complementary intrastrand base pairing during initiation of herpes simplex virus type 1 DNA replication. Proc. Natl Acad. Sci. USA. 2001;98(13):7194–7199. doi: 10.1073/pnas.121177198.. ▪ Identification of a critical DNA hairpin formed at an inverted repeat within the essential region of HSV OriLyt.
- 153.Aslani A, Olsson M, Elias P. ATP-dependent unwinding of a minimal origin of DNA replication by the origin-binding protein and the single-strand DNA-binding protein ICP8 from herpes simplex virus type I. J. Biol. Chem. 2002;277(43):41204–41212. doi: 10.1074/jbc.M208270200. [DOI] [PubMed] [Google Scholar]
- 154.Olsson M, Tang KW, Persson C, Wilhelmsson LM, Billeter M, Elias P. Stepwise evolution of the herpes simplex virus origin binding protein and origin of replication. J. Biol. Chem. 2009;284(24):16246–16255. doi: 10.1074/jbc.M807551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marintcheva B, Weller SK. Helicase motif Ia is involved in single-strand DNA-binding and helicase activities of the herpes simplex virus type 1 origin-binding protein, UL9. J. Virol. 2003;77(4):2477–2488. doi: 10.1128/JVI.77.4.2477-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tsurumi T, Kobayashi A, Tamai K, et al. Epstein–Barr virus single-stranded DNA-binding protein: purification, characterization, and action on DNA synthesis by the viral DNA polymerase. Virology. 1996;222(2):352–364. doi: 10.1006/viro.1996.0432. [DOI] [PubMed] [Google Scholar]
- 157.Quinn JP, McGeoch DJ. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985;13(22):8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Mapelli M, Panjikar S, Tucker PA. The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding. J. Biol. Chem. 2005;280(4):2990–2997. doi: 10.1074/jbc.M406780200.. ▪▪ Crystal structure of the herpesvirus ssDNA-binding protien.
- 159.Mumtsidu E, Makhov AM, Konarev PV, Svergun DI, Griffith JD, Tucker PA. Structural features of the single-stranded DNA-binding protein of Epstein–Barr virus. J. Struct. Biol. 2008;161(2):172–187. doi: 10.1016/j.jsb.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 160.Gupte SS, Olson JW, Ruyechan WT. The major herpes simplex virus type-1 DNA-binding protein is a zinc metalloprotein. J. Biol. Chem. 1991;266(18):11413–11416. [PubMed] [Google Scholar]
- 161.Lee CK, Knipe DM. An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J. Virol. 1985;54(3):731–738. doi: 10.1128/jvi.54.3.731-738.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boehmer PE, Craigie MC, Stow ND, Lehman IR. Association of origin binding protein and single strand DNA-binding protein, ICP8, during herpes simplex virus type 1 DNA replication in vivo. J. Biol. Chem. 1994;269(46):29329–29334. [PubMed] [Google Scholar]
- 163.Boehmer PE, Lehman IR. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc. Natl Acad. Sci. USA. 1993;90(18):8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.He X, Lehman IR. An initial ATP-independent step in the unwinding of a herpes simplex virus type I origin of replication by a complex of the viral origin-binding protein and single-strand DNA-binding protein. Proc. Natl Acad. Sci. USA. 2001;98(6):3024–3028. doi: 10.1073/pnas.061028298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Reuven NB, Staire AE, Myers RS, Weller SK. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 2003;77(13):7425–7433. doi: 10.1128/JVI.77.13.7425-7433.2003.. ▪ Suggests a similarity between herpesvirus replication proteins and the λ RED recombination machinery.
- 166.Boehmer PE, Lehman IR. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J. Virol. 1993;67(2):711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bortner C, Hernandez TR, Lehman IR, Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J. Mol. Biol. 1993;231(2):241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 168.Dutch RE, Bruckner RC, Mocarski ES, Lehman IR. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J. Virol. 1992;66(1):277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nimonkar AV, Boehmer PE. The herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) promotes strand invasion. J. Biol. Chem. 2003;278(11):9678–9682. doi: 10.1074/jbc.m212555200. [DOI] [PubMed] [Google Scholar]
- 170.Nimonkar AV, Boehmer PE. On the mechanism of strand assimilation by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) Nucleic Acids Res. 2003;31(18):5275–5281. doi: 10.1093/nar/gkg740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nimonkar AV, Boehmer PE. Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1. Proc. Natl Acad. Sci. USA. 2003;100(18):10201–10206. doi: 10.1073/pnas.1534569100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dutch RE, Lehman IR. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J. Virol. 1993;67(12):6945–6949. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tsurumi T, Kishore J, Yokoyama N, et al. Overexpression, purification and helix-destabilizing properties of Epstein–Barr virus ssDNA-binding protein. J. Gen. Virol. 1998;79(Pt 5):1257–1264. doi: 10.1099/0022-1317-79-5-1257. [DOI] [PubMed] [Google Scholar]
- 174.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 175.Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 176.Boehmer PE, Nimonkar AV. Herpes virus replication. IUBMB Life. 2003;55(1):13–22. doi: 10.1080/1521654031000070645. [DOI] [PubMed] [Google Scholar]
- 177.Lehman IR, Boehmer PE. Replication of herpes simplex virus DNA. J. Biol. Chem. 1999;274(40):28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 178.Strang BL, Stow ND. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 2005;79(19):12487–12494. doi: 10.1128/JVI.79.19.12487-12494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.LaFemina RL, Hayward GS. Replicative forms of human cytomegalovirus DNA with joined termini are found in permissively infected human cells but not in non-permissive Balb/c-3T3 mouse cells. J. Gen. Virol. 1983;64(Pt 2):373–389. doi: 10.1099/0022-1317-64-2-373. [DOI] [PubMed] [Google Scholar]
- 180.Marks JR, Spector DH. Fusion of the termini of the murine cytomegalovirus genome after infection. J. Virol. 1984;52(1):24–28. doi: 10.1128/jvi.52.1.24-28.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McVoy MA, Adler SP. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 1994;68(2):1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sato H, Takimoto T, Tanaka S, Tanaka J, Raab-Traub N. Concatameric replication of Epstein–Barr virus: structure of the termini in virus-producer and newly transformed cell lines. J. Virol. 1990;64(11):5295–5300. doi: 10.1128/jvi.64.11.5295-5300.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cho MS, Tran VM. A concatenated form of Epstein–Barr viral DNA in l ymphoblastoid cell lines induced by transfection with BZLF1. Virology. 1993;194(2):838–842. doi: 10.1006/viro.1993.1327. [DOI] [PubMed] [Google Scholar]
- 184.Garber DA, Beverley SM, Coen DM. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197(1):459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 185.Ben-Porat T, Kaplan AS, Stehn B, Rubenstein AS. Concatemeric forms of intracellular herpesvirus DNA. Virology. 1976;69(2):547–560. doi: 10.1016/0042-6822(76)90484-0. [DOI] [PubMed] [Google Scholar]
- 186.Severini A, Morgan AR, Tovell DR, Tyrrell DL. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200(2):428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 187.Jacob RJ, Morse LS, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 1979;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jongeneel CV, Bachenheimer SL. Structure of replicating herpes simplex virus DNA. J. Virol. 1981;39(2):656–660. doi: 10.1128/jvi.39.2.656-660.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ben-Porat T, Tokazewski SA. Replication of herpesvirus DNA. II. Sedimentation characteristics of newly synthesized DNA. Virology. 1977;79(2):292–301. doi: 10.1016/0042-6822(77)90356-7. [DOI] [PubMed] [Google Scholar]
- 190.Hirsch I, Roubal J, Vonka V. Replicating DNA of herpes simplex virus type 1. Intervirology. 1976;7(3):155–175. doi: 10.1159/000149949. [DOI] [PubMed] [Google Scholar]
- 191.Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 1985;53(2):587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Skaliter R, Lehman IR. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc. Natl Acad. Sci. USA. 1994;91(22):10665–10669. doi: 10.1073/pnas.91.22.10665.. ▪▪ Reconstitution of rolling-circle replication by herpesvirus core lytic replication proteins in vitro.
- 193.Skaliter R, Makhov AM, Griffith JD, Lehman IR. Rolling circle DNA replication by extracts of herpes simplex virus type 1-infected human cells. J. Virol. 1996;70(2):1132–1136. doi: 10.1128/jvi.70.2.1132-1136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 1996;70(5):3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J. Virol. 1977;23(2):394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pfuller R, Hammerschmidt W. Plasmid-like replicative intermediates of the Epstein–Barr virus lytic origin of DNA replication. J. Virol. 1996;70(6):3423–3431. doi: 10.1128/jvi.70.6.3423-3431.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;203(2):384–388. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- 198.Honess RW, Buchan A, Halliburton IW, Watson DH. Recombination and linkage between structural and regulatory genes of herpes simplex virus type 1: study of the functional organization of the genome. J. Virol. 1980;34(3):716–742. doi: 10.1128/jvi.34.3.716-742.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Umene K. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 1985;66(Pt 12):2659–2670. doi: 10.1099/0022-1317-66-12-2659. [DOI] [PubMed] [Google Scholar]
- 200.Brown SM, Subak-Sharpe JH, Harland J, MacLean AR. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 1992;73(Pt 2):293–301. doi: 10.1099/0022-1317-73-2-293. [DOI] [PubMed] [Google Scholar]
- 201.Poffenberger KL, Tabares E, Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc. Natl Acad. Sci. USA. 1983;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jenkins FJ, Roizman B. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. J. Virol. 1986;59(2):494–499. doi: 10.1128/jvi.59.2.494-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mocarski ES, Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc. Natl Acad. Sci. USA. 1982;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zimmermann J, Hammerschmidt W. Structure and role of the terminal repeats of Epstein–Barr virus in processing and packaging of virion DNA. J. Virol. 1995;69(5):3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dutch RE, Bianchi V, Lehman IR. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J. Virol. 1995;69(5):3084–3089. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bataille D, Epstein AL. Herpes simplex virus type 1 replication and recombination. Biochimie. 1995;77(10):787–795. doi: 10.1016/0300-9084(96)88197-1. [DOI] [PubMed] [Google Scholar]
- 207. Weber PC, Challberg MD, Nelson NJ, Levine M, Glorioso JC. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0.. ▪▪ Demonstrates the nonspecific recombination potential of herpesvirus core lytic-replication proteins.
- 208.Weber PC, Levine M, Glorioso JC. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J. Virol. 1990;64(1):300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.McVoy MA, Ramnarain D. Machinery to support genome segment inversion exists in a herpesvirus which does not naturally contain invertible elements. J. Virol. 2000;74(10):4882–4887. doi: 10.1128/jvi.74.10.4882-4887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zeng Y, Middeldorp J, Madjar JJ, Ooka T. A major DNA binding protein encoded by BALF2 open reading frame of Epstein–Barr virus (EBV) forms a complex with other EBV DNA-binding proteins: DNAase, EA-D, and DNA polymerase. Virology. 1997;239(2):285–295. doi: 10.1006/viro.1997.8891. [DOI] [PubMed] [Google Scholar]
- 211.Buisson M, Geoui T, Flot D, et al. A bridge crosses the active-site canyon of the Epstein–Barr virus nuclease with DNase and RNase activities. J. Mol. Biol. 2009 doi: 10.1016/j.jmb.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 212.Stolzenberg MC, Ooka T. Purification and properties of Epstein–Barr virus DNase expressed in Escherichia coli. J. Virol. 1990;64(1):96–104. doi: 10.1128/jvi.64.1.96-104.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu MT, Hu HP, Hsu TY, Chen JY. Site-directed mutagenesis in a conserved motif of Epstein–Barr virus DNase that is homologous to the catalytic centre of type II restriction endonucleases. J. Gen. Virol. 2003;84(Pt 3):677–686. doi: 10.1099/vir.0.18739-0. [DOI] [PubMed] [Google Scholar]
- 214.Lin SF, Hsu TY, Liu MY, et al. Characterization of Epstein–Barr virus DNase and its interaction with the major DNA binding protein. Virology. 1995;208(2):712–722. doi: 10.1006/viro.1995.1203. [DOI] [PubMed] [Google Scholar]
- 215.Feederle R, Bannert H, Lips H, Muller-Lantzsch N, Delecluse HJ. The Epstein–Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 2009;83(10):4952–4962. doi: 10.1128/JVI.00170-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Weitzman MD, Carson CT, Schwartz RA, Lilley CE. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst.) 2004;3(8–9):1165–1173. doi: 10.1016/j.dnarep.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 217.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15(3):119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 218.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55(8):451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 219.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl Acad. Sci. USA. 2005;102(16):5844–5849. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Shirata N, Kudoh A, Daikoku T, et al. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 2005;280(34):30336–30341. doi: 10.1074/jbc.M500976200. [DOI] [PubMed] [Google Scholar]
- 221. Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 2004;78(9):4783–4796. doi: 10.1128/JVI.78.9.4783-4796.2004.. ▪▪ Cellular recombination/repair proteins implicated in herpesvirus replication.
- 222.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 2004;78(11):5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Murata LB, Dodson MS, Hall JD. A human cellular protein activity (OF-1), which binds herpes simplex virus type 1 origin, contains the Ku70/Ku80 heterodimer. J. Virol. 2004;78(14):7839–7842. doi: 10.1128/JVI.78.14.7839-7842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 1999;73(1):650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Li H, Baskaran R, Krisky DM, et al. Chk2 is required for HSV-1 ICP0-mediated G2/M arrest and enhancement of virus growth. Virology. 2008;375(1):13–23. doi: 10.1016/j.virol.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wilkinson DE, Weller SK. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 2006;119(Pt 13):2695–2703. doi: 10.1242/jcs.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Luo MH, Rosenke K, Czornak K, Fortunato EA. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 2007;81(4):1934–1950. doi: 10.1128/JVI.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Casavant NC, Luo MH, Rosenke K, Winegardner T, Zurawska A, Fortunato EA. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 2006;80(17):8390–8401. doi: 10.1128/JVI.00505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Kudoh A, Fujita M, Zhang L, et al. Epstein–Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 2005;280(9):8156–8163. doi: 10.1074/jbc.M411405200.. ▪ Demonstrates that EBV activates the ATM DNA damage response pathway.
- 230. Kudoh A, Iwahori S, Sato Y, et al. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein–Barr virus replication compartments. J. Virol. 2009;83(13):6641–6651. doi: 10.1128/JVI.00049-09.. ▪ Demonstrates that cellular recombination proteins are recruited to EBV replication compartments.
- 231.Wiedmer A, Wang P, Zhou J, et al. Epstein–Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J. Virol. 2008;82(9):4647–4655. doi: 10.1128/JVI.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bailey SG, Verrall E, Schelcher C, Rhie A, Doherty AJ, Sinclair AJ. Functional interaction between Epstein–Barr virus replication protein Zta and host DNA-damage response protein 53BP1. J. Virol. 2009;83(21):11116–11122. doi: 10.1128/JVI.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Toita N, Hatano N, Ono S, et al. Epstein–Barr virus-associated B-cell lymphoma in a patient with DNA ligase IV (LIG4) syndrome. Am. J. Med. Genet. A. 2007;143(7):742–745. doi: 10.1002/ajmg.a.31644.. ▪ EBV implicated as a causative agent in the formation of lymphomas in patients with DNA ligsase IV syndrome.
- 234. Daikoku T, Kudoh A, Sugaya Y, et al. Postreplicative mismatch repair factors are recruited to Epstein–Barr virus replication compartments. J. Biol. Chem. 2006;281(16):11422–11430. doi: 10.1074/jbc.M510314200.. ▪ Demonstrates that cellular mismatch repair proteins are recruited to EBV replication compartments.
- 235.Wang P, Rennekamp AJ, Yuan Y, Lieberman PM. Topoisomerase I and RecQL1 function in Epstein–Barr virus lytic reactivation. J. Virol. 2009;83(16):8090–8098. doi: 10.1128/JVI.02379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lu CC, Chen YC, Wang JT, Yang PW, Chen MR. Xeroderma pigmentosum C is involved in Epstein Barr virus DNA replication. J. Gen. Virol. 2007;88(Pt 12):3234–3243. doi: 10.1099/vir.0.83212-0. [DOI] [PubMed] [Google Scholar]
- 237.Gershburg E, Raffa S, Torrisi MR, Pagano JS. Epstein–Barr virus-encoded protein kinase (BGLF4) is involved in production of infectious virus. J. Virol. 2007;81(10):5407–5412. doi: 10.1128/JVI.02398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tarakanova VL, Leung-Pineda V, Hwang S, et al. γ-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe. 2007;1(4):275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Liang X, Pickering MT, Cho NH, et al. Deregulation of DNA damage signal transduction by herpesvirus latency-associated M2. J. Virol. 2006;80(12):5862–5874. doi: 10.1128/JVI.02732-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89(6–7):744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lee HR, Toth Z, Shin YC, et al. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J. Virol. 2009;83(13):6739–6747. doi: 10.1128/JVI.02353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang Y, Li H, Tang Q, Maul GG, Yuan Y. Kaposi’s sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J. Virol. 2008;82(6):2867–2882. doi: 10.1128/JVI.01319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Quadt I, Gunther AK, Voss D, Schelhaas M, Knebel-Morsdorf D. TATA-binding protein and TBP-associated factors during herpes simplex virus type 1 infection: localization at viral DNA replication sites. Virus Res. 2006;115(2):207–213. doi: 10.1016/j.virusres.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 244.Rice SA, Long MC, Lam V, Spencer CA. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 1994;68(2):988–1001. doi: 10.1128/jvi.68.2.988-1001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Leopardi R, Ward PL, Ogle WO, Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, viral DNA requires posttranslational modification by the U(L)13 proteinkinase. J. Virol. 1997;71(2):1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kohzaki H, Murakami Y. Transcription factors and DNA replication origin selection. Bioessays. 2005;27(11):1107–1116. doi: 10.1002/bies.20316. [DOI] [PubMed] [Google Scholar]
- 247.Khalil MI, Hay J, Ruyechan WT. Cellular transcription factors Sp1 and Sp3 suppress varicella-zoster virus origin-dependent DNA replication. J. Virol. 2008;82(23):11723–11733. doi: 10.1128/JVI.01322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lieberman PM, Hardwick JM, Hayward SD. Responsiveness of the Epstein–Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J. Virol. 1989;63(7):3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hubenthal-Voss J, Starr L, Roizman B. The herpes simplex virus origins of DNA synthesis in the S component are each contained in a transcribed open reading frame. J. Virol. 1987;61(11):3349–3355. doi: 10.1128/jvi.61.11.3349-3355.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Voss JH, Roizman B. Properties of two 5′-coterminal RNAs transcribed part way and across the S component origin of DNA synthesis of the herpes simplex virus 1 genome. Proc. Natl Acad. Sci. USA. 1988;85(22):8454–8458. doi: 10.1073/pnas.85.22.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Xue SA, Griffin BE. Complexities associated with expression of Epstein–Barr virus (EBV) lytic origins of DNA replication. Nucleic Acids Res. 2007;35(10):3391–3406. doi: 10.1093/nar/gkm170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lee DY, Clayton DA. Properties of a primer RNA–DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J. Biol. Chem. 1996;271(39):24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 253.Norseen J, Johnson FB, Lieberman PM. A role for G-quadruplex RNA binding by Epstein–Barr virus nuclear antigen 1 (EBNA1) in DNA replication and metaphase chromosome attachment. J. Virol. 2009;83(20):10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM. RNA-dependent recruitment of the origin recognition complex. EMBO J. 2008;27(22):3024–3035. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Asai R, Kato A, Kato K, et al. Epstein–Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J. Virol. 2006;80(11):5125–5134. doi: 10.1128/JVI.02674-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang JT, Yang PW, Lee CP, Han CH, Tsai CH, Chen MR. Detection of Epstein–Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 2005;86(Pt 12):3215–3225. doi: 10.1099/vir.0.81313-0. [DOI] [PubMed] [Google Scholar]
- 257.Kudoh A, Daikoku T, Ishimi Y, et al. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4–MCM6–MCM7 complex during Epstein–Barr virus productive replication. J. Virol. 2006;80(20):10064–10072. doi: 10.1128/JVI.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Tsurumi T. In vivo dynamics of EBNA1–OriP interaction during latent and lytic replication of Epstein–Barr virus. J. Biol. Chem. 2004;279(52):54817–54825. doi: 10.1074/jbc.M405911200. [DOI] [PubMed] [Google Scholar]
- 259.Zhu J, Liao G, Shan L, et al. Protein array identification of substrates of the Epstein–Barr virus protein kinase BGLF4. J. Virol. 2009;83(10):5219–5231. doi: 10.1128/JVI.02378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lu CC, Huang HT, Wang JT, et al. Characterization of the uracil-DNA glycosylase activity of Epstein–Barr virus BKRF3 and its role in lytic viral DNA replication. J. Virol. 2007;81(3):1195–1208. doi: 10.1128/JVI.01518-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pyles RB, Thompson RL. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 1994;68(8):4963–4972. doi: 10.1128/jvi.68.8.4963-4972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Courcelle CT, Courcelle J, Prichard MN, Mocarski ES. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J. Virol. 2001;75(16):7592–7601. doi: 10.1128/JVI.75.16.7592-7601.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Focher F, Verri A, Verzeletti S, Mazzarello P, Spadari S. Uracil in OriS of herpes simplex 1 alters its specific recognition by origin binding protein (OBP): does virus induced uracil-DNA glycosylase play a key role in viral reactivation and replication? Chromosoma. 1992;102(1 Suppl.):S67–S71. doi: 10.1007/BF02451788. [DOI] [PubMed] [Google Scholar]
- 264.Bogani F, Chua CN, Boehmer PE. Reconstitution of uracil DNA glycosylase-initiated base excision repair in herpes simplex virus-1. J. Biol. Chem. 2009;284(25):16784–16790. doi: 10.1074/jbc.M109.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Prichard MN, Lawlor H, Duke GM, et al. Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA. Virol. J. 2005;2:55. doi: 10.1186/1743-422X-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ranneberg-Nilsen T, Dale HA, Luna L, et al. Characterization of human cytomegalovirus uracil DNA glycosylase (UL114) and its interaction with polymerase processivity factor (UL44) J. Mol. Biol. 2008;381(2):276–288. doi: 10.1016/j.jmb.2008.05.028. [DOI] [PubMed] [Google Scholar]