Abstract
Breast cancers are highly heterogeneous but can be grouped into subtypes based on several criteria, including level of expression of certain markers. Claudin-low breast cancer (CLBC) is associated with early metastasis and resistance to chemotherapy, while gene profiling indicates it is characterized by the expression of markers of epithelial-mesenchymal transition (EMT) — a phenotypic conversion linked with metastasis. Although the epigenetic program controlling the phenotypic and cellular plasticity of EMT remains unclear, one contributor may be methylation of the E-cadherin promoter, resulting in decreased E-cadherin expression, a hallmark of EMT. Indeed, reduced E-cadherin often occurs in CLBC and may contribute to the early metastasis and poor patient survival associated with this disease. Here, we have determined that methylation of histone H3 on lysine 9 (H3K9me2) is critical for promoter DNA methylation of E-cadherin in three TGF-β–induced EMT model cell lines, as well as in CLBC cell lines. Further, Snail interacted with G9a, a major euchromatin methyltransferase responsible for H3K9me2, and recruited G9a and DNA methyltransferases to the E-cadherin promoter for DNA methylation. Knockdown of G9a restored E-cadherin expression by suppressing H3K9me2 and blocking DNA methylation. This resulted in inhibition of cell migration and invasion in vitro and suppression of tumor growth and lung colonization in in vivo models of CLBC metastasis. Our study not only reveals a critical mechanism underlying the epigenetic regulation of EMT but also paves a way for the development of new treatment strategies for CLBC.
Introduction
Human breast cancer is a heterogeneous disease with respect to molecular alterations, incidence, survival, and response to therapy. Based on gene expression profiling, 6 different subtypes of breast cancer have been established, including luminal A, luminal B, HER2-enriched, basal-like, claudin-low, and normal breast-like groups (1–4). Claudin-low breast cancer (CLBC), which was identified in 2007 and initially grouped into basal-like subtype (5), has drawn great attention recently, as it is believed to originate from mammary stem cells (MSCs) and has poor clinical outcome associated with early metastasis and resistance to chemotherapy (2, 4). This distinct subtype is characterized by the expression of EMT markers and stem cell–associated genes and has low expression of the tight junction protein claudin and adherens junction molecule E-cadherin (2, 4).
EMT is an essential phenotypic conversion during embryonic development, tissue remodeling, wound healing, and metastasis (6–8). In these EMT processes, epithelial cells acquire fibroblast-like properties and exhibit reduced intercellular adhesion and increased motility. In addition, cells in which EMT is activated have stem cell–like features, which provides a distinct advantage in tumor progression and metastasis (9, 10). EMT is a dynamic and reversible process that mainly occurs at the invasive front of the tumor and is provoked by signals that cells receive from their microenvironment (11–13), such as TGF-β, Wnt, and TNF-α (14, 15). When cancer cells disseminate to distant sites of the body, they no longer encounter the signals that they received in the primary tumor, and they can revert to an epithelial state via mesenchymal-epithelial transition (MET) (7). The ability of EMT to convert cells from one state of differentiation to another suggests that EMT activates an epigenetic program to block MSC differentiation toward epithelial or luminal lineage in CLBC, resulting in the poor clinical outcome of this disease.
This phenotypic and cellular plasticity of EMT is determined by a unique gene expression pattern and can be memorized and passed on to daughter cells by epigenetic mechanisms through DNA methylation and histone modifications (16–18). DNA methylation, which commonly associates with gene repression and formation of heterochromatin, is defined by the addition of a methyl group to the cytosine of a CpG dinucleotide commonly occurring in the promoter region of genes (19). Histone modifications, in particular acetylation and methylation, extend the information content of the underlying DNA sequence and confer unique transcriptional potential (16). The most well-characterized modifications are the methylation of the Lys9 and Lys27 residues of histone H3 (H3K9me2/3 and H3K27me3), which repress gene expression, and acetylation of H3K4 (H3K4Ac) and H3K9 (H3K9Ac), which are associated with gene activation (20). Together, these epigenetic modifications create unique promoter architectures that control gene expression. The reversibility of these epigenetic modifications provides a rapid switch for regulating gene expression during differentiation while retaining cellular plasticity in response to developmental and microenvironmental signals (20, 21).
A hallmark of EMT is the loss of E-cadherin expression (7). Several transcription factors, such as Snail, Twist, and ZEB1, have been implicated in the transcriptional repression of E-cadherin and the induction of EMT (6, 7). Snail is a transcriptional repressor that controls large-scale cell movement during the formation of the mesoderm and neural crest (22). Expression of Snail induces EMT in MDCK and breast cancer cells by suppressing E-cadherin expression (23–25). Although E-cadherin expression is often reduced in triple-negative breast cancer (which includes CLBC) and inversely correlated with tumor grade and stage (26, 27), the epigenetic mechanism by which Snail controls E-cadherin silencing remains unclear.
In this study, we examined the epigenetic program of EMT in CLBC by focusing on the transcriptional regulation of E-cadherin. We found that H3K9 methylation and the corresponding G9a methyltransferase are required for EMT-induced E-cadherin promoter DNA methylation in three model cell lines and CLBC. Snail interacts with G9a and DNA methyltransferases (DNMTs) and is required for the association of these two proteins with the E-cadherin promoter. Knockdown of G9a restores E-cadherin expression by suppressing H3K9me2 and DNA methylation, and thus results in the inhibition of cell migration and invasion in vitro and suppression of tumor growth and lung colonization in vivo. Our study not only reveals a critical mechanism underlying metastasis but also has important implications for the development of treatment strategies for CLBC.
Results
TGF-β–induced EMT requires G9a-mediated H3K9 methylation.
The majority of the CLBC cells are “locked” in the mesenchymal state by DNA methylation at the E-cadherin promoter, which makes it difficult to study the initial and dynamic event of chromatin modification leading to E-cadherin silencing in this disease (28). To overcome this technical issue, we began by investigating the mechanism underlying histone modification and DNA methylation during TGF-β–induced EMT in three model cell lines, NMuMG (mouse), MCF10A (human), and HMLE (human), all commonly used for studying EMT in vitro (14, 29–31). As expected, TGF-β treatment resulted in the induction of Snail, the acquisition of fibroblastic mesenchymal morphology, downregulation of epithelial markers (E-cadherin and claudin-3 and -7), and upregulation of mesenchymal markers (vimentin and N-cadherin) in these three model cell lines (Figure 1, A and B). The timing of Snail induction correlated well with the downregulation of E-cadherin and upregulation of vimentin and N-cadherin in these cells, with full induction of EMT occurring at 2, 9, and 12 days in NMuMG, MCF10A, and HMLE cells, respectively (Figure 1, A and B).
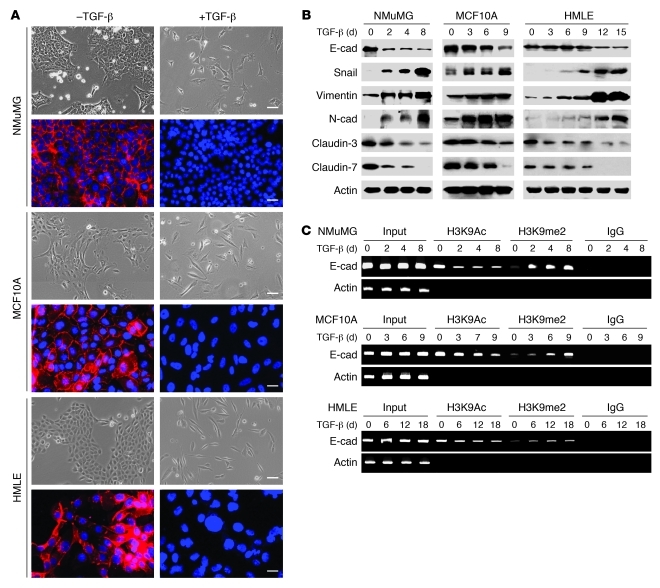
Figure 1. H3K9 methylation at the E-cadherin promoter is associated with TGF-β–induced EMT in three model cell lines.
(A) NMuMG, MCF10A, and HMLE cells were treated with TGF-β1 (5 ng/ml) for 3, 9, and 12 days, respectively; cell morphological changes associated with EMT are shown in the phase contrast images. Expression of E-cadherin (red) was analyzed by immunofluorescence staining. Nuclei were visualized with DAPI staining (blue). Scale bars: 50 μm. (B) NMuMG, MCF10A, and HMLE cells were treated with TGF-β1 (5 ng/ml) for the indicated time periods, and expression of E-cadherin (E-cad), claudin-3, claudin-7, Snail, N-cadherin, and vimentin in these cells was analyzed by Western blotting. (C) NMuMG, MCF10A, and HMLE cells were treated with TGF-β1 (5 ng/ml) for different time periods, and H3K9me2 and H3K9Ac at the E-cadherin promoter in these cell lines were analyzed by ChIP assay.
Because chromatin modifications play a critical role in controlling gene expression, and because H3K9me2 and H3K27me3 are generally linked to gene repression in euchromatin/facultative heterochromatin (32, 33), we examined the histone modifications of the E-cadherin promoter using ChIP assays. We did not observe any significant variation of H3K27me3 after TGF-β treatment (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI57349DS1). However, we found that H3K9me2 was significantly increased at the E-cadherin promoter after TGF-β treatment in all three cell lines tested (Figure 1C), and this increased H3K9me2 was accompanied by decreased H3K9 acetylation of the E-cadherin promoter in these cells (Figure 1C). Interestingly, the timing of increased H3K9me2 correlated well with the downregulation of E-cadherin and the induction of EMT in these cells (Figure 1, A–C). Together, these results suggest that the cooperative downregulation of H3K9 acetylation and upregulation of H3K9me2 at the E-cadherin promoter play a critical role in silencing the expression of E-cadherin.
Because H3K9me2 contributes significantly to DNA methylation (17) and because E-cadherin suppression is commonly associated with CpG island methylation within its promoter, we examined DNA methylation at the E-cadherin promoter in these cell lines treated with TGF-β. In the absence of TGF-β treatment, the E-cadherin promoter was completely unmethylated in NMuMG cells. However, de novo DNA methylation occurred at the E-cadherin promoter following TGF-β treatment as measured by bisulfate sequencing (Figure 2A). We also performed methylation-specific PCR (MSP) analysis and found that DNA methylation started at day 2 and reached a maximum at day 8 after TGF-β treatment in NMuMG cells (Figure 2B). A similar pattern of DNA methylation occurred in MCF10A and HMLE cells at day 6 and a maximal level was reached at day 18 after TGF-β treatment (Figure 2B).
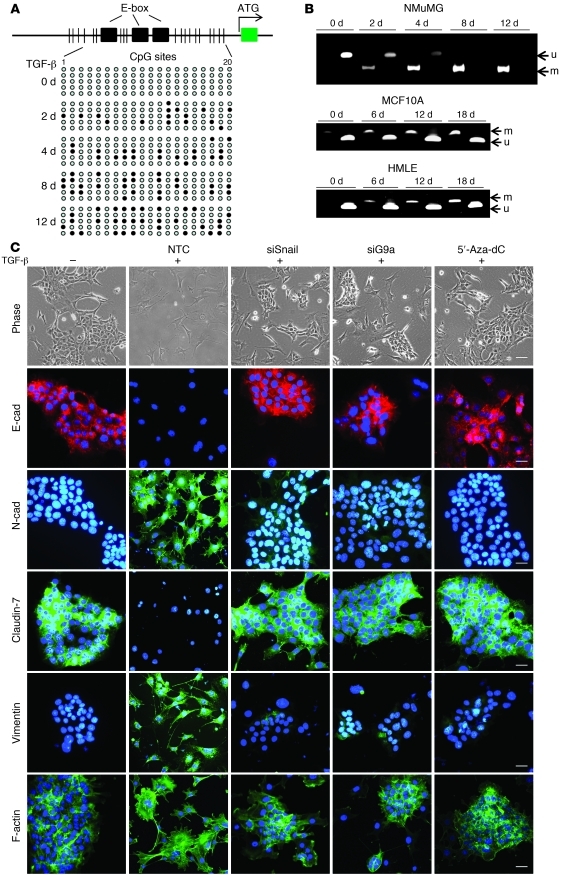
Figure 2. G9a is critical for TGF-β–induced EMT.
(A) Schematic diagram showing the position of 3 E-box and CpG dinucleotides at the promoter region of E-cadherin. NMuMG cells were treated with TGF-β1 (5 ng/ml) for different time periods, and EMT-mediated methylation of the E-cadherin promoter was analyzed by bisulfite sequencing. (B) NMuMG, MCF10A, and HMLE cells were treated with TGF-β1 (5 ng/ml) for the indicated time periods, and EMT-mediated E-cadherin promoter methylation was examined by MSP. m, methylated; u, unmethylated. (C) G9a, Snail, or non-target control (NTC) siRNA was expressed in NMuMG cells followed by TGF-β treatment for 2 days or cells were treated with 5′-Aza-dC. Plus and minus signs indicate treatment with and without TGF-β1, respectively. The morphological changes are shown in the phase contrast images. Expression of E-cadherin, N-cadherin, claudin-7, vimentin, and alternation of lamellipodia (staining with actin-phalloidin) was analyzed by immunofluorescence staining. Scale bars: 30 μm.
EMT has been proposed to be a dynamic process, as NMuMG cells undergo EMT after exposure to TGF-β for 2 days, whereas the process can be reversed after withdrawal of TGF-β (14). We noticed that the timing of E-cadherin promoter DNA methylation fell slightly behind the increase in H3K9me2 in the model cell lines (Figure 1, B and C, and Figure 2B). For example, downregulation of E-cadherin and increased H3K9me2 were obvious at day 2 of TGF-β treatment in NMuMG cells (Figure 1, B and C), whereas E-cadherin promoter DNA methylation reached a maximal level at day 8 (Figure 2, A and B). As DNA methylation is known to be a relatively stable repression marker, we speculate that mesenchymal cells are difficult to reverse to the epithelial state when the E-cadherin promoter becomes DNA methylated. To test this idea, we treated NMuMG cells with TGF-β for 2 days (DNA methylation is minimal) or 12 days (DNA methylation is maximal), followed by withdrawal of TGF-β for an additional 2 days. We found that mesenchymal cells with 2 days of TGF-β treatment completely reversed to the epithelial state (Supplemental Figure 2A). H3K9me2 and DNA methylation on the E-cadherin promoter had almost completely disappeared in these cells after TGF-β withdrawal (Supplemental Figure 2, B and C), consistent with the notion that EMT is a reversible event. However, mesenchymal cells with 12 days of TGF-β treatment did not reverse to the epithelial state after TGF-β withdrawal for 2 days (Supplemental Figure 2A). Interestingly, H3K9me2 and DNA methylation on the E-cadherin promoter in these cells remained unchanged (Supplemental Figure 2, B and C), suggesting that E-cadherin promoter DNA methylation is a relatively stable memory marker in cells undergoing long-term EMT through continuous TGF-β exposures.
G9a is a euchromatin-associated methyltransferase responsible for mono- and dimethylation of H3K9, which is tightly associated with DNA methylation (34). To further establish the causal relationship between H3K9me2 and the de novo DNA methylation of the E-cadherin promoter in TGF-β–induced EMT, we knocked down the expression of Snail or G9a or inhibited the function of DNMTs with the specific inhibitor 5′-Aza-dC in NMuMG cells (35). We found that knockdown of Snail or G9a or inhibition of DNMTs blocked TGF-β–induced changes of EMT markers and the formation of lamellipodia (staining with F-actin) in these cells (Figure 2C; Western blotting data are presented on Supplemental Figure 3). In addition, we found that knockdown of Snail or G9a expression significantly suppressed TGF-β–induced H3K9me2 and restored the H3K9 acetylation at the E-cadherin promoter (Figure 3A). Quantitative real-time PCR using E-cadherin promoter primers on ChIP samples also confirmed this result (Figure 3A). These data suggest that Snail and G9a are involved in TGF-β–mediated H3K9me2 within the E-cadherin promoter.
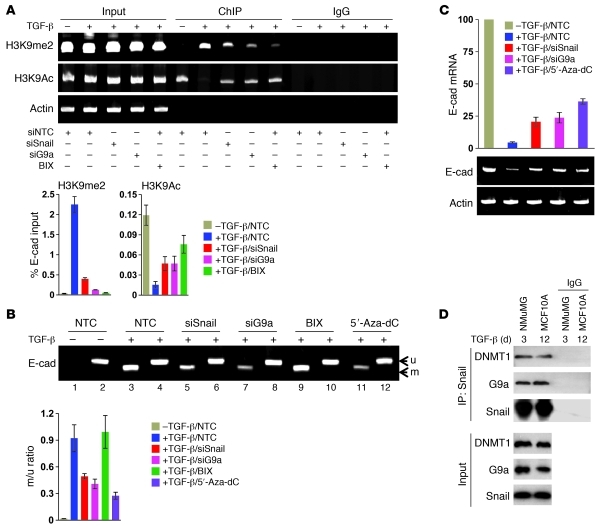
Figure 3. G9a is required for H3K9me2 and DNA methylation at the E-cadherin promoter.
(A) G9a, Snail, or NTC siRNA was expressed in, or BIX01294 (BIX; 2.5 μM) was added to NMuMG cells followed by treatment with or without TGF-β1 (5 ng/ml) for 3 days. H3K9me2 and H3K9 acetylation at the E-cadherin promoter was analyzed by ChIP. ChIP samples were also analyzed by quantitative real-time PCR (mean ± SD from 3 separate experiments; bottom panel). (B) NMuMG cells were treated as described in A. DNA methylation at the E-cadherin promoter was analyzed by MSP. Samples from MSP analyses were also analyzed by quantitative real-time PCR, and the ratio of methylated to unmethylated DNA was plotted (mean ± SD from 3 separate experiments; bottom panel). (C) NMuMG cells were treated as described in B. The expression of E-cadherin mRNA was analyzed by either semi-quantitative RT-PCR (bottom panel) or quantitative real-time PCR (top panel) (mean ± SD from 3 separate experiments). (D) NMuMG and MCF10A cells were treated with TGF-β1 for 3 and 12 days, respectively. After immunoprecipitation of endogenous Snail, associated endogenous G9a and DNMT1 were analyzed by Western blotting.
We also examined DNA methylation at the E-cadherin promoter. As a positive control, treatment with 5′-Aza-dC led to a dramatic suppression of TGF-β–mediated DNA methylation at the E-cadherin promoter (lane 11 vs. lane 3, Figure 3B; and ref. 36). In accordance with the finding above, knockdown of Snail or G9a expression greatly inhibited de novo DNA methylation within the E-cadherin promoter (lanes 5 and 7 vs. lane 3, Figure 3B). Similar results were obtained by quantitative real-time PCR analysis (Figure 3B). Intriguingly, the G9a inhibitor BIX01294 did not affect the DNA methylation status of the E-cadherin promoter (lane 9 vs. lane 3, Figure 3B), although it did inhibit H3K9me2 (lane 10, Figure 3A). This result was unexpected, because H3K9me2 is tightly associated with DNA methylation. Epsztejn-Litman et al. showed that a methylase-inactive G9a mutant could interact with and recruit DNMTs directly for DNA methylation (37). Thus, our results suggest that the H3K9me2 generated by G9a as well as the physical interaction between G9a and DNMTs are two distinct routes for DNA methylation at the E-cadherin promoter. In line with the changes in DNA methylation at the E-cadherin promoter, knockdown of Snail or G9a expression or inhibition of DNMTs significantly blocked the downregulation of E-cadherin mRNA mediated by TGF-β (lanes 3–5 vs. lane 2, Figure 3C; quantitative real-time PCR results are also shown). The observation that knockdown of Snail or G9a expression or inhibition of DNMTs could not completely rescue the downregulation of E-cadherin mRNA mediated by TGF-β suggests that an additional route, such as miR-200, is involved in TGF-β–mediated E-cadherin downregulation (38, 39). Taken together, these data indicate that both G9a and Snail are required for H3K9me2 and de novo DNA methylation of the E-cadherin promoter at EMT.
G9a interacts with Snail and forms a complex with Snail and DNMTs.
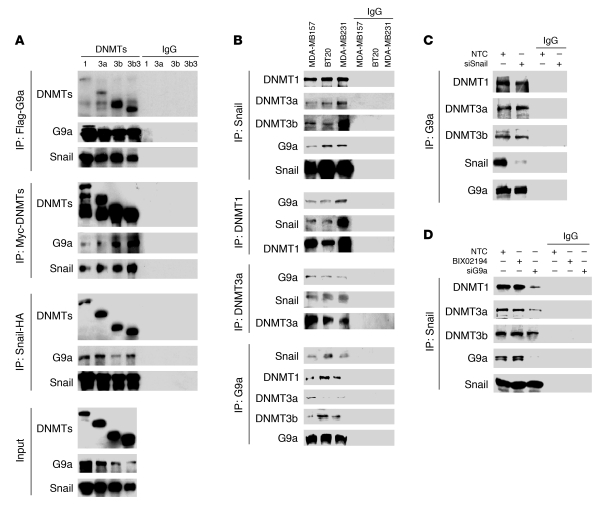
DNA methylation is catalyzed by DNMT1, DNMT3a, and DNMT3b (19). It has been shown that G9a can associate with DNMT1, DNMT3a, and DNMT3b (37, 40). The involvement of Snail, G9a, and DNMTs in H3K9me2 and DNA methylation of the E-cadherin promoter suggests that these molecules may interact with each other. To test this idea, we treated NMuMG and MCF10A cells with TGF-β for 3 and 12 days, respectively, to induce the expression of Snail and induction of EMT. After immunoprecipitating Snail, we found the association of G9a and DNMT1 (Figure 3D). We also coexpressed Flag-tagged G9a, HA-tagged Snail, and Myc-tagged DNMTs in HEK293 cells. After immunoprecipitating G9a, DNMTs, or Snail, we detected the association of the other two molecules, which indicated that these three proteins can interact mutually as a complex (Figure 4A). To further extend our finding in CLBC cell lines, we immunoprecipitated endogenous Snail, DNMT1, DNMT3a, DNMT3b, and G9a individually from breast cancer MDA-MB157, MDA-MB231, and BT20 cells, and we found the association of the other components of this complex, confirming the formation of the G9a-Snail-DNMTs complex in vivo (Figure 4B and Supplemental Figure 4A). To define the relationship between these molecules, we knocked down the expression of Snail in MDA-MB157 cells. After immunoprecipitating G9a, we found that it did not affect the interaction of G9a with DNMTs (Figure 4C and Supplemental Figure 4B). However, when G9a was knocked down, the DNMTs bound with immunoprecipitated Snail were dramatically reduced, indicating that G9a is a bridge molecule that connects Snail with DNMTs (Figure 4D and Supplemental Figure 4C).
Figure 4. G9a forms a complex with Snail and DNMTs.
(A) HEK293 cells were transiently coexpressed with Flag-tagged G9a, HA-tagged Snail, and Myc-tagged DNMTs. Cell extracts were immunoprecipitated separately with Flag, HA, or Myc antibodies, and the associated G9a, Snail, and DNMTs were examined by Western blotting, respectively. (B) Endogenous Snail, G9a, and DNMTs were immunoprecipitated from MDA-MB157, BT20, and MDA-MB231 cells, and bound endogenous Snail, G9a, and DNMTs were examined by Western blotting. (C) Snail or NTC siRNA was expressed in MDA-MB157 cells, and after immunoprecipitation of endogenous G9a, bound Snail and DNMTs was subjected to Western blotting. (D) G9a or NTC siRNA was expressed in MDA-MB157 cells, or cells were treated with the G9a inhibitor BIX01294 (2.5 μM), and after immunoprecipitating endogenous Snail, bound DNMTs and G9a were subjected to Western blotting.
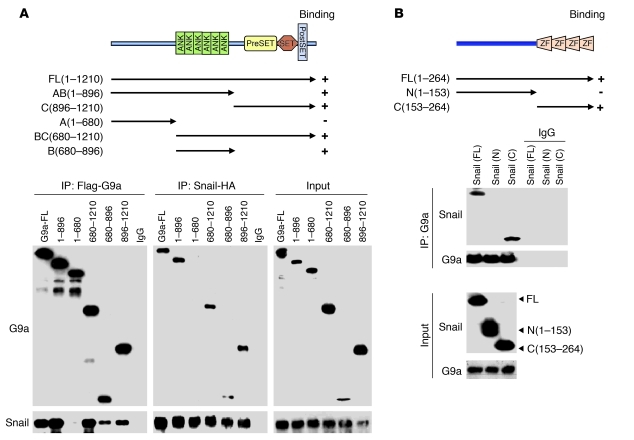
G9a contains several functional domains, including a cysteine-rich region, 6 centrally located ankyrin repeats, and a C-terminal enzymatic SET domain (Figure 5A). To identify the region responsible for the interaction with Snail, we generated several G9a deletion mutants (Figure 5A) and coexpressed them with Snail in HEK293 cells. We found that the ankyrin repeats and the C-terminal SET domain retained the ability to interact with Snail (Figure 5A). The N-terminal region of G9a, however, was unable to interact with Snail (Figure 5A). We also purified full-length Snail from a GST-Snail fusion protein and subsequently cleaved and removed the GST portion with the tobacco etch virus (TEV) protease. When purified Snail was incubated with various deletion mutants of GST-G9a, the ankyrin repeat, the SET domain, or the ankyrin repeat plus the SET domain of G9a, but not GST, immunoprecipitated with Snail (Supplemental Figure 5A). Conversely, when Snail was immunoprecipitated, we found the presence of GST-G9a (ankyrin repeat), GST-G9a (SET domain), or GST-G9a (ankyrin repeat plus SET domain) (Supplemental Figure 5B), indicating that G9a interacts directly with Snail both in vitro and in vivo. BIX01294, which binds to the SET domain of G9a, did not affect the interaction of Snail with DNMTs (Supplemental Figure 6). This observation is in accordance with the finding that Snail interacts with the ankyrin repeat and SET domain of G9a and that inhibition of G9a activity does not affect DNA methylation at the E-cadherin promoter (lane 10, Figure 3B).
Figure 5. G9a interacts with Snail directly.
(A) Schematic diagram showing the structure of G9a and the different deletion constructs (top panel). HEK293 cells were transiently coexpressed with plasmids encoding Flag-tagged full-length (FL) or deletion mutants (designated A, B, and C) of G9a and HA-tagged Snail. Extracts were immunoprecipitated with Flag or HA antibodies, and bound G9a or Snail was examined by Western blotting. (B) Schematic diagram showing the structure of Snail and two deletion mutants (top panel). Full-length and deletion mutants of Snail were coexpressed with G9a in HEK293 cells. After immunoprecipitation of G9a, associated Snail was analyzed by Western blotting. ZF, zinc finger.
To identify the region in Snail that associates with G9a, we generated two deletion mutants of Snail (41): the N-terminal region of Snail (ΔC-Snail; amino acids 1–153), which contains the SNAG domain of Snail; and the C-terminal region of Snail (ΔN-Snail; amino acids 153–264), which includes the conserved zinc finger motif (Figure 5B). When these two deletion mutants of Snail were coexpressed with G9a in HEK293 cells, we found that ΔN-Snail was able to interact with G9a, indicating that the C-terminal region of Snail was responsible for its interaction with G9a (Figure 5B). Taken together, our results indicate that Snail, G9a, and DNMTs form a complex in vivo and that the binding of Snail with DNMTs is mediated, at least in part, through the association with G9a.
Loss of E-cadherin expression is associated with elevated H3K9me2 and DNA methylation at the E-cadherin promoter in CLBC.
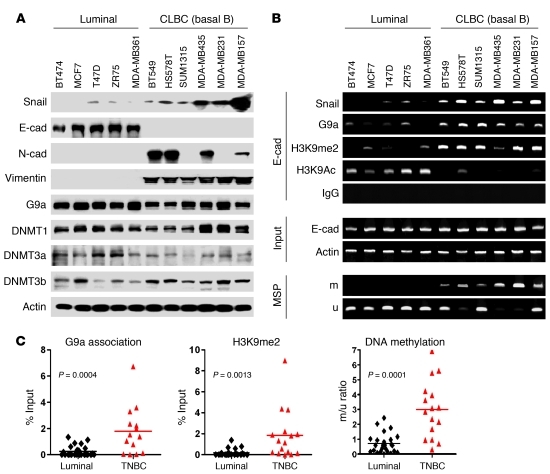
CLBC is associated with a poor clinical outcome, as it has many EMT characteristics and a significantly reduced level of E-cadherin expression (1, 27, 42). Having established that the Snail-G9a-DNMT complex is required for E-cadherin silencing in three model cell lines, we hypothesized that this complex may also be responsible for the loss of E-cadherin expression in CLBC. To test this idea, we compared the expression of Snail, G9a, DNMTs, and EMT markers in 5 luminal and 6 CLBC cell lines (previously referred as basal B subtype) (2, 43). Consistent with previous reports, 6 of these CLBC cell lines lost E-cadherin expression and gained the expression of Snail, N-cadherin, and vimentin (Figure 6A). However, we did not observe any significant alterations in the protein level of G9a and DNMTs when comparing the luminal and CLBC cell lines (Figure 6A). We also examined the global level of H3K9 methylation and H3K9 acetylation in these cell lines and did not find any significant variations (Supplemental Figure 7). However, when we performed ChIP analysis on the E-cadherin promoter, we detected a dramatic elevation of H3K9me2 at the E-cadherin promoter in most of the CLBC cell lines (Figure 6B; quantitative real-time PCR results are presented in Supplemental Figure 8). In line with these findings, H3K9 acetylation at the E-cadherin promoter was lower in CLBC cell lines than in luminal breast cancer cell lines (Figure 6B). The elevation of H3K9me2 at the E-cadherin promoter is likely due to the association of the Snail-G9a-DNMTs complex, because the occupancy of Snail and G9a at the E-cadherin promoter was also significantly higher in CLBC cell lines in comparison to that of luminal breast cancer cell lines (Figure 6B and Supplemental Figure 8). We also assessed E-cadherin promoter DNA methylation in both subtypes using MSP. Again, all CLBC cell lines showed DNA methylation at the E-cadherin promoter, whereas no methylation at the E-cadherin promoter was observed in any of the luminal breast cancer cell lines (Figure 6B). To further extend our observation in vivo, we collected fresh-frozen breast tumor tissues from 25 patients with luminal breast cancer and 16 patients with triple-negative breast cancer (triple-negative for the expression of ERα, progesterone receptor (PR), and HER2/neu; commonly referred to as basal-like breast cancer) with pathological grading (Supplemental Table 1). Expression of E-cadherin, ERα, and claudin-3 on these samples correlated with the information from pathological examination (Supplemental Figure 9). Although G9a expression did not vary substantially between luminal and triple-negative breast cancers, expression of Snail was greatly elevated in triple-negative breast cancer (Supplemental Figure 9). We performed ChIP assays on these tumor tissues and found that the association of G9a, the level of H3K9me2, and DNA methylation at the E-cadherin promoter were significantly elevated in triple-negative breast cancer in comparison to luminal breast cancer (Figure 6C). These results in human breast cancer tissues confirm our observations in breast cancer cell lines, which lends further support to our finding that G9a and H3K9me2 are critical for the epigenetic silencing of E-cadherin expression in CLBC.
Figure 6. G9a-related repressive marks are enriched at the E-cadherin promoter in CLBC cell lines and tumor samples.
(A) Cell extracts were prepared from 5 luminal and 6 claudin-low subtypes of human breast cancer cell lines, and expression of Snail, G9a, DNMT1, and other EMT markers was analyzed by Western blotting. (B) The association of G9a, Snail, and the level of H3K9me2 and H3K9 acetylation at the E-cadherin promoter in various cell lines was analyzed with the ChIP assay. Methylation of the E-cadherin promoter in various breast cell lines was examined by MSP (bottom 2 panels). (C) The association of G9a, H3K9me2, and DNA methylation at the E-cadherin promoter in fresh frozen human tumor tissues of luminal (25 cases) and triple-negative (16 cases) breast cancer was analyzed by ChIP and MSP, respectively. Statistical analyses (mean ± SD) for the association of G9a (0.26 ± 0.08 versus 1.68 ± 0.44), H3K9me2 (0.18 ± 0.07 versus 1.83 ± 0.6), and DNA methylation (0.70 ± 0.13 versus 3.00 ± 0.48) is shown in the distribution plots. TNBC, triple negative breast cancer.
G9a is recruited to the E-cadherin promoter by Snail and results in the suppression of E-cadherin expression.
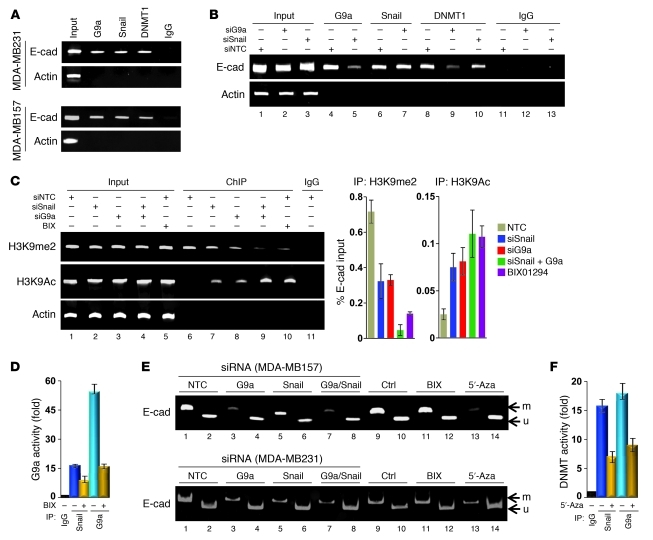
To further validate that the G9a-Snail-DNMT complex is associated with the E-cadherin promoter and mediates the transcriptional repression of the E-cadherin gene in CLBC, we performed ChIP assays in MDA-MB231 and MDA-MB157 cells. As anticipated, G9a, Snail, and DNMT1 were all bound to the E-cadherin promoter (Figure 7A). Interestingly, the occupancy of the E-cadherin promoter by G9a was significantly decreased when Snail was specifically knocked down in MDA-MB157 cells (lane 5 vs. lane 4, Figure 7B; quantitative real-time PCR results are presented in Supplemental Figure 10), whereas knockdown of G9a expression did not affect the binding of Snail to the E-cadherin promoter (lane 7 vs. lane 6, Figure 7B), indicating that the association of G9a with the E-cadherin promoter is mediated by its interaction with Snail. Consistent with these findings, knockdown of Snail or G9a reduced the level of H3K9me2 and increased H3K9 acetylation at the E-cadherin promoter (lanes 7 and 8 vs. lane 6, Figure 7C; quantitative real-time PCR results are also presented on the right panel). Knockdown of both molecules further reduced the level of H3K9me2, similar to what occurred in BIX01294-treated cells (lanes 9 and 10 vs. lane 6, Figure 7C). We also immunoprecipitated the Snail complex and performed G9a enzymatic assays. The Snail complex contained H3K9me2 methyltransferase activity in vitro, and this enzymatic activity could be suppressed by BIX01294 (Figure 7D). These results further support the findings that Snail interacts with G9a and is required to recruit G9a to the E-cadherin promoter for H3K9me2.
Figure 7. G9a is recruited to the E-cadherin promoter for epigenetic silencing of E-cadherin expression.
(A) The association of endogenous G9a, Snail, and DNMT1 at the E-cadherin promoter was analyzed by ChIP. (B) G9a, Snail, or NTC siRNA was expressed in MDA-MB157 cells, and the association of endogenous G9a, Snail, and DNMT1 at the E-cadherin promoter was analyzed with the ChIP assay. Results of quantitative real-time PCR are presented on Supplemental Figure 10. (C) G9a, Snail, or NTC siRNA was expressed in MDA-MB157 cells, or cells were treated with BIX01294 (2.5 μM); H3K9me2 and H3K9 acetylation at the E-cadherin promoter was analyzed by the ChIP assay. Results of quantitative real-time PCR are presented in the right panels (mean ± SD from 3 separate experiments). (D) Statistical analysis of the in vitro G9a methylation assay, mean ± SD from 3 independent experiments, is shown. (E) G9a, Snail, or NTC siRNA was expressed in MDA-MB157 and MDA-MB231 cells, or these cells were treated with the DNMT inhibitor 5′-Aza-dC (5′-Aza; 10 μM), and DNA methylation at the E-cadherin promoter was analyzed by MSP. Ctrl, control. (F) Statistical analysis of the in vitro DNA methylation assay, mean ± SD from 3 independent experiments, is shown.
We noticed that knockdown of G9a expression remarkably decreased the association of DNMT1 at the E-cadherin promoter (lane 9 vs. lane 8, Figure 7B), and knockdown of Snail expression also decreased the binding of DNMT1 at the E-cadherin promoter (lane 10 vs. lane 8, Figure 7B). The relatively strong DNMT1 association with knockdown of Snail compared with knockdown of G9a (lane 10 vs. lane 9, Figure 7B) was due to the large amount of Snail expression (lane 11, Figure 6A) and the inefficiency of knockdown of Snail expression in MDA-MB157 cells (Supplemental Figure 11). As H3K9me2 is tightly linked to DNA methylation, we examined the role of Snail and G9a on E-cadherin promoter DNA methylation in MDA-MB157 and MDA-MB231 cells. We found that knockdown of Snail or G9a expression reduced the level of DNA methylation at the E-cadherin promoter (lanes 3 and 5 vs. lane 1, Figure 7E) in both MDA-MB157 and MDA-MB231 cells. Knockdown of the expression of both molecules further reduced DNA methylation to levels similar to those observed after treatment with 5′-Aza-dC (lanes 7 and 13 vs. lane 1, Figure 7E). When the Snail complex was immunoprecipitated and assayed for DNMT enzymatic activity, we found that the Snail complex also contained DNMT activity and this activity could be inhibited by 5′-Aza-dC (Figure 7F). Together, these results indicate that Snail is required for the recruitment of G9a, which promotes a subsequent association of DNMTs and results in the methylation of the E-cadherin promoter in vivo.
To further establish the functional relationship of G9a and Snail in vivo, we knocked down the expression of G9a, Snail, or both in MDA-MB157 cells and measured E-cadherin mRNA levels by quantitative real-time PCR. Knockdown of either G9a or Snail expression enhanced the mRNA levels of E-cadherin (Supplemental Figure 12A). Knockdown of the expression of both molecules further enhanced E-cadherin mRNA levels. Similar results were obtained when we used E-cadherin promoter luciferase as a reporter (Supplemental Figure 12B). These data indicate a cooperative role for G9a and Snail in the suppression of E-cadherin expression and induction of EMT.
G9a is required for CLBC cell migration and invasion in vitro and tumor growth and lung colonization in vivo.
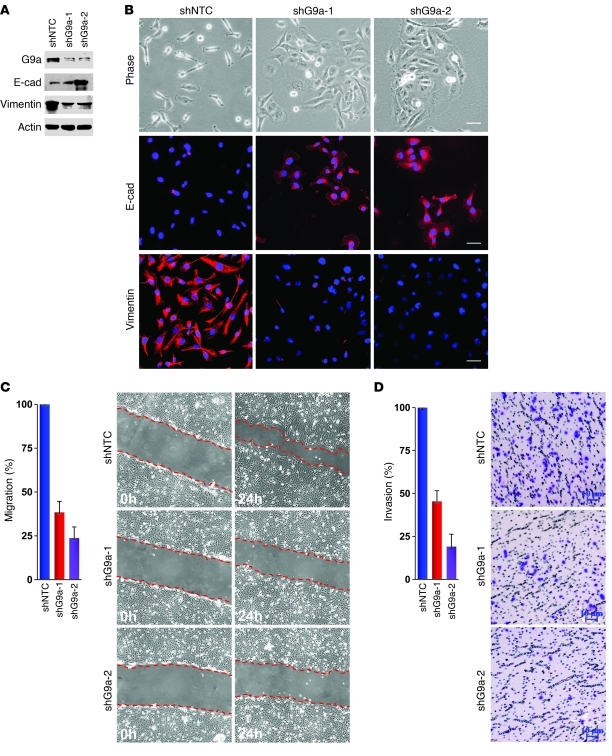
Suppression of E-cadherin expression is critical to EMT induction and cancer metastasis. Because G9a, when associated with Snail, induced H3K9me2 and resulted in the DNA methylation of the E-cadherin promoter in three model cell lines and CLBC, we hypothesized that G9a is critical for breast cancer cell migration and invasion in vitro. To test this hypothesis, we established stable transfectants with knockdown of G9a expression in MDA-MB231 cells. We achieved about 80%–90% knockdown efficiency of endogenous G9a using two independent shRNAs (Figure 8A). Knockdown of G9a expression in both clones caused a morphological change; cells became clustered together, accompanied by a significant restoration of E-cadherin expression and a dramatic downregulation of vimentin expression (Figure 8, A and B). Knockdown of G9a expression also reduced lamellipodia formation as evidenced by actin-phalloidin staining (Supplemental Figure 13). Although we did not observe notable changes in cell growth or proliferation in vitro as measured by the cell count and MTT assay, respectively (Supplemental Figure 14, A and B), knockdown of G9a expression greatly inhibited the migratory ability and invasiveness of MDA-MB231 cells (Figure 8, C and D). Similar results were obtained when we knocked down G9a expression in MDA-MB157 cells (Supplemental Figure 15, A and B). These results clearly support our finding that G9a is the major factor controlling the silencing of E-cadherin in EMT and cancer metastasis.
Figure 8. Knockdown of G9a expression inhibits breast cancer cell migration and invasion in vitro.
(A) MDA-MB231 cells stably expressing control vector or G9a shRNA were examined for the expression of G9a, E-cadherin, and vimentin by Western blotting. (B) Morphological changes in MDA-MB231 cells and stable transfectants with knockdown of G9a are shown in the phase contrast images. Expression of E-cadherin and vimentin in these cells was analyzed by immunofluorescence staining. Nuclei were stained with DAPI (blue). Scale bars: 25 μm. (C) The migratory ability of MDA-MB231 cells and the corresponding stable transfectants with knockdown of G9a expression was analyzed by wound healing assay. Statistical analysis for the cell migration is shown in the bar graph (mean ± SD from 3 independent experiments), and a representative experiment is shown in the right panel. Scale bars: 100 μm. (D) The invasiveness of MDA-MB231 cells stably expressing control vector or G9a shRNA was analyzed with a modified Boyden chamber invasion assay. The percentage of invasive cells is shown in the bar graph (mean ± SD from 3 separate experiments), and a representative experiment is shown in the right panel. Scale bars: 10 μm.
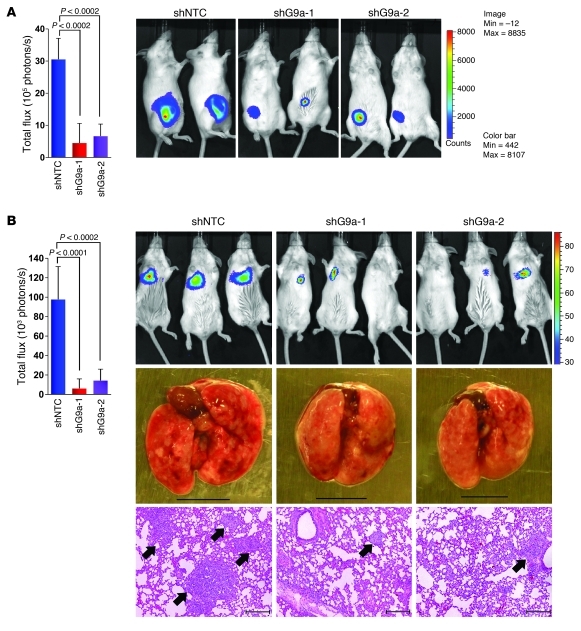
We also extended our findings in a xenograft metastasis model in which MDA-MB231 cells were used to generate pulmonary metastases. First, we used a spontaneous metastatic model by injecting MDA-MB231 cells and the corresponding clones with knockdown of G9a expression into the mammary fat pad of SCID mice. Although knockdown of G9a did not affect the growth of MDA-MB231 cells in vitro, knockdown of G9a expression significantly suppressed tumor formation in vivo (Figure 9A). When tumors from the control group (shNTC) reached 2 cm3 in size, we removed the tumors and allowed these mice to grow an additional 4 weeks to examine lung metastases. We noticed that the control mice had large numbers of lung metastases, whereas mice with knockdown of G9a expression did not have any sign of lung metastases (data not shown). Although this result is intriguing and indicates the critical function of G9a in tumor growth and metastasis, we cannot determine whether the lack of lung metastases from cells with knockdown of G9a expression is due to the slow growth of the primary tumor or to the inability of these cells to invade and disseminate in primary tumors. Thus, we used an experimental metastasis model in which the tumor cells were directly injected into the tail veil of SCID mice. Although an equal number of cells (1 × 106) was injected and these cells had migrated to the lung, as determined by comparable intensity of bioluminescence (Supplemental Figure 16), we found that knockdown of G9a expression in both stable clones suppressed lung colonization in these mice (Figure 9B). To further extend these observations, we established two stable clones with knockdown G9a expression in another breast cancer cell line, MDA-MB435 (Supplemental Figure 17A). Similar to the observation in MDA-MB231 cells, knockdown of G9a expression suppressed migration and invasion of these cells in vitro (Supplemental Figure 17, B and C) and inhibited both breast tumor growth and lung colonization in vivo (Supplemental Figure 17, D and E). Together, our results demonstrate that G9a is critical for cell migration, invasion, and tumor growth and colonization through its interaction with Snail and DNMTs to suppress E-cadherin expression.
Figure 9. Knockdown of G9a expression suppresses breast tumor growth and lung colonization in vivo.
(A) MDA-MB231 cells stably expressing control vector or G9a shRNA were injected into the mammary fat pad of ICR-SCID mice. The growth of breast tumors was monitored every 3 days. After 9 weeks, the size of tumors from each group was recorded by using bioluminescence imaging and quantified by measuring photon flux. Values are the mean of 6 animals ± SEM. (B) Cells from A were also injected into the tail vein of ICR-SCID mice. After 9 weeks, the development of lung metastases was recorded using bioluminescence imaging and quantified by measuring photon flux (mean of 6 animals ± SEM). Results for 3 representative mice from each group are shown. Mice were sacrificed, and lung metastatic nodules were examined macroscopically or detected in paraffin-embedded sections stained with H&E. Scale bars: 100 μm. Arrowheads indicate lung metastases.
Knockdown of G9a expression alters the expression of epithelial and mesenchymal markers associated with EMT.
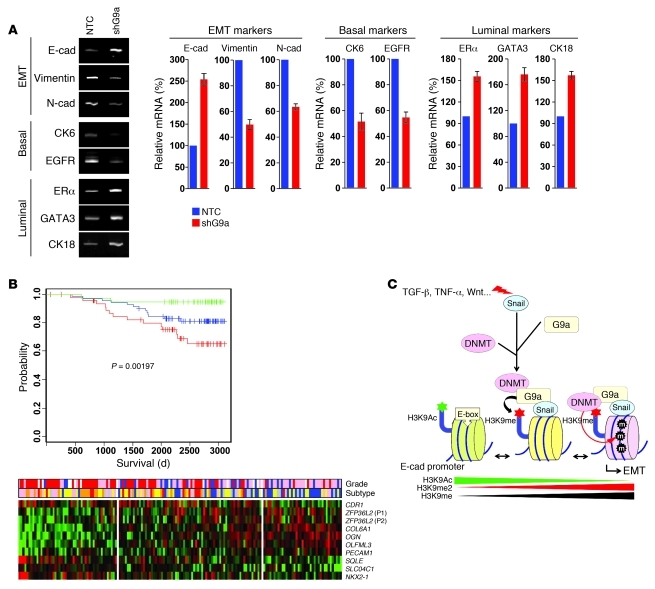
Breast cancer is categorized into 6 clinically relevant subtypes based upon molecular gene signatures. These subtypes fit into the broader groupings of either “basal” or “luminal” types because of their molecular similarity to the basal or luminal cells of the normal mammary gland. The basal-type breast cancers (including basal-like, claudin-low, and normal-like) express high levels of basal cell markers (cytokeratins 5, 6, and 14), whereas luminal-type breast cancers (including luminal A and luminal B) are defined by high expression of luminal cell markers (ERα, GATA3, FoxA1, and cytokeratin 18) (44). To examine whether G9a regulation is specific to E-cadherin or the EMT differentiation program in general, we performed gene expression array analysis on MDA-MB231 cells and the corresponding stable transfectants with knockdown of G9a expression. Of the 433 differentially expressed genes identified (P < 0.01), 183 were upregulated and 250 were downregulated. Among these differentially expressed genes, we noticed the downregulation of EMT markers (such as vimentin and N-cadherin) and basal markers (such as CK6 and EGFR) and upregulation of luminal molecules (ERα, GATA3, and CK18) (Figure 10A; quantitative real-time PCR results are also presented). Strikingly, the expression of many claudin genes was also upregulated in G9a-knockdown cells (Supplemental Figure 18). These findings support our observation that G9a is critical in controlling the switch between epithelial and mesenchymal states of the cells. Ingenuity pathway analysis (IPA) of these 433 transcripts revealed networks and pathways related to TGF-β, Wnt, and cell adhesion signaling (data not shown). We measured the activation of these pathways in MDA-MB231 cells and the corresponding stable transfectants with knockdown of G9a expression using the TGF-β responsive element TOP/FOP and E-cadherin promoter reporter luciferase. Consistent with the IPA analysis, knockdown of G9a expression inhibited the activation of TGF-β and β-catenin pathways and upregulated E-cadherin promoter luciferase activity (Supplemental Figure 19).
Figure 10. Knockdown of G9a expression alters the expression of epithelial and mesenchymal markers associated with EMT.
(A) The differentially expressed markers for EMT and basal and luminal breast cancer from MDA-MB231 cells and the corresponding stable clone with knockdown of G9a expression were analyzed by quantitative RT-PCR. (B) Kaplan-Meier overall survival curve separates the tumors (from GSE1456) into 3 groups with expression of a 9-gene prognostic signature (top panel). Expression of the 9-gene signature in 159 breast cancer patients is shown in the heatmap (bottom panel). Top bars: tumor grade (1: blue, 2: pink, 3: red) and tumor subtypes (normal-like: blue, luminal A: yellow, luminal B: orange, basal: pink, HER2-positive: red]). P1, probe 1; P2, probe 2. (C) A proposed model to illustrate the interaction of Snail with G9a and DNMTs leading to E-cadherin promoter methylation and EMT induction (see Discussion).
Having identified genes regulated by G9a in EMT, CLBC, and metastasis, we then sought to elucidate their clinical relevance in breast cancer. If genes associated with G9a regulation are biologically meaningful, we expect that a subset of the genes regulated by G9a may confer prognostic value in breast cancer. To this end, we chose GSE11121 (comprises 200 node-negative breast cancer patients) as a training dataset to stratify genes whose expression is potentially associated patient survival and overlapped with genes affected by G9a knockdown. We identified a 9-gene signature (10-probe set) (OLFML3, ZFP36L2, OGN, PECAM1, COL6A1, which were upregulated after knockdown of G9a; NKX21, SQLE, CDR1, SLCO4C1, which were downregulated after knockdown of G9a) that effectively separated patients into poor and good prognosis groups (Supplemental Figure 20A, P = 1.999 × 10–8). This signature also separated patients by grade, with low-score group having more grade 1 and high-score group having more grade 3 tumors (Supplemental Figure 20B). To validate this 9-gene signature, we applied it to GSE2034, consisting of 286 node-negative breast patients. It can separate these patients into different prognosis groups (Supplemental Figure 21A, P = 0.00469), although this separation seems to be independent of their ER status (Supplemental Figure 21B). In addition, we applied this signature to GSE1456, which includes 159 breast patients whose lymph node status was not reported. Again, the 9-gene signature was able to separate patients into three prognostic groups, using either disease-free survival or overall survival (Figure 10B and Supplemental Figure 22A; P = 0.00919 and P = 0.00197). This corresponded well with grades and breast tumor subtypes, with the low score group having more grade 1 and normal-like subtypes, and the high score group having more grade 3 and basal-like as well as HER2 subtypes (Figure 10B and Supplemental Figure 22B). The clinical validation of genes associated with G9a regulation supports the finding that G9a is critical in controlling EMT in CLBC and metastasis.
Discussion
Our study provides several insights into the epigenetic program in EMT, CLBC, and metastasis. First, we have identified a key mechanism underlying the epigenetic regulation of EMT and reinforce the notion that EMT is an epigenetically regulated program. To uncover the mechanism of epigenetic regulation of EMT, we examined the dynamic chromatin modifications at the promoter of E-cadherin, a typical epithelial molecule and trait marker of EMT, in three model cell lines that are commonly used for EMT induction. The gradual increase in H3K9me2 and decrease in H3K9 acetylation correlated with the timing of Snail induction, the morphological changes indicative of EMT, and the de novo DNA methylation of the E-cadherin promoter. These findings suggest that H3K9me2 plays a critical role in silencing the expression of E-cadherin. G9a, a key methyltransferase responsible for H3K9me2 at euchromatin and facultative heterochromatin, does not contain a DNA binding sequence. We found that Snail interacted with G9a both in vitro and in vivo and was required for G9a recruitment to the E-cadherin promoter (Figure 10C). Consistent with this finding, the immunoprecipitated Snail complex contained G9a methyltransferase activity, and knockdown of Snail expression disrupted the association of G9a with the E-cadherin promoter. The domains responsible for their interaction were mapped to the ankyrin repeat and SET domains of G9a and the C-terminal zinc finger region of Snail, respectively. This is consistent with the observation that the interaction of Snail with G9a is independent of the catalytic activity of G9a. The transcriptional repressive activity of Snail requires both the N-terminal SNAG domain and the C-terminal zinc finger region (7). We and others showed previously that the SNAG domain of Snail interacted with LSD1 and Sin3A/HDAC for H3K4 demethylation and histone deacetylation, respectively (45, 46). However, H3K4 demethylation is known to be an initial step in gene repression (47), suggesting that an intermediate step is required to bridge H3K4 demethylation to the DNA methylation on the E-cadherin promoter. Here we identified that the C terminus of Snail interacted with G9a directly, which recruited DNMT to the E-cadherin promoter for DNA methylation. Thus, demethylation on H3K4 and methylation on H3K9 by LSD1 and G9a, respectively, provide great synergy in gene repression and DNA methylation (17, 48). Consistent with this observation, a zinc finger transcriptional repressor, REST, also recruits LSD1 and G9a through its C-terminal domain and middle region, respectively, in repressing neuronal gene expression (49).
Histone methylation is intimately linked to DNA methylation, which provides reinforcement as well as establishment of gene silencing. DNA methylation is executed by a family of highly related DNA methyltransferase enzymes (DNMT1, DNMT3a, and DNMT3b) that transfer a methyl group to the cytosine in a CpG dinucleotide, which commonly occurs in the promoter region of genes (17, 19). Typically, the maintenance of DNA methylation in somatic cells is attributed to DNMT1, whereas de novo DNA methylation during embryonic development is credited to DNMT3a and DNMT3b. However, there is overlap in the function of these two types of DNMTs, as DNMT1 can also contribute to de novo DNA methylation both in vitro and in vivo, and the maintenance of methylation in certain regions of the genome requires DNMT3a and DNMT3b (17, 19). We found that Snail can interact with DNMT1, DNMT3a, and DNMT3b, and this interaction is likely to be indirect, as knockdown of G9a expression disrupted the interaction of Snail with DNMTs. Thus, G9a provides a platform for the efficient assembly of the Snail-G9a-DNMTs complex in vivo (Figure 10C).
Although the causal relationship between H3K9me2 and DNA methylation can be bidirectional, both processes are tightly associated in heterochromatin and transcriptionally repressed euchromatic regions. For example, H3K9me2, catalyzed by G9a, is absolutely required for DNA methylation in fungi, plants, and mammals (50, 51). Conversely, reactivation of silenced tumor suppressor genes in response to 5′-Aza-dC–induced DNA demethylation is accompanied by a decrease in H3K9me2, but not other silencing markers such as H3K9me3 or H3K27me3 (52). In this study, we found that H3K9me2 coincided with DNA methylation at the E-cadherin promoter in three model cell lines and CLBC. Knockdown of G9a expression significantly inhibited DNA methylation at the E-cadherin promoter and reactivated E-cadherin expression in MDA-MB231 cells. Our study suggests that G9a-mediated H3K9me2 is one of the key events in the maintenance of transcriptionally silent gene promoters in cancer. In line with our findings, G9a is enriched at the promoters of aberrantly methylated genes in cancer cells, and co-recruitment of G9a, DNMT1, and HP1 to the promoter of the survivin gene stimulates H3K9me2 and DNA hypermethylation (53). Intriguingly, G9a seems to use two distinct modes for generating DNA methylation at the E-cadherin promoter (Figure 10C). In the first mode, G9a creates H3K9me2 via its catalytic activity, which subsequently recruits HP1 and DNMTs for DNA methylation (17). In the second mode, G9a interacts with DNMTs directly and recruits them to the E-cadherin promoter through the association with Snail. This explains why the interaction of the Snail-G9a-DNMT complex does not require G9a activity and indicates that the inhibition of G9a activity does not significantly alter DNA methylation at the E-cadherin promoter.
E-cadherin downregulation is commonly associated with DNA methylation of its promoter (28, 54), which provides a relative stable “memory” marker for gene silencing. Using a classic TGF-β–induced EMT model in NMuMG cells, which was first identified elegantly by Derynck’s group in 1994 (14), we found that DNA methylation on the E-cadherin promoter can be reversed (Supplemental Figure 2). Two mechanisms of DNA demethylation have been proposed (19, 55). The first one is passive DNA demethylation, in which the activity of DNMT is suppressed and DNA methylation cannot be maintained during DNA replication. This results in the loss of DNA methylation after cell propagation (19). The second mechanism involves active DNA demethylation, in which the 5′-methylcytosine (5 mC) is recognized and removed by DNA mismatch and repair enzymes (55). The recent identification of ten-eleven translocation 1 (TET1) supports this notion (56). TET1 converts 5 mC to 5-hydroxymethylcytosine (5 hmC), which precludes the preferentially binding of methyl-CpG–binding proteins (MBDs and MeCP) that commonly associate with histone deacetylases (HDACs) in gene repression (55). In addition, 5 hmC facilitates DNA demethylation through a process that requires the base excision repair pathway mediated by cytidine deaminases or thymine-DNA glycosylase (57). In our study, the decreased DNA methylation on the E-cadherin promoter after TGF-β withdrawal suggests that a passive mechanism is involved. This may be due to the downregulation of Snail after TGF-β withdrawal and the consequent loss of G9a and DNMT recruitment to the E-cadherin promoter.
Second, our study has delineated a critical role of the Snail-G9a-DNMT complex in CLBC. Ample evidence supports an epithelial hierarchy within the human breast. This cellular differentiation process starts with an undifferentiated ERα-negative MSC that either maintains itself through self-renewal or differentiates into committed progenitors (4). These progenitors ultimately give rise to progeny that consist of mature ductal and alveolar cells, which belong to the luminal epithelial cell lineage and line the lumen of the mammary gland, and mature myoepithelial cells, which surround the luminal epithelium and contact the basement membrane (4). Because the MSC signature was enriched in CLBC, which is characterized by the expression of mesenchymal and stem cell–associated genes and the lack of expression of claudin and E-cadherin, it is speculated that CLBC may originate from the transformation of MSCs that arrest at an early stage of differentiation (2, 4). Alternatively, this can be mediated by a de-differentiation process, as proposed by Weinberg and colleagues (10, 58). Both of these phenotypic and cellular conversions require the activation of EMT. Intriguingly, CLBC has many EMT characteristics (1, 9, 27, 59–62), suggesting that the activation of EMT blocks cellular differentiation by repressing epithelial molecules, such as E-cadherin in this case. Consistent with this idea, we found that the G9a is critical for EMT in both three model cell lines and CLBC through its interaction with Snail. Although the protein level of G9a and DNMTs remained unchanged between luminal and CLBC cell lines, the association of Snail, G9a, and the level of H3K9me2 at the E-cadherin promoter were dramatically elevated in both CLBC cell lines and tumor samples, indicating the critical role of G9a and H3K9me2 in the epigenetic silencing of the E-cadherin promoter in CLBC. In line with this finding, knockdown of G9a expression downregulated EMT and basal markers (vimentin, N-cadherin, CK6, and EGFR) and upregulated luminal epithelial molecules (ERα, GATA3, CK18, and E-cadherin) and claudin. Furthermore, knockdown of G9a expression suppressed cell migration and invasion in vitro, inhibited metastasis in vivo, and predicted increased survival for patients with breast cancer. Thus, we speculate that G9a is involved in the control of a common epigenetic EMT program to block differentiation toward epithelial or luminal lineage (Figure 10C).
Third, our study indicates that the cellular plasticity of CLBC represents a potential therapeutic target. Because they are characterized by acquisition of an EMT phenotype and loss of E-cadherin expression, CLBC cells have a distinct advantage in terms of invasion and metastasis to distant organs or tissues during neoplastic development. This EMT program is also indicative of stem cell–like characteristics, making CLBC resistant to apoptosis mediated by standard chemotherapeutics (10, 63). It is likely that CLBC becomes “addicted” to this program for the advantage of survival and metastasis. Activation of this program can be achieved by genetic mutation/deletion of the E-cadherin gene or by epigenetic reprogramming of gene expression in such a way that abnormal silencing becomes the default state and is inherited by progeny upon cell division. However, while gene mutation is rare in invasive breast ductal carcinoma, promoter DNA methylation is a common mechanism causing the loss of E-cadherin expression in this disease (54, 64). We found that knockdown of G9a led to the restoration of E-cadherin gene expression and suppression of cell migration, invasion, and metastasis in CLBC. These data suggest that the machinery for E-cadherin expression in these tumor cells remains intact and functional and can mediate reexpression if the repressive signal regulating histone and/or DNA methylation is removed. Thus, the epigenetic program in CLBC may represent a therapeutic target for treating this aggressive and metastatic disease.
In summary, our study highlights the importance of G9a-mediated epigenetic modification in EMT, CLBC, and metastasis. Blocking the binding in Snail-G9a-DNMTs may pave the way for the development of novel therapeutic approaches that target metastatic CLBC.
Methods
Plasmids, siRNA, and antibodies.
G9a shRNA expression plasmids were purchased from MISSION shRNA at Sigma-Aldrich. Smartpool siRNA against human G9a (siRNA-1) and Snail (siRNA-1) were from Dharmacon. Expression plasmids for DNMT1, DNMT3a, DNMT3b, and DNMT3b3 were provided by Arthur D. Riggs (City of Hope, Duarte, California, USA). Human G9a was amplified from a HeLa cDNA library and subcloned into pcDNA3-Flag. Deletion mutants of G9a were constructed as described previously (41, 45). The expression plasmid for human Snail was described previously (15, 25, 41, 45).
Antibodies against Myc, HA, Flag, and actin were purchased from Sigma-Aldrich. Antibodies for E-cadherin and β-actin were from BD Transduction Laboratories. N-cadherin and G9a antibodies were from Upstate and Abcam, respectively.
Cell culture.
All cancer cell lines were grown in DMEM/F12 supplemented with 10% FBS, except breast cancer cell lines T47D and ZR75, which were grown in RPMI-1640 plus 10% FBS. HMLE cells were provided by Robert A. Weinberg (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA). For establishing stable transfectants with knockdown of G9a expression, MDA-MB231-Luc-D3H1 (with stable expression of luciferase, from Xenogene Corp.) and MDA-MB435 cells were transfected with G9a shRNA; stable clones were selected with puromycin (300 ng/ml) for 4 weeks.
Immunostaining, immunoprecipitation, and immunoblotting.
Experiments were performed as described previously (25). For immunofluorescence staining, cells were grown on chamber slides, fixed with 4% paraformaldehyde, and incubated with primary antibodies. Secondary antibodies used were Texas red–conjugated goat anti-mouse, FITC-conjugated goat anti-rabbit, or Alexa Fluor 350–goat anti-rabbit (Molecular Probes, Invitrogen).
DNA methylation analysis.
Genomic DNA (~0.75 μg) was treated with sodium bisulfite using the EpiTect system (QIAGEN) by following the manufacturer’s protocol. Bisulfite-converted DNAs (~50 ng) were used as templates for PCR amplification of the CpG islands in the CDH1 promoter. All PCR products were purified from 1.5% agarose gels using a Gel Extraction Kit (QIAGEN) and cloned into the pGEM-T Easy vector (Promega). Five randomly selected clones from each sample were selected for sequencing. MSP was performed on bisulfate-modified DNA as described by Herman et al. (65).
Quantitative real-time PCR.
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Specific quantitative real-time PCR experiments were performed using SYBR Green Power Master Mix following the manufacturer’s protocol (Applied Biosystems).
GST pull-down assay.
GST proteins were expressed as described previously (45). The pull-down complexes were examined by Western blotting.
Invasion assay.
Invasion assays were performed as described previously (15, 45). All experiments were performed at least twice in triplicate. Statistical analysis was performed using the Student’s t test; a P value of less than 0.05 was considered significant.
Luciferase reporter assay.
Invasion assays were performed as described previously (15, 25, 45). All experiments were performed 3 times in triplicate.
RT-PCR.
Total RNA was prepared using the TRIzol reagent (Invitrogen) following the manufacturer’s protocol. RT-PCR was performed using a SuperScript One-Step RT-PCR Kit (Invitrogen). The PCR products were analyzed by electrophoresis on a 2% agarose gel containing ethidium bromide, and visualized and photographed under UV light.
ChIP.
ChIP assays were performed as described previously (45). The primers for the E-cadherin promoter were: 5′-ACTCCAGGCTAGAGGGTCACC-3′ and 5′-CCGCAAGCTCACAGGTGCTTTGCAGTTCC-3′.
The frozen fresh tumor samples were collected from resected breast tumors from patients at the University of Texas MD Anderson Cancer Center with the approval of the IRB. Data regarding the stage, grade, and expression of ERα, PR, and HER2 are presented in Supplemental Table 1. These frozen samples were snap-frozen in liquid nitrogen and stored at –80°C. Each sample was examined histologically with H&E-stained sections. Regions from tumor samples were microdissected and examined, and only samples with a consistent tumor cell content of more than 75% in tissues were used for ChIP analysis. The tumor samples were prepared using the Imprint ChIP Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, 100 mg of breast tumor tissue was cross-linked with 1% formaldehyde (Pierce). The sample was then homogenized using 20 strokes of a Dounce homogenizer in 1 ml of homogenizing buffer. Following centrifugation, the pellet was resuspended in 0.25 ml lysis buffer and processed for ChIP assay.
G9a and DNMT methyltransferase assay.
HEK293T cells were cotransfected with constructs encoding G9a and Snail. After immunoprecipitation of Snail1 or G9a, the complexes were analyzed for DNA methyltransferase activity and H3K9 methyltransferase activity using the DNA Methyltransferase Assay Kit and the H3K9 Methyltransferase Assay Kit (Epigentek) following the manufacturer’s protocols. The enzymatic activities of DNMTs and G9a were detected with a microplate reader at 450 nm and fluorescent plate reader at 450 nm, respectively.
Experimental lung metastasis model.
Female ICR-SCID mice (6–8 weeks old) were purchased from Taconic and maintained and treated under specific pathogen–free conditions. Mice were injected with MDA-MB231-luc-D3H1 (1 × 106 cells/mouse) or MDA-MB435 (2 × 106 cells/mouse) cells and their corresponding stable clones with knockdown of G9a expression via mammary fat pad or tail vein (6 mice/group). Mice with MDA-MB231 cells were imaged from dorsal and ventral views once per week. All mice (6 of 6) injected with MDA-MB231 cells developed the expected lung metastatic lesions, with luciferase signals beginning to appear at week 5. For MDA-MB435 cells, lung metastatic lesions were developed in 12 weeks. Visible lung metastatic nodules were examined macroscopically or detected in paraffin-embedded sections stained with H&E. Data were analyzed using Student’s t test; a P value less than 0.05 was considered significant.
Microarray profiling and G9a prognostic signature.
Expression profiling analysis was performed on MDA-MB231 cells and the corresponding stable transfectants with knockdown of G9a expression (performed in triplicate for each sample) using Affymetrix U133A microchips (dataset was deposited in the NCBI GEO database, with accession number GSE34925). The most differentially expressed probesets (as defined by fold change in median expression values) were subjected to SAM (significance analysis of microarrays) analysis (66) using a minimum of a 1.33-fold change in differential expression in comparison with control. This analysis yielded 183 genes that were upregulated and 250 genes that were downregulated after knockdown of G9a expression in MDA-MB231 cells, with a predicted false discovery rate (FDR) of 0.85%.
To identify genes potentially regulated by the G9a signaling and playing a role in breast cancer patient survival, we first chose the GSE11121 dataset (from the NCBI GEO website; http://www.ncbi.nlm.nih.gov/geo/) comprising 200 node-negative breast cancer patients as the training dataset and performed a univariate Cox proportional hazards regression analysis to find genes whose expression was potentially associated with patient survival. This gene list was then overlapped with the G9a-knockdown gene signature to find common genes, and further limited by clustering analysis to 9 genes that are coexpressed in breast cancers (OLFML3, ZFP36L2, OGN, PECAM1, COL6A1, which were upregulated after G9a knockdown, and NKX21, SQLE, CDR1, SLCO4C1, which were downregulated after G9a knockdown). We next constructed a multi-gene score using the following algorithm. Normalized probeset log2 values were first subtracted with median values from all samples for one particular probeset. The multi-gene score is the averaged sum from 5 upregulated genes minus that from 4 downregulated ones. Based on the gene expression heatmap, an arbitrary cutoff was set at ±0.45 to stratify patients into three groups having low, medium, and high 9-gene scores (Supplemental Figure 20B). This effectively separated patients into poor and good prognosis groups (Supplemental Figure 20A, P = 1.999 × 10-8). In addition, it separated grade 1 and grade 3 patients from the extreme groups (Supplemental Figure 20B).
To validate the 9-gene signature, we first used GSE2034, another dataset with 286 node-negative breast patients, and applied the same 9-gene score stratification procedure to separate these patients into different prognosis groups (Supplemental Figure 21A, P = 0.00469). The separation of the patients seemed to be independent of their ER status (Supplemental Figure 21B). We next used GSE1456, which included 159 breast patients whose lymph node status was not reported. Again, the 9-gene signature was able to separate patients into three prognostic groups, using either disease-free survival or overall survival (Supplemental Figure 22A, P = 0.00919 and 0.00197). This corresponded well with grades and tumor subtypes, with the low-score group having more grade 1 and normal-like tumors, and the high-score group having more grade 3 and basal as well as HER2 tumors (Supplemental Figure 22B).
Statistics.
The experiments were repeated at least 2 times. Results are expressed as mean ± SD or SEM as indicated. An independent Student’s t test was performed to analyze the luciferase assay; a 2-tailed Student’s t test was used for intergroup comparisons. A P value less than 0.05 was considered statistically significant.
Study approval.
All procedures for animal study were approved by the Institutional Animal Care and Use Committee of the University of Kentucky College of Medicine and conform to the legal mandates and federal guidelines for the care and maintenance of laboratory animals.
Supplementary Material
Acknowledgments
We thank Nathan L. Vanderford for critical reading and editing of the manuscript. This work was supported by grants from the NIH (R01CA125454), the Susan G. Komen Foundation (KG081310), and the Mary Kay Ash Foundation (to B.P. Zhou).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(4):1469–1486. doi:10.1172/JCI57349.
References
- 1.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107(35):15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23(22):2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Zhou BP. Snail: more than EMT. Cell Adh Migr. 2010;4(2):199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 11.Christofori G. New signals from the invasive front. Nature. 2006;441(7092):444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Novoa JM, Nieto MA. Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1(6–7):303–314. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Zhou BP. Inflammation: a driving force speeds cancer metastasis. Cell Cycle. 2009;8(20):3267–3273. doi: 10.4161/cc.8.20.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127(6 pt 2):2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15(5):416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 17.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 18.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 19.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15(12):3927–3937. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8(11):829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 22.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3(3):155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 23.Batlle E, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 24.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 25.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 26.Mahler-Araujo B, Savage K, Parry S, Reis-Filho JS. Reduction of E-cadherin expression is associated with non-lobular breast carcinomas of basal-like and triple negative phenotype. J Clin Pathol. 2008;61(5):615–620. doi: 10.1136/jcp.2007.053991. [DOI] [PubMed] [Google Scholar]
- 27.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 28.Lombaerts M, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94(5):661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bindels S, et al. Regulation of vimentin by SIP1 in human epithelial breast tumor cells. Oncogene. 2006;25(36):4975–4985. doi: 10.1038/sj.onc.1209511. [DOI] [PubMed] [Google Scholar]
- 30.Xie L, et al. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res. 2003;5(6):R187–R198. doi: 10.1186/bcr640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 33.Mohn F, Schubeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 2009;25(3):129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Collins R, Cheng X. A case study in cross-talk: the histone lysine methyltransferases G9a and GLP. Nucleic Acids Res. 2010;38(11):3503–3511. doi: 10.1093/nar/gkq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25(3):473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 36.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15(11):1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iliopoulos D, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2(92):ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteve PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20(22):3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem. 2009;284(1):640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreike B, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24(1):306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudolph T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26(1):103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 48.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49(14):2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8(7):544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 50.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416(6880):556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 51.Ikegami K, et al. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12(1):1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 52.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66(7):3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 53.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21(10):1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nabel CS, Kohli RM. Molecular biology. Demystifying DNA demethylation. Science. 2011;333(6047):1229–1230. doi: 10.1126/science.1211917. [DOI] [PubMed] [Google Scholar]
- 58.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15(9):1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 59.Honeth G, et al. The CD44+/CD24– phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reiman JM, Knutson KL, Radisky DC. Immune promotion of epithelial-mesenchymal transition and generation of breast cancer stem cells. Cancer Res. 2010;70(8):3005–3008. doi: 10.1158/0008-5472.CAN-09-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Storci G, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol. 2008;214(1):25–37. doi: 10.1002/path.2254. [DOI] [PubMed] [Google Scholar]
- 63.Li X, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 64.Cheng CW, et al. Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene. Oncogene. 2001;20(29):3814–3823. doi: 10.1038/sj.onc.1204505. [DOI] [PubMed] [Google Scholar]
- 65.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.