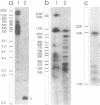
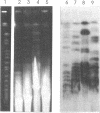
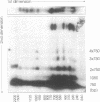
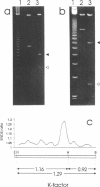
Abstract
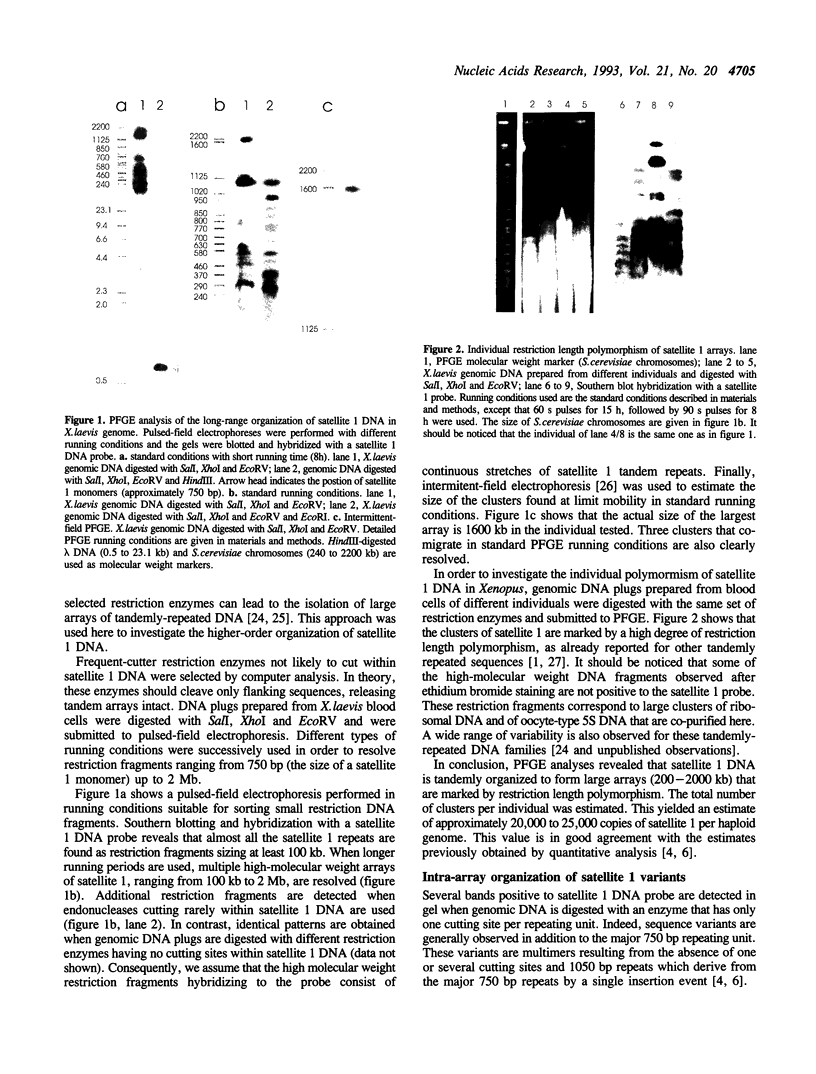
We have investigated the long-range organization and the intrinsic curvature of satellite 1 DNA, an unusual tandemly-repeated DNA family of Xenopus laevis presenting sequence homologies to SINEs. PFGE was used in combination with frequent-cutter restriction enzymes not likely to cut within satellite 1 DNA and revealed that almost all the repeating units are tandemly organized to form large arrays (200 kb to 2 Mb) that are marked by restriction length polymorphism and contain intra-array domains of sequence variation. Besides that, we have analysed the secondary structure of satellite 1 DNA by computer modelling. Theoretical maps of curvature obtained from three independent models of DNA bending (the dinucleotide wedge model of Trifonov, the junction model of Crothers and the model of de Santis) showed that satellite 1 DNA is intrinsically curved and these results were confirmed experimentally by polyacrylamide gel electrophoresis. Moreover, we observed that this bending element is highly conserved among all the members of the satellite 1 DNA family that are accessible to analysis. A potential genetic role for satellite 1 DNA based on this unusual structural feature is discussed.
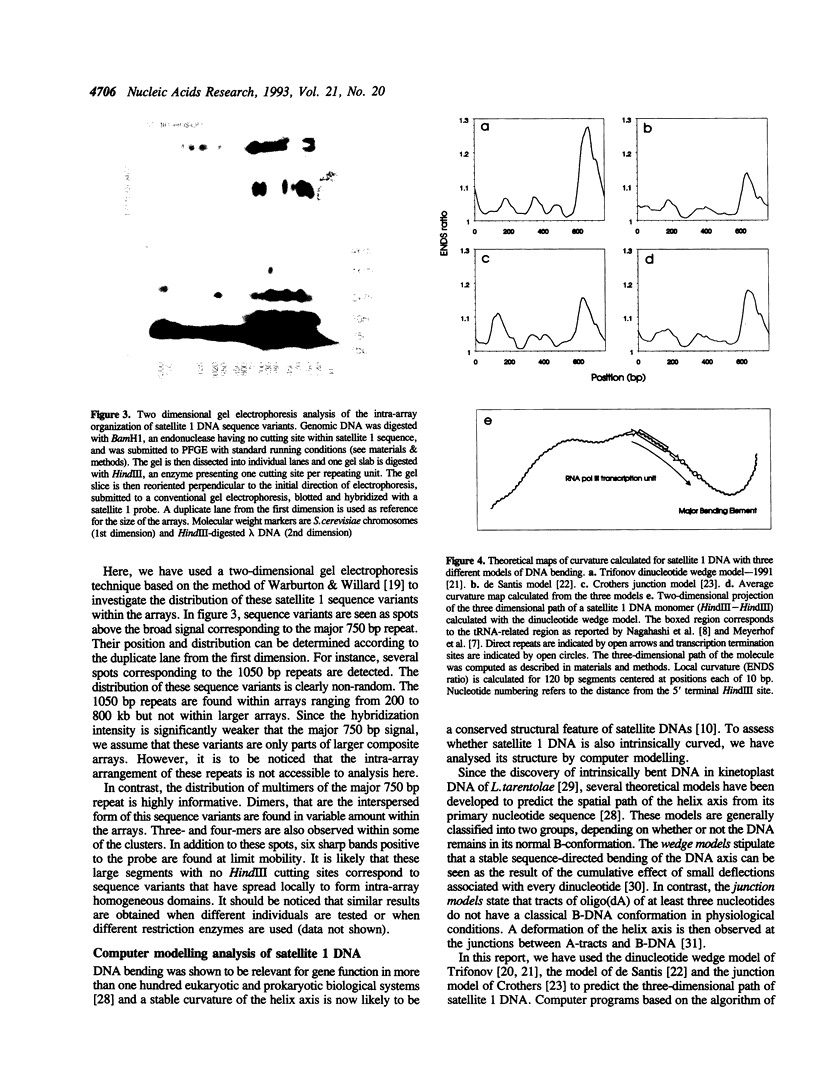
Full text
PDF

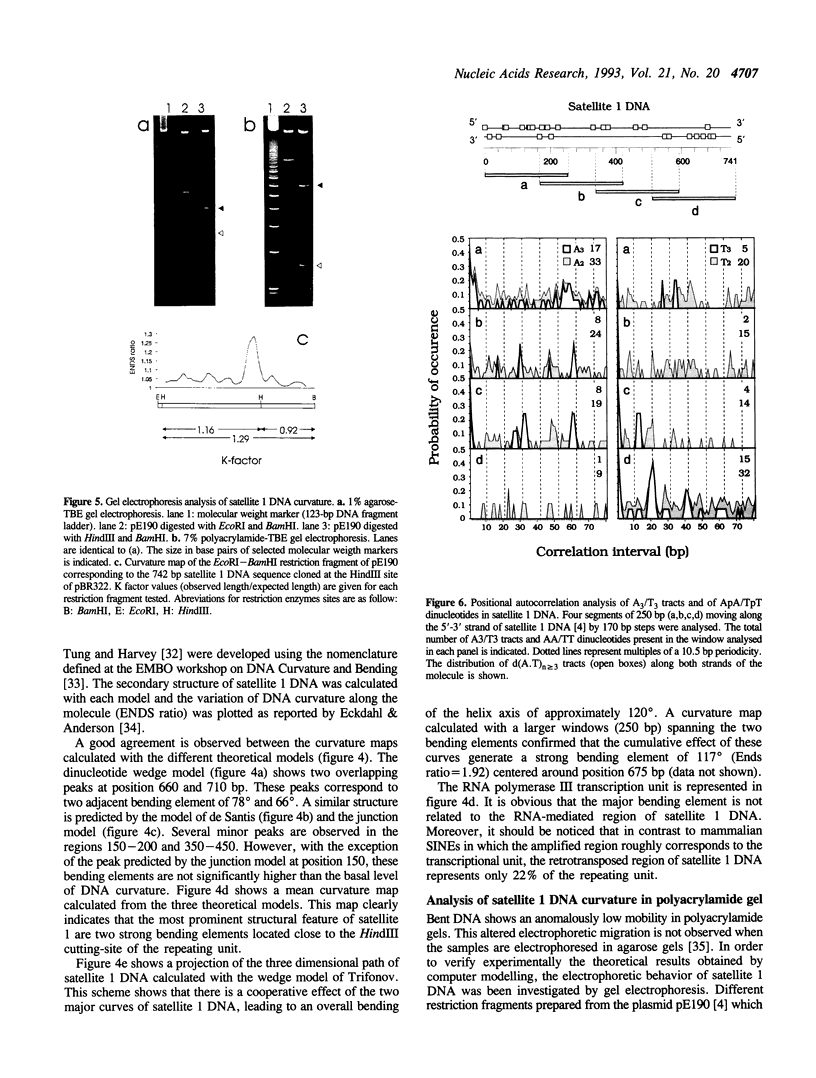


Images in this article
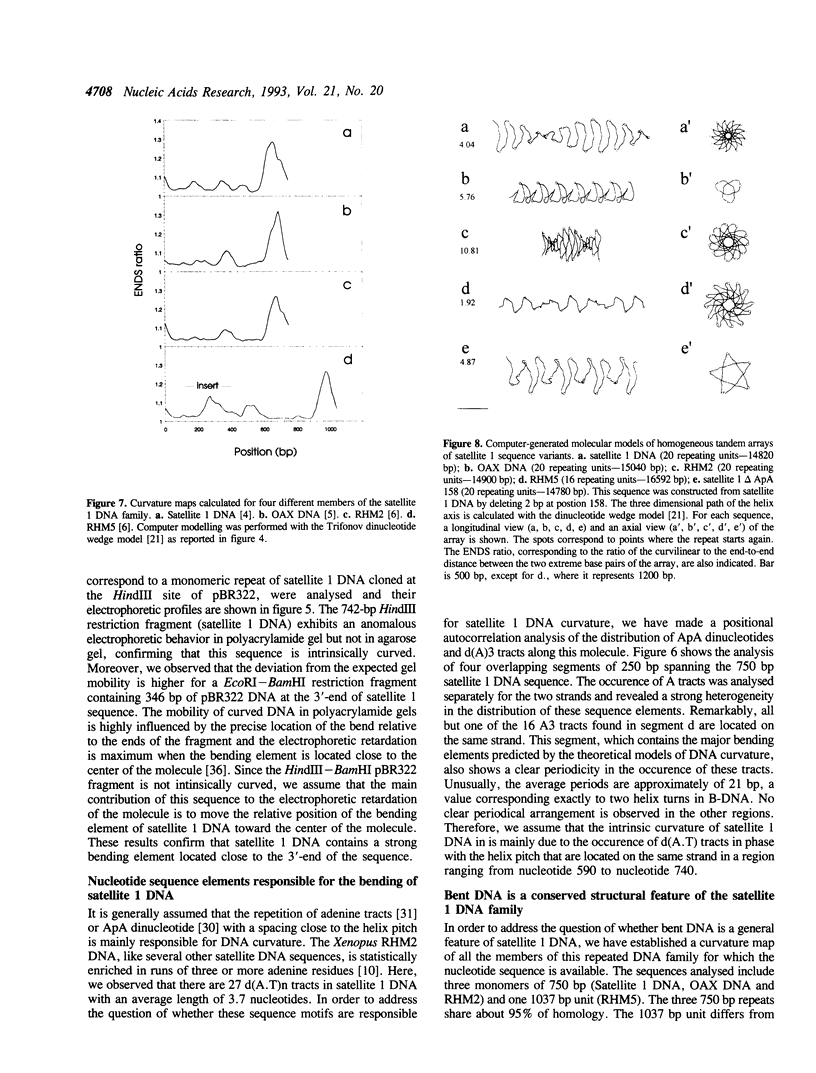
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman E. J. Molecular cloning and sequencing of OAX DNA: an abundant gene family transcribed and activated in Xenopus oocytes. EMBO J. 1983;2(8):1417–1422. doi: 10.1002/j.1460-2075.1983.tb01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila J., Montejo de Garcini E., Wandosell F., Villasante A., Sogo J. M., Villanueva N. Microtubule-associated protein MAP2 preferentially binds to a dA/dT sequence present in mouse satellite DNA. EMBO J. 1983;2(8):1229–1234. doi: 10.1002/j.1460-2075.1983.tb01574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., De Santis P., Palleschi A., Risuleo G., Savino M. A theoretical method to predict DNA permutation gel electrophoresis from the sequence. FEBS Lett. 1992 Mar 30;300(2):175–178. doi: 10.1016/0014-5793(92)80190-r. [DOI] [PubMed] [Google Scholar]
- Bolshoy A., McNamara P., Harrington R. E., Trifonov E. N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Carrera P., Martínez-Balbás M. A., Portugal J., Azorín F. Identification of sequence elements contributing to the intrinsic curvature of the mouse satellite DNA repeat. Nucleic Acids Res. 1991 Oct 25;19(20):5639–5644. doi: 10.1093/nar/19.20.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- De Santis P., Palleschi A., Savino M., Scipioni A. A theoretical model of DNA curvature. Biophys Chem. 1988 Dec;32(2-3):305–317. doi: 10.1016/0301-4622(88)87016-9. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Analyzing DNA curvature in polyacrylamide gels. Methods Enzymol. 1992;212:30–46. doi: 10.1016/0076-6879(92)12004-a. [DOI] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Computer modelling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987 Oct 26;15(20):8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D. S., Kopka M. L., Cascio D., Dickerson R. E. Crystal structure of CATGGCCATG and its implications for A-tract bending models. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Straightening out the bends in curved DNA. Biochim Biophys Acta. 1992 Jun 15;1131(2):125–132. doi: 10.1016/0167-4781(92)90066-9. [DOI] [PubMed] [Google Scholar]
- Hibino Y., Nakamura K., Asano S., Sugano N. Affinity of a highly repetitive bent DNA for nuclear scaffold proteins from rat liver. Biochem Biophys Res Commun. 1992 Apr 30;184(2):853–858. doi: 10.1016/0006-291x(92)90668-b. [DOI] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell. 1988 Feb 26;52(4):535–544. doi: 10.1016/0092-8674(88)90466-7. [DOI] [PubMed] [Google Scholar]
- Jamrich M., Warrior R., Steele R., Gall J. G. Transcription of repetitive sequences on Xenopus lampbrush chromosomes. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3364–3367. doi: 10.1073/pnas.80.11.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C., Trifonov E. N. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982 Feb 11;10(3):1097–1104. doi: 10.1093/nar/10.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam B. S., Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat). J Mol Biol. 1983 Apr 25;165(4):567–585. doi: 10.1016/s0022-2836(83)80267-8. [DOI] [PubMed] [Google Scholar]
- Marilley M., Gassend-Bonnet G. Supercoiled loop organization of genomic DNA: a close relationship between loop domains, expression units, and replicon organization in rDNA from Xenopus laevis. Exp Cell Res. 1989 Feb;180(2):475–489. doi: 10.1016/0014-4827(89)90074-8. [DOI] [PubMed] [Google Scholar]
- Marilley M., Pasero P., Got C. Molecular dissection of a specific nuclear domain: the chromatin region of the ribosomal gene cluster in Xenopus laevis. Exp Cell Res. 1992 Sep;202(1):87–97. doi: 10.1016/0014-4827(92)90407-y. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L. LINEs. Curr Opin Genet Dev. 1991 Dec;1(4):505–508. doi: 10.1016/s0959-437x(05)80199-6. [DOI] [PubMed] [Google Scholar]
- Martínez-Balbás A., Rodríguez-Campos A., García-Ramírez M., Sainz J., Carrera P., Aymamí J., Azorín F. Satellite DNAs contain sequences that induced curvature. Biochemistry. 1990 Mar 6;29(9):2342–2348. doi: 10.1021/bi00461a019. [DOI] [PubMed] [Google Scholar]
- Meyerhof W., Tappeser B., Korge E., Knöchel W. Satellite DNA from Xenopus laevis: comparative analysis of 745 and 1037 base pair Hind III tandem repeats. Nucleic Acids Res. 1983 Oct 25;11(20):6997–7009. doi: 10.1093/nar/11.20.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W., Wittig B., Tappeser B., Knöchel W. Transcription termination and processing of transcripts from tRNA-related Xenopus satellite DNA sequences. Eur J Biochem. 1987 Apr 15;164(2):287–293. doi: 10.1111/j.1432-1033.1987.tb11056.x. [DOI] [PubMed] [Google Scholar]
- Muzard G., Théveny B., Révet B. Electron microscopy mapping of pBR322 DNA curvature. Comparison with theoretical models. EMBO J. 1990 Apr;9(4):1289–1298. doi: 10.1002/j.1460-2075.1990.tb08238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi S., Endoh H., Suzuki Y., Okada N. Characterization of a tandemly repeated DNA sequence family originally derived by retroposition of tRNA(Glu) in the newt. J Mol Biol. 1991 Nov 20;222(2):391–404. doi: 10.1016/0022-2836(91)90218-u. [DOI] [PubMed] [Google Scholar]
- Neuer-Nitsche B., Lu X. N., Werner D. Functional role of a highly repetitive DNA sequence in anchorage of the mouse genome. Nucleic Acids Res. 1988 Sep 12;16(17):8351–8360. doi: 10.1093/nar/16.17.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Pasero P., Marilley M. Size variation of rDNA clusters in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol Gen Genet. 1993 Jan;236(2-3):448–452. doi: 10.1007/BF00277147. [DOI] [PubMed] [Google Scholar]
- Radic M. Z., Lundgren K., Hamkalo B. A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987 Sep 25;50(7):1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. DNA in profile. Trends Biochem Sci. 1991 Dec;16(12):467–470. doi: 10.1016/0968-0004(91)90181-t. [DOI] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. Computer graphics program to reveal the dependence of the gross three-dimensional structure of the B-DNA double helix on primary structure. Nucleic Acids Res. 1986 Jan 10;14(1):381–387. doi: 10.1093/nar/14.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel C., Brassard E., Slater G. W., Noolandi J. Molecular detrapping and band narrowing with high frequency modulation of pulsed field electrophoresis. Nucleic Acids Res. 1990 Feb 11;18(3):569–575. doi: 10.1093/nar/18.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Vogt P. Code domains in tandem repetitive DNA sequence structures. Chromosoma. 1992 Oct;101(10):585–589. doi: 10.1007/BF00360534. [DOI] [PubMed] [Google Scholar]
- Vogt P. Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved "chromatin folding code". Hum Genet. 1990 Mar;84(4):301–336. doi: 10.1007/BF00196228. [DOI] [PubMed] [Google Scholar]
- Vollrath D., Davis R. W. Resolution of DNA molecules greater than 5 megabases by contour-clamped homogeneous electric fields. Nucleic Acids Res. 1987 Oct 12;15(19):7865–7876. doi: 10.1093/nar/15.19.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton P. E., Willard H. F. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J Mol Biol. 1990 Nov 5;216(1):3–16. doi: 10.1016/s0022-2836(05)80056-7. [DOI] [PubMed] [Google Scholar]
- Willard H. F. Evolution of alpha satellite. Curr Opin Genet Dev. 1991 Dec;1(4):509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]