Abstract
Pathogenic bacteria frequently cloak themselves with a capsular polysaccharide layer. Escherichia coli group 1 capsules are formed from repeat-unit polysaccharides with molecular weights exceeding 100 kDa. The export of such a large polar molecule across the hydrophobic outer membrane in Gram-negative bacteria presents a formidable challenge, given that the permeability barrier of the membrane must be maintained. We describe the 2.26 Å structure of Wza, an integral outer membrane protein, that is essential for capsule export. Wza is an octamer, with a composite molecular weight of 340 kDa, and it forms an “amphora”-like structure. The protein has a large central cavity 100 Å long and 30 Å wide. The transmembrane region is a novel α-helical barrel, and is linked to three additional novel periplasmic domains, marking Wza as the representative of a new class of membrane protein. Although Wza is open to the extracellular environment, a flexible loop in the periplasmic region occludes the cavity and may regulate the opening of the channel. The structure defines the route taken by the capsular polymer as it exits the cell, using the structural data we propose a mechanism for the translocation of the large polar capsular polysaccharide.
Introduction
Many bacteria produce extracellular polysaccharides (EPSs). Some are secreted polymers and show only limited association with the cell surface, while others are firmly attached to the cell surface and form a discrete structural layer enveloping the cell, known as the capsule. In pathogens, the role of EPS is protective; capsules provide essential virulence determinants that allow the bacteria to evade or counteract the host immune response1. EPSs play critical roles in forming biofilms and in the colonization of surfaces2-4, such as epithelia and medical implants. Therefore, the synthesis and translocation of EPSs represents a promising target for therapeutic intervention. Further, some EPSs have important commercial and medical applications5 in their own right.
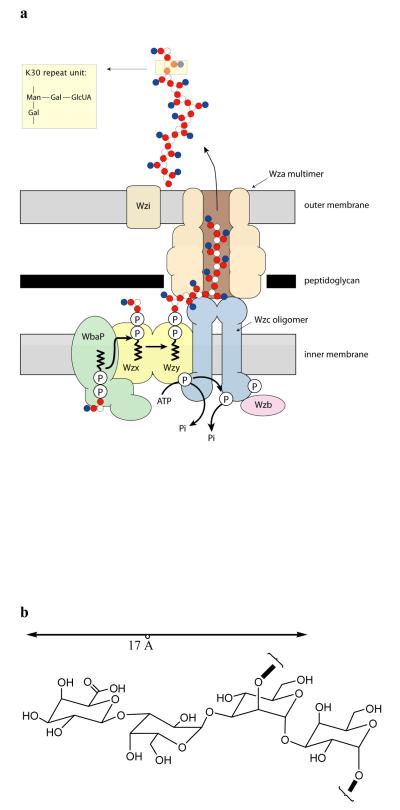
EPSs show immense diversity in monomer composition, linkage sequence and type, and substitution with non-carbohydrate residues. They are linear or branched polymers with reported sizes typically ranging from 104-106 daltons. EPSs therefore represent one of the largest and most polar molecules to be transported across a biological membrane and the desolvation of hundreds of carbohydrate rings during passage across the cell envelope seemingly presents an insurmountable kinetic barrier to export. Escherichia coli has provided the model for EPS biosynthesis and assembly6. There are two assembly pathways in E. coli and these are fundamentally different in terms of the mechanism and membrane topology of the polymerization process. The defining characteristic of group 1 (Wzy dependent) pathway is that individual lipid-linked polymer repeat units are synthesized and exported to the periplasm, where a putative polymerase (Wzy) assembles the polymer in a block-wise elongation process. E. coli capsular (K antigen) serotype K30 is the prototype for the group 1 capsule assembly system and a model depicting current understanding of the pathway is shown in Fig. 1a. The K30 polysaccharide is assembled and exported by dedicated proteins encoded by a 12-gene operon7 and includes a member of the OMA (outer membrane auxiliary) protein family8 for the export (or translocation) of nascent polymer across the outer membrane. The best studied OMA family member is Wza from the E. coli group 1 K30 system.
Figure 1. Group 1 capsular polysaccharide export in Gram-negative bacteria.
a, Model and proposed activities of a hypothetical biosynthetic complex carrying out coordinated synthesis and export of serotype K30 group 1 capsule in E. coli6. Individual tetrasaccharide repeat units of the polymer47 are assembled by a series of enzymes including integral or peripheral membrane proteins on a lipid (undecaprenol diphosphate; und-PP) acceptor, using sugar nucleotide precursors available in the cytoplasm. The und-PP-linked repeat units are “flipped” across the inner membrane by a process involving an integral membrane protein (Wzx). Polymerization occurs at the periplasmic face and is dependent on another integral membrane protein, the putative polymerase, Wzy. High-level Wzy-dependent polymerization requires the activity of the tetrameric Wzc protein12. To be active in capsule assembly, Wzc must undergo autophosphorylation13. Dephosphorylation of Wzc by the Wzb phosphatase is also crucial for capsule synthesis, suggesting a need for cycling of the phosphorylation state of Wzc14. Export of polymer to the surface requires the outer membrane Wza octameric complex9,10,16. Biochemical10 and genetic11 data indicate that the Wza and Wzc proteins interact to form a complex that potentially spans the periplasm. b, A simple molecular model of the carbohydrate polymer, assuming the most extended conformation of the sugar, the maximum width that the channel has to accommodate at any point is approximately 17 Å.
Mature Wza is a 359 amino acid protein that is synthesized as a precursor with a cleavable 20 residue N-terminal signal sequence. Wza is a lipoprotein9. Cys 21 is modified by a thioether-linked diacylglyceryl group and its amino group is also acylated. Wza forms oligomers that are stable even in SDS9. Preliminary electron microscopy (EM) studies of two-dimensional crystals of Wza protein in lipid bilayers revealed octameric ring-like structures, leading to the proposal that Wza may form a channel to facilitate translocation of nascent polymer9. “Knock-out” mutants lacking wza have no capsular polysaccharide and are unable to synthesize detectable intracellular polymer9,10. This suggests a feedback process in which synthesis and export are coupled. Interestingly, Wza interacts with an inner membrane tyrosine autokinase protein, Wzc10,11. Negatively stained cryo-EM reveals that Wzc forms a tetrameric complex12. Mutations that eliminate Wzc, or that compromise its phosphorylation, also turn off capsular polymer biosynthesis13,14. A non-acylated Wza mutant (Cys21Ala) forms oligomers with poor stability and polymer accumulates in the periplasm10. One interpretation is that the non-acylated oligomer is not fully competent for polymer translocation but does not compromise those interactions with Wzc that are required to sustain polymer synthesis.
The overall sequence relationships between OMA proteins are limited9, but family members do share some regions of similarity that contain the polysaccharide biosynthesis/export (PES) motif (Pfam02563)15 (Supplementary Fig. S1b, Fig. S2). Single particle Wza octamers visualized by negative staining cryo-EM (15.5 Å resolution) reveal a barrel-like structure of dimensions 90 Å × 90 Å × 100 Å16. Although the study identified a large central cavity, it did not locate a route for the polysaccharide or an unambiguous orientation of the barrel with respect to the membrane. As a result two key questions remain unanswered. How does Wza specifically export a large polar molecule through the outer membrane? What structural feature allows the channel to facilitate export of a large branched polysaccharide? The 2.26 Å resolution crystal structure reported here begins to address these questions and provides insight into processes that may be involved in the translocation of other large polar molecules across bacterial outer membranes.
The structure of Wza
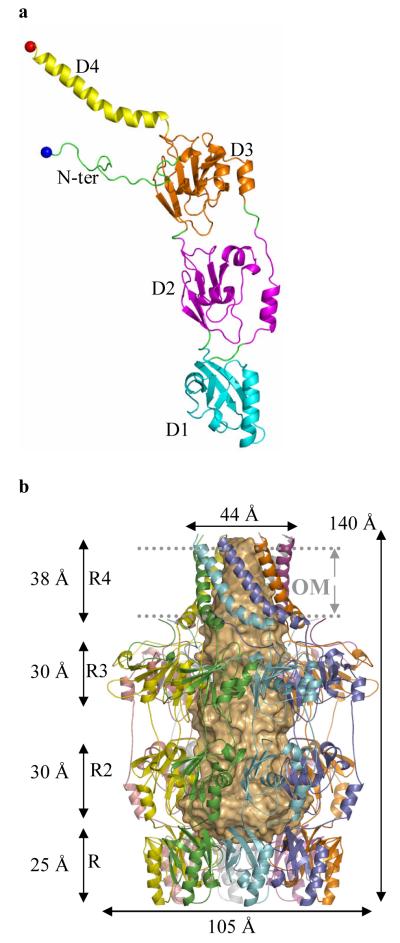
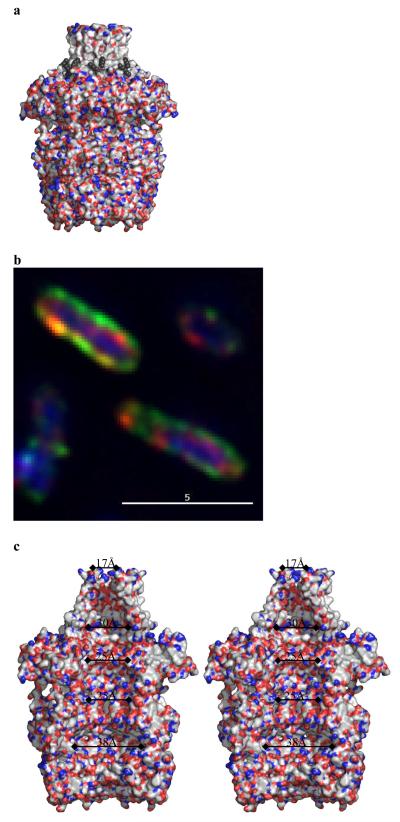
The structure of mature acylated Wza from E. coli K30 was solved using single-wavelength anomalous diffraction with selenomethionine labeling (Fig. 2a). The protein crystallizes in space group P212121 with eight monomers in the asymmetric unit. From the location of the selenium atoms, it was clear the asymmetric unit contained a molecule with eight-fold rotational symmetry, consistent with an octamer of molecular weight 320 kDa. The high quality of the electron density map allowed 2848 amino acid residues, 16 lipid fragments, 16 sulfate ions and 1264 water molecules to be built. The octameric structure is best described as having the shape of a classical “amphora” without the handles (Fig. 2b). Wza has a large internal cavity which is opens at a narrow “neck” and is closed at its base. The long axis of the molecule is approximately 140 Å and the diameter at the widest point is 105 Å (Fig. 2b,c). The eight-fold rotational axis runs parallel to the long axis of molecule. Comparison of the crystal structure to that derived from negatively-stained cryo EM16 reveals that the “neck” is missing in the EM structure. This region may be unstructured in the conditions of the EM experiment, or it may interact poorly with the stain. The dimensions of the remainder of the structure match well; in particular the large central cavity is seen. The EM structure derived from single-particle analysis had only four-fold rotational symmetry suggesting conformational change induced by negative staining16, although positively-stained samples of 2-dimensional crystals in proteolipid bilayers showed eight-fold symmetry10.
Figure 2. The structure of Wza.
a, The monomer of Wza can be decomposed into four domains; these are labeled as D1 - 4 and colored differently. Cys 21 (fatty acid modified N-terminus in mature Wza) is shown as a blue ball and Arg 376 (the last ordered residue) as a red ball. A stereo figure is shown in Supplementary Fig. S1a. b, The Wza octamer is shown in ribbon format. In this view the large central cavity is highlighted by space-filling light orange shape. The octamer is described as an amphora made of four rings (labeled R1-4). Ring 1 is formed by 8 copies of domain 1, ring 2 by 8 copies of domain 2 and so on. The helical barrel (R4) forms the “neck” of the structure and ring 1 (R1) the “base”. The predicted position of the outer membrane (OM) is marked and is found at the neck of the structure. The C-terminus of protein is predicted to be exposed on the cell surface and rings 1,2 and 3 located inside the periplasm. c, A view of the octamer such that one looks down into the central cavity from outside the cell through the helical barrel. The separation of loops which close the cavity in ring 1 are marked on the diagram. d, Wza is shown as a space filling model colored according to electrostatic charge and is rotated 180° from the view in Fig. 2c, such that one is now looking towards the cavity from periplasm. The concave surface of ring 1 can be seen and is closed by the loop at Tyr 110. There is a band of negative charge on the base of the structure. This may provide the association interface for protein-protein interactions with Wzc.
In describing the protein monomer, we reduce it to four domains (Fig. 2a). Domain 1 (residues 89-169) is an anti-parallel β-sandwich with an α-helix at one edge and represents a novel fold. Domain 1 contains the conserved PES motif (Supplementary Fig. S1b, S2). The eight copies of domain 1 combine to form a ring structure (ring 1) at the bottom of the Wza, with an eight-fold axis through the centre (Fig. 2b,c). Ring 1 presents a concave surface at the base of the structure and the centre is filled by eight loops (residues 105 to 112 of each domain). Measuring from the main chain of these loops, the channel is narrowed to 15 Å. Tyr 110 is located at the tip of the loop but is not clearly visible in the experimental map, suggesting the side chain is flexible (Supplementary Fig. S3a). Once past Tyr 110, the ring has an internal diameter of over 25 Å (Fig. 2b,c). Domain 2 (residues 68-84 and 175-252) also has a novel structure, despite having similar overall dimensions to domain 1. This domain has a central five-stranded mixed β-sheet with three α-helices on one face. The eight copies of domain 2 form an eight-fold symmetric ring structure (ring 2) with an inner diameter of approximately 25 Å. Domain 3 (residues 46-64 and 255-344) is a structural duplication of domain 2 (Supplementary Fig. S1c). The eight copies of domain 3 form a ring structure (ring 3, Fig. 2b). In domain 3, a longer loop connects two β-strands and there is an extra β-strand (residues 49 to 54) compared to domain 2 (Supplementary Fig. S1c). As a result, the third ring has a significantly larger external diameter of 105 Å than that of ring 2. The three rings sit one atop the other with a 10 Å spacing between rings 2 and 3. These three rings form the body of the amphora. A ribbon representation of the structure (Fig. 2b) gives the appearance of side holes being present, but these are filled by side chains (Fig. 3a). Domain 4 comprises the C-terminus of the monomer (residues 345-376) (Fig. 2a) and is an amphipathic helix. As a result of the eight-fold rotational symmetry, the helices create an α-helical barrel (Fig. 2b) at the “neck” of the molecule. The axis of each helix is offset by approximately 35° to the long axis of the molecule and is kinked at Pro 359. The barrel is tapered such that part attached to the third ring has an internal diameter of approximately 30 Å. At the open end the internal diameter is 17 Å. The N-terminus (residues 22 to 45) is long loop which is wrapped around the top of ring 3 (Fig. 2a, b).
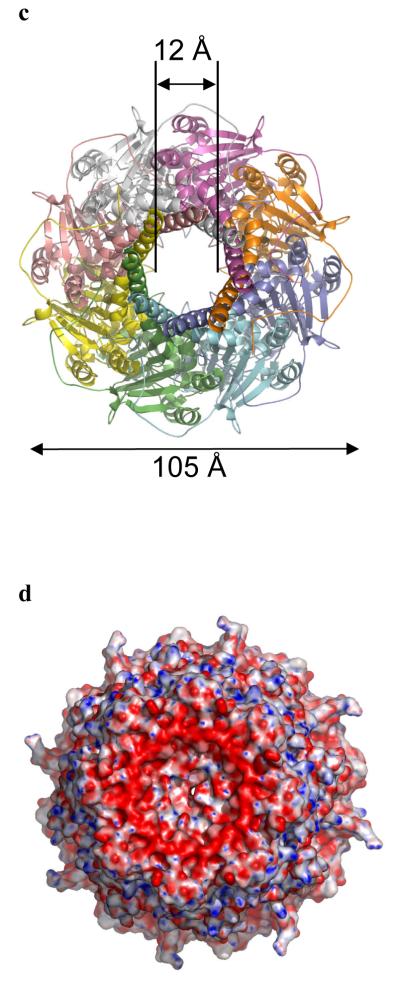
Figure 3. The central cavity of Wza.
a, Wza is shown as a surface with polar atoms colored (colors are Fig. 2c.). Wza is oriented as the Fig. 2b. The lipid molecules are shown as black spheres and are located near the top of the structure. There are no gaps in the through the walls of the structure. The helical barrel is clearly non-polar and a band of tryptophan residues is exposed at the base of helical barrel (Fig S4b). b Green fluorescent anti-PK antibody binds to the surface of the cells which express Wza with a PK tag added to the C-terminus (Wza-PK). The periplasm is located by a red anti- alkaline phosphatase antibody and the nucleus is stained blue by DAPI. This confirms the C-terminus of Wza-PK is exposed on the surface of cells and thus the orientation of Wza is as shown in Fig. 2b. c, A stereo diagram of the internal cavity. The surface is colored according to polarity, (oxygen red, nitrogen blue and carbon, selenium, sulfur white) and reveals the interior of Wza is polar. The cavity is open at top through the helical barrel.
Wza has a large central cavity
The central cavity of the protein is open to the solvent (Fig. 2b, Fig. 3a) and has an internal volume of approximately 15000 Å3. This large volume is comparable to the 84000 Å3 reported for chaperone GroEL GroES17 and 44000 Å3 for the outer membrane drug efflux protein TolC18. In Wza, the protein surface which encloses the central cavity is polar (Fig. 3a) and lined by residues which exhibit little sequence conservation. Those conserved residues that do exist are principally structural, or are located at subunit interfaces (Supplementary Fig. S2). Examining the exterior surface of Wza reveals that it is almost all polar (Fig. 3a). The exception is the helical barrel which has a markedly hydrophobic exterior (Fig. 3a). Notably Trp 350 is exposed on the surface of the barrel (Supplementary Fig S4b). Exposed tryptophan residues are commonly observed in integral membrane proteins. The lipid modified N-terminii (Cys 21) are adjacent to the helical barrel, placing the acyl chains in an ideal position to intercalate into the inner leaflet of the outer membrane. We have identified lipid molecules bound to the outside of the barrel (Fig 3a. and Supplementary Fig. S3b), at least one of which may be connected to Cys 21. We identify the helical barrel as the transmembrane region with bulk of the structure periplasmic and the C-terminus exposed on the cell surface. This orientation has been experimentally confirmed by light microscopy. Wza, modified by addition of the 14 residue PK epitope to the C-terminus, was expressed in E. coli. Addition of fluorescent anti-PK antibodies to these cells lead to the outside of the E. coli becoming fluorescent (Fig 3b.). Analysis by flow cytometry showed around 12% of Wza-PK transformed cells bind fluorescently labeled anti-PK antibody, consistent with direct visualization (Supplementary Fig. S5). The relatively low proportion of cells expressing the protein is a known feature of the arabinose inducible pBADD expression system.
While typical cell membranes (including the bacterial inner membrane) are composed of a phospholipid bilayer, the outer membrane is distinct and asymmetric. The inner leaflet is composed of glycerophospholipids and the outer leaflet contains the unique glycolipid, lipopolysaccharide19. To date, all structures of integral outer membrane proteins have contained a trans-membrane β-barrel20,21. Wza is the first example of an α-helical barrel which crosses the outer membrane of bacteria. Although transmembrane α-helices are well known in bacterial inner membrane proteins and eukaryotic membrane proteins, there is no corollary for the transmembrane helical barrel observed in Wza. Wza, therefore expands the repertoire of known trans-membrane protein structures. Interestingly, many antimicrobial peptides are known to form amphipathic helices22. The C-terminus of Wza may serve as a structural model of how antimicrobial peptides aggregate in the membrane to disrupt outer membrane integrity and create pores.
The route for export of polysaccharide
The cross-sectional area of the carbohydrate polymer is difficult to estimate reliably, because of the inherent conformational flexibility of the glycosidic linkages. We have constructed a model of the repeating tetrasaccharide unit of the K30 capsule and, assuming the most extended conformation, the model gives an estimated width of 17 Å (Fig. 1b). The helical barrel with its diameter of 17 Å would be sufficient to accommodate the exit of the polymer from the large central cavity (Fig. 2c, Fig. 3a) without structural change. However, a continually open 17 Å wide channel would present a problem in terms of maintaining outer membrane barrier function. Therefore some form of regulation of the channel opening would be essential. Isolated Wza multimers do not allow transmission of ions across model lipid bilayers suggesting the channel is normally closed (C. Whitfield and R.E.W. Hancock, unpublished data). This is consistent with the crystal structure which shows no portal between the central cavity and periplasm. In one conformer, the disordered Tyr 110 can completely seal the central cavity at the base of the structure in the periplasm (Fig. S3c). In another conformer, a portal of diameter of around 8 Å could be created (Fig. S3d). An 8 Å portal could be wide enough to accommodate the carbohydrate, although this would require a very compressed conformation of the polymer. The trimeric β-barrel outer membrane porin proteins facilitate diffusion of small hydrophilic compounds (typically nutrients)23 by creating permanently open channels of around 8 Å24 diameter, but they do not export large polysaccharides. In Wza, the loop to which Tyr 110 is attached (Fig. 2c, Fig. S3a,b,c) could undergo a conformation change creating a significantly wider portal. The trigger for such hypothetical conformation changes is unknown but the binding of Wzc seems a logical candidate. Wza can be cross linked to Wzc in both solution and cells, suggesting their periplasmic structures do interact10. Looking at the base of Wza, the circle of negative charge and a concave surface are suggestive of a site for protein-protein interaction (Fig. 2d). The use of conformational changes to gate large channels is seen elsewhere in outer membrane proteins. For example, FhuA which imports an iron siderophore25, relies on a conformational change in a “cork” domain26. TolC18 forms an integral membrane β-barrel with a diameter of 35 Å which is closed at one end. Conformational changes in TolC’s periplasmic domain are thought to mediate an “iris-like” mechanism to open the channel18. Both TolC and Wza are efflux proteins which would appear to use a periplasmic gating mechanism. Wza may represent an excellent model for the mechanism of export of toxins, enzymes and effector proteins via multimeric “secretin” protein complexes found in type II, III and IV secretion systems in bacteria27. A particularly intriguing example is the export of single-stranded DNA (a large polar molecule) via type IV systems28. EM structures of several secretins have been reported29-31 and these form ring-like structures which are capable of accommodating larger substrates.
The structural basis for carbohydrate export by Wza
The lack of conserved residues lining the cavity indicates there is little specific recognition between protein and carbohydrate. This is consistent with the observation that the wza genes are essentially identical in the chromosomal loci responsible for expression different E. coli group 1 capsule serotypes32. Moreover, related Wza homologues from other polysaccharide systems can complement Wza-deficient E. coli K309,11. EPS is essential for survival of the bacterium in its particular environmental niche and is subject to selective pressure from host and other environmental factors. This is reflected in the enormous diversity in capsule composition. By utilizing a non-selective translocase, serotype divergence can be achieved by modifying only the glycosyltransferases and polymerase.
In the transport of potassium ions, one has to consider the desolvation penalty for a single ion. The structure of potassium transporter revealed that specific recognition by acidic side chains inside the channel33 mediates transport. The ammonia/ammonium transporter uses a series of histidine residues to bind its substrate34 during transit. Carbohydrates make a very large number of hydrogen bonds; one glucose molecule, for example, can make 17 hydrogen bonds. The desolvation of hydroxyl groups is energetically unfavorable. Lactose permease solves this problem by making hydrogen bonds to the two carbohydrate rings inside a central cavity35 which then undergoes a conformational change releasing the encapsulated sugar on the opposite side of the membrane. A similar model has been proposed for the ATP-dependent lipopolysaccharide lipid A flippase, MsbA, in which the carbohydrate head group is sequestered inside a central cavity whilst the protein undergoes conformational change36. For an EPS chain, which has hundreds of carbohydrate rings, making hydrogen bonds with the protein is a formidable challenge. Although extended carbohydrate binding sites in proteins are well known37, the characteristic of such extended sites is their specificity. Whilst beneficial in binding a single repeat unit of the sugar, such recognition would hinder export in which the polymer is constantly moving, effectively changing the spatial configuration of polar atoms in contact with the protein. Efficient export must avoid both low and high energy intermediates. Using protein side chains to recognize multiple carbohydrates and all their possible intermediates would seem an almost impossible task. We propose that Wza has evolved a particularly elegant solution to this problem. Its large internal polar cavity (Fig. 3c) would allow the water molecules to remain inside whilst the carbohydrate is present. During the export of the polysaccharide, we propose that water molecules mediate protein carbohydrate hydrogen bonds, in effect solvating both. As the polymer moves through the Wza, the water molecules, akin to ball bearings, “roll” across the protein surface. As the movement of water across a polar surface is a relatively low energy process the export process avoids any significant kinetic barriers. The structure of Wza defines a new class of integral membrane protein and suggests a mechanism for the transport of the capsular polysaccharide across the bacterial outer membrane. This work lays the structural foundation for the development of novel antimicrobial strategies aimed at blocking the translocation of capsule. Transport processes across membranes are fundamental in biology and the structure Wza suggests a novel mechanism for the translocation of large polar molecules.
Methods and Materials
Structural Biology
Full details of the purification and crystallization of the native protein have been published elsewhere 38. Briefly, the crystals were grown from hanging drops containing 6 mg/ml protein in 0.008 % DDM, 100 mM NaCl and crystallization condition 8 % (w/v) PEG4000, 100 mM sodium citrate pH 4.6, 50 mM NaCl and 50 mM MgSO4. A two-step protocol was used to make selemethionine labeled Wza. E. coli LE392 cells were transformed with plasmid pWQ126 (pBAD24 expressing Wza9). The cells were grown in M9 minimal medium containing 2 % (w/v) casamino acids until the OD600nm reached to 0.8. The cells were then harvested and washed twice using M9 medium, and then transferred into the same amount of the M9 medium containing a mixture of all L-amino acids (except methionine and cysteine) each at a final concentration of 50 mg/L, together with 50 mg/L selenomethonine. The selenomethionine-labeled Wza was over-expressed by induction using 0.008 % (w/v) L-arabinose and incubation overnight at 30 °C. Purification of the labeled protein was the same as for the native protein. However, the crystallization condition was optimized to 7 % PEG4000, 0.1 M sodium acetate pH 4.6 and 0.2 M (NH4)2SO4 with 10 mg/ml protein containing 80 mM NaCl, 0.008 % (w/v) DDM and 20 mM Tris pH 8.0. Mass spectrometry confirmed Wza has full incorporation of selenium and does have the classical diacylgycerol and N-acylation lipoprotein modification (Supplementary Fig. S6). The crystals of native Wza and, especially the selenomethionine derivative, are highly variable and even within the same crystal some regions give better data than others. The 2.26 Å data set was collected on ID29 of the ESRF on a single selenomethionine crystal protected with 25 % (w/v) glycerol in the mother liquid and cooled to 100 K. Data were recorded on an ASDC 3 × 3 tiled array CCD detection. Six data sets were collected at three wavelengths using different crystals and reduced with MOSFLM / SCALA39. In the end, only one peak wavelength data set allowed location of 78 (out of 80) selenium ions by SHELX40 using data to a resolution up to 3.2 Å. The correct solution had a frequency of 1 in 600 trials. Once located however, the phases from the selenium atoms were of a high enough quality to begin to trace the structure. Refinement of the sites, density modification and eight-fold averaging gave a map that was of exceptionally high quality. Arp/Warp41 built the entire structure automatically when the resolution extended to 2.26 Å. The structure was refined using REFMAC42,43, waters and sulfate ions added. Two bands of sulfate ions are bound inside the first ring, water molecules are found on both the internal and external surface. Only the final three C-terminal residues are not located. Interestingly, unlike other membrane proteins which pack as type I or II44, the crystal packing of the Wza resembles that of soluble proteins. This may reflect the novel structure of its membrane spanning region. During refinement density became visible for portions of the lipid molecules; these were fitted conservatively as aliphatic chains. Full statistics are given in Table S1, coordinates and data have been deposited with the Protein Data Bank.
Imaging (fluorescence microscopy) and flow cytometry
The paramyxovirus SV5 “PK” tag was added to the carboxyl terminus of Wza to make Wza-PK. The residues were added by the polymerase chain reaction using the forward primer AGGAGGAATTCATGAAGAAAAAACTTGTTAGATTTTCG (the EcoRI site is underlined). The reverse primer has a PstI site (underlined) and codes for the additional 14 amino acids (Gly Lys Pro Ile Pro Asn Pro Leu Leu Gly Leu Asp Ser Thr) (shown in bold and underlined in the primer sequence) which code for the “PK” epitopte. GCATGCTGCAGTTAGGTGGAGTCCAATCCCAGCAAAGGGTTTGGGATCGGCTTTCC GTTGGGCCATCTCTTAATGTATC The addition of the tag was confirmed by DNA sequencing and its integrity of the tag by western blotting using PK monoclonal antibody as primary antibody (mouse) (kindly provided by R. Randall) and second antibody (anti -mouse) as described45. (Hanke T et al, J Gen Virol 73:653-60(1992))(Fig. S5a). Expression of Wza-PK was carried out using the same cells and protocol as for crystallization and the cells were harvested after 2 hour induction. Controls of induced wild type Wza (Wt-Wza) expressing cells and non-induced PK-tagged Wza expressing cells (PK-control) were grown in parallel. The cells were directly visualized with a Delta Vision restoration microscope (Fig. 3.b, S5b,c). For direct visualization the cells, the cells were washed twice by PBS and incubated with green fluorescent FITC-conjugated PK monoclonal antibody overnight at 4°C, followed three times washes by PBS following a previously described method46. The cells were plated on the poly-Lys coated slides and then fixed with 4% paraformaldehyde. The cytoplasm was stained with 4′,6-diamidino-2-phenylindole (DAPI) which stains DNA blue. The cells were thoroughly washed and permeablised. They were then incubated with red fluorescent anti-alkaline phosphatase antibody (Abcam Ltd) to visualize the periplasm. For flow cytometry the same three populations of cells were examined. They were incubated with FITC-conjugated PK antibody) as described as above and fixed in 4% paraformaldehyde, and the cells were then stained by 7-aminoactinomycin D which stains DNA and enables bacteria to be distinguished from other particulates in the sample and machine. Samples were analyzed on an LSR fitted with beam-switch (Becton Dickinson) using CellQuest acquisition software and FlowJo software (Treestar inc) at the Dundee University FACS unit (Fig. S5d).
Supplementary Material
Acknowledgements
JHN is a Biotechnology and Biology Sciences Research Council (BBSRC) Career Development Fellow. CW holds a Canada Research Chair and acknowledges funding from the Canadian Institutes of Health Research. The experimental structural biology was performed by the Scottish Structural Proteomics Facility which is funded by the Scottish Higher Education Funding Council and the BBSRC. We acknowledge the use of ESRF beamlines and are grateful for assistance with data collection. We thank Leslie Cuthbertson for assistance with bioinformatic analyses, Dominique Bourgeois for help with data collection on ID-29, Margaret Taylor for confocal imaging, Rosemary Clarke for FACS, Garry Taylor, Malcolm White and Bill Hunter for a critical review of the manuscript.
References
- 1.Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 2.Donlan RM, Costerton JW. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland IW. Novel and established applications of microbial polysaccharides. Trends Biotechnol. 1998;16:41–46. doi: 10.1016/S0167-7799(97)01139-6. [DOI] [PubMed] [Google Scholar]
- 6.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 7.Drummelsmith J, Whitfield C. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a: K30) Mol. Microbiol. 1999;31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 8.Paulsen IT, Park JH, Choi PS, Saier MH. A family of Gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from Gram-negative bacteria. FEMS Microbiol. Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 9.Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO. J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesper J, et al. Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30. Structural and functional analysis of the outer membrane lipoprotein Wza. J Biol Chem. 2003;278:49763–72. doi: 10.1074/jbc.M308775200. [DOI] [PubMed] [Google Scholar]
- 11.Reid AN, Whitfield C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: Evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 2005;187:5470–5481. doi: 10.1128/JB.187.15.5470-5481.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins RF, et al. Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J. Biol. Chem. 2006;281:2144–2150. doi: 10.1074/jbc.M508078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wugeditsch T, et al. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J. Biol. Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 14.Paiment A, Hocking J, Whitfield C. Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J. Bacteriol. 2002;184:6437–6447. doi: 10.1128/JB.184.23.6437-6447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman A, et al. The Pfam Protein Families Database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beis K, et al. Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J. Biol. Chem. 2004;279:28227–28232. doi: 10.1074/jbc.M402913200. [DOI] [PubMed] [Google Scholar]
- 17.Xu ZH, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)(7) chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 18.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 19.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 21.Schulz GE. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta, Biomembr. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 22.Bulet P, Stocklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. Restoring permeability barrier function to outer membrane. Chem. Biol. 2005;12:507–509. doi: 10.1016/j.chembiol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Cowan SW, et al. Crystal-Structures Explain Functional-Properties of 2 Escherichia coli Porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 26.Eisenhauer HA, Shames S, Pawelek PD, Coulton JW. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J. Biol. Chem. 2005;280:30574–30580. doi: 10.1074/jbc.M506708200. [DOI] [PubMed] [Google Scholar]
- 27.Bayan N, Guilvout I, Pugsley AP. Secretins take shape. Mol. Microbiol. 2006;60:1–4. doi: 10.1111/j.1365-2958.2006.05084.x. [DOI] [PubMed] [Google Scholar]
- 28.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linderoth NA, Simon MN, Russel M. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science. 1997;278:1635–1638. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 30.Nouwen N, et al. Secretin PulD: Association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins RF, et al. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 angstrom resolution. J. Biol. Chem. 2004;279:39750–39756. doi: 10.1074/jbc.M405971200. [DOI] [PubMed] [Google Scholar]
- 32.Rahn A, Drummelsmith J, Whitfield C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: Relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 1999;181:2307–2313. doi: 10.1128/jb.181.7.2307-2313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou YF, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 angstrom resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 34.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.3.5 angstrom. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 35.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 36.Reyes CL, Chang G. Structure of the ABC transporter MsbA in complex with ADP-vanadate and lipopolysaccharide. Science. 2005;308:1028–1031. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- 37.Moothoo DN, Naismith JH. Concanavalin A distorts the β-GlcNAc-(1-2)-Man linkage of β-GlcNAc-(1-2)-α-Man-(1-3)-[β-GLcNAc-(1-2)-α-Man-(1-6)]-Man upon binding. Glycobiology. 1998;8:173–181. doi: 10.1093/glycob/8.2.173. [DOI] [PubMed] [Google Scholar]
- 38.Beis K, Nesper J, Whitfield C, Naismith JH. Crystallisation and preliminary X-ray diffraction analysis of Wza- outer membrane lipoprotein from E. coli serotype O9a:K30. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003;60:558–560. doi: 10.1107/S0907444903029494. [DOI] [PubMed] [Google Scholar]
- 39.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB newsletter on protein crystallography. 1992;26:1–10. [Google Scholar]
- 40.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 41.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 43.CCP4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Ostermeier C, Michel H. Crystallization of membrane proteins. Curr. Opin. Struct. Biol. 1997;7:697–701. doi: 10.1016/s0959-440x(97)80080-2. [DOI] [PubMed] [Google Scholar]
- 45.Hanke T, Szawlowski P, Randall RE. Construction of Solid Matrix Antibody Antigen Complexes Containing Simian Immunodeficiency Virus-P27 Using Tag-Specific Monoclonal-Antibody and Tag-Linked Antigen. J. Gen. Virol. 1992;73:653–660. doi: 10.1099/0022-1317-73-3-653. [DOI] [PubMed] [Google Scholar]
- 46.McNulty C, et al. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: the role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol. Microbiol. 2006;59:907–922. doi: 10.1111/j.1365-2958.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 47.Chakraborty AK, Friebolin H, Stirm S. Primary structure of the Escherichia coli serotype K30 capsular polysaccharide. J. Bacteriol. 1980;141:971–972. doi: 10.1128/jb.141.2.971-972.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.