During evolution, the yeast coactivator SAGA complex, which is involved in the regulation of gene expression and histone modification, has diverged into numerous distinct protein complexes in metazoans, including conserved SAGA variants and more distantly related ATAC variants. The functions of metazoan SAGA/ATAC have now been linked to stress response pathways. In this review by Timmers and colleagues, novel insights into distinct chromatin-modifying functions, target gene regulation, and interplay with signaling pathways are discussed and placed in the context of evolutionarily conserved domains and enzymatic activities of SAGA/ATAC subunits.
Keywords: chromatin, histone acetylation, SAGA, TFIID, transcription regulation
Abstract
The yeast SAGA (Spt–Ada–Gcn5–acetyltransferase) coactivator complex exerts functions in gene expression, including activator interaction, histone acetylation, histone deubiquitination, mRNA export, chromatin recognition, and regulation of the basal transcription machinery. These diverse functions involve distinct modules within this multiprotein complex. It has now become clear that yeast SAGA has diverged during metazoan evolution into two related complexes, SAGA and ATAC, which exist in two flavors in vertebrates. The compositions of metazoan ATAC and SAGA complexes have been characterized, and functional analyses indicate that these complexes have important but distinct roles in transcription, histone modification, signaling pathways, and cell cycle regulation.
Almost two decades ago, Allis and coworkers (Brownell and Allis 1995; Brownell et al. 1996) discovered that an ∼55-kDa protein purified from the macronuclei of the ciliated protozoan Tetrahymena thermophila exhibited acetyltransferase activity toward histones. This protein was orthologous to the yeast transcriptional coactivator Gcn5 that was found to be present in a protein complex with Ada2. The Ada2 protein was identified in genetic screens for transcriptional coactivators (Berger et al. 1992; Marcus et al. 1994). These two key discoveries provided the basis for the concept that histone acetylation and transcription are intimately connected. Within chromatin, DNA is wrapped around histone octamers consisting of the histone H3–H4 tetramer associated with two H2A–H2B dimers to form discrete nucleosome units. These nucleosomes are the basic repeating unit of chromatin and restrict access by protein complexes involved in DNA-mediated cellular transactions. The N termini of these core histones protrude from the nucleosome particle and are subjected to a variety of post-translational modifications, including acetylation (Berger 2007). Histone acetylation has been one of the most studied histone modifications, and the histone acetyltransferase (HAT) complexes involved have become major research topics. Acetylation modifies the physical–chemical properties of the chromatin fiber, but it also provides interaction sites for a myriad of binding proteins. These “readers” typically use a bromodomain for acetyl-lysine interactions (Mujtaba et al. 2007). In cellular systems, histone acetylation is a dynamic process, which has attracted particular interest from a clinical perspective (Rodriguez-Paredes and Esteller 2011). Drugs inhibiting the enzymes known as the “erasers” of acetylation, the histone deacetylases (HDACs), are effective in particular types of lymphomas. Very recently, molecules inhibiting acetyl-lysine binding of the bromodomain-containing Brd2 and Brd4 proteins have been shown to be effective suppressors of inflammation and leukemic growth (Nicodeme et al. 2010; Dawson et al. 2011; Delmore et al. 2011). Clearly, this interface between gene regulation and chromatin modification has expanded enormously and has revolutionized molecular models of gene expression, genome structure, (epi)genetic inheritance, and human disease.
A few years after their characterization, the highly conserved Gcn5 HAT and Ada2 proteins were found to be part of a multiprotein complex named SAGA (Spt–Ada–Gcn5–acetyltransferase) (Grant et al. 1997). The unveiling of the modular architecture of the SAGA complex has permitted deciphering of its multifunctional role (Sterner et al. 1999). Characterization of the yeast SAGA complex revealed several layers of transcription-related functions, including transcription initiation and elongation, histone ubiquitination, and TATA-binding protein (TBP) interactions. In addition, SAGA has nontranscriptional roles, including mRNA export (Rodriguez-Navarro et al. 2004) and maintenance of DNA integrity (Atanassov et al. 2009; Guo et al. 2011). Recent evidence indicates that the yeast SAGA complex has diverged during evolution in several distinct protein complexes in metazoans, which include conserved SAGA variants and more distantly related ATAC variants (Guelman et al. 2006a). We refer to these complexes collectively as SAGA-like complexes because they share a common catalytic core. Many aspects of their transcriptional and nontranscriptional functions have been covered in excellent reviews (Nagy and Tora 2007; Rodriguez-Navarro 2009; Koutelou et al. 2010; Rodriguez-Navarro and Hurt 2011; Weake and Workman 2011).
Here, we focus on the evolutionary divergence of SAGA-like complexes in the context of transcriptional regulation, chromatin modification, and cellular signaling.
Compositions and post-translational modifications of SAGA-like complexes
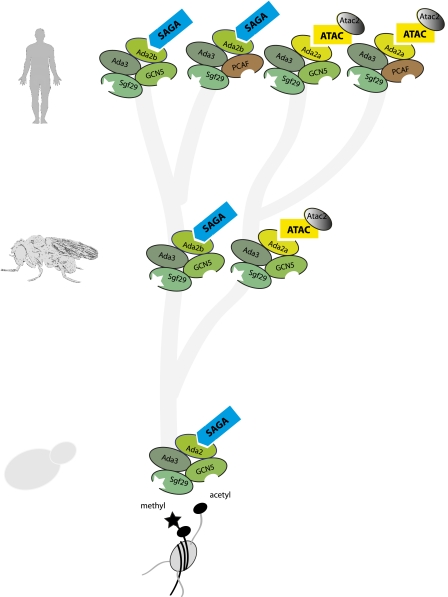
We first describe how the yeast SAGA complex has diverged with respect to composition and conservation of protein domains. The subunit comparison indicates two main points. First, SAGA architecture is strongly conserved between yeast, flies, and mammals (Fig. 1). Second, the number of SAGA-like complexes increased during evolution (Fig. 2). Yeast contains one SAGA complex (and its subcomplexes/proteolytic derivatives) (see below), insects contain two SAGA-like complexes (SAGA itself and ATAC), and mammals contain four main SAGA-like complexes (SAGA itself with Gcn5 or the highly related HAT Pcaf, and ATAC with Gcn5 or Pcaf).
Figure 1.
Composition and domain organization of SAGA and ATAC. Protein domains have been assigned based on published data and using SMART (http://smart.embl-heidelberg.de). Homologous domains of published domains were in some cases refined by alignments using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2) or BLAST. The prefix “y” indicates Saccharomyces cerevisiae, “d” indicates Drosophila melanogaster, and “h” indicates Homo sapiens.
Figure 2.
Divergence and multiplication of SAGA-like complexes during evolution. Incorporation of Ada2a or Ada2b determines assembly of ATAC or SAGA, respectively. Some of the chromatin interaction modules are indicated: The Tudor domain in Sgf29 binds H3K4me3, and the bromodomain in Gcn5 and in Pcaf binds acetylated histones. HAT activity is provided by Gcn5 or Pcaf. The second HAT in ATAC, Atac2, is also indicated.
It is well known that diversification of protein function can be accomplished by the duplication and mutation of ancestor genes (Soskine and Tawfik 2010). Diversification of protein complexes seems to be a common theme for increasing functional complexity during evolution. For example, the six Set1/MLL complexes in mammals have one ancestor complex, termed Set1C/COMPASS, in yeast (Miller et al. 2001; Roguev et al. 2001; Smith et al. 2011). Also, the Sin3 complex involved in transcriptional repression and histone deacetylation exists in one complex in yeast, while mammals contain two related but distinct complexes (Sin3A and Sin3B) (Hayakawa and Nakayama 2011). In many cases, overall architecture has been conserved for related protein complexes, and the composition of modules and domains has diversified. This seems particularly relevant for SAGA-like complexes, which underwent important changes during evolution, and we hypothesize that these changes and expansions are used for functional cellular specialization involved in development and homeostasis of metazoan organisms. In addition, most SAGA and ATAC subunits are subjected to extensive post-translational modification, including phosphorylation and acetylation, which are listed in Supplemental Table S1. In most cases, the functions of these modifications have not been determined, but we provide them as resources for future investigations, as these modifications could be end points of signal transduction pathways.
Diversification of SAGA and links to other protein complexes
The yeast SAGA complex is composed of 19 subunits (Mischerikow et al. 2009; Lee et al. 2011) with a total mass of ∼2 MDa, which are organized into a five-domain structure as indicated by a three-dimensional (3D) model reconstructed from (immuno-)EM images (Wu et al. 2004). Each domain harbors specific SAGA functions (Timmers and Tora 2005; Lee et al. 2011). The transcription factor interaction domain localizes to the foot of the structure containing the Tra1 subunit. Together with Spt20 and Spt7, the subunits Ada1, Taf5, Taf6, Taf9, Taf10, and Taf12 are believed to form the structural core of SAGA. The Taf subunits received particular interest, as they are also integral subunits of the basal transcription factor TFIID. Interaction with the TBP resides with Spt3 and Spt8 (Eisenmann et al. 1992). The HAT function resides in a domain containing Gcn5, Ada2, Ada3, and Sgf29, whereas the histone H2B deubiquitination (DUB) module harbors Usp8, Sgf11, Sgf73, and Sus1. Interestingly, Sus1 is also part of the mRNA export complex TREX-2 (in yeast)/AMEX (in metazoans). This property connects SAGA to a process called gene gating, in which certain transcribed genes are located near nuclear pore complexes to increase nuclear mRNA export. While first identified in yeast (Rodriguez-Navarro et al. 2004) and confirmed in Drosophila (Kurshakova et al. 2007), the relevance of this process for mammalian gene expression remains to be established (Garcia-Oliver et al. 2011).
An important change in SAGA function stems from the fact that no ortholog of the SPT8 gene is present in the genomes of metazoans. In addition, higher eukaryotes contain a truncated homolog of yeast Spt7 lacking the C terminus. In yeast, a variant of SAGA exists, termed SLIK/SALSA (Belotserkovskaya et al. 2000; Pray-Grant et al. 2002; Sterner et al. 2002; Wu and Winston 2002), in which the C terminus of Spt7 is proteolytically cleaved by Pep4 (Spedale et al. 2010), resulting in the removal of the Spt8-binding domain. SLIK/SALSA has altered properties compared with SAGA, in particular with regard to the TBP interaction module. This consists of Spt3 and Spt8 in SAGA (Eisenmann et al. 1992) and has now been weakened in SLIK/SALSA due to the absence of Spt8 (Sermwittayawong and Tan 2006; Laprade et al. 2007; Mischerikow et al. 2009). As noted previously (Nagy et al. 2009), metazoan SAGA more closely resembles the yeast SLIK/SALSA complex because it contains a C-terminally truncated version of Spt7 and lacks Spt8. This may have consequences for the TBP-interacting properties of metazoan SAGA, which may be weaker compared with yeast SAGA. Indeed, while TBP can be easily detected in preparations of yeast SAGA, it is absent from human SAGA (Wieczorek et al. 1998).
Most domains in SAGA subunits are remarkably conserved (Fig. 1). Exceptions are listed hereafter. First and as stated above, metazoan homologs of Spt7 are C-terminally truncated. This has led to loss of the histone fold (HF) and bromodomain in the fly ortholog. The HF is a conserved motif that is present in the core nucleosomal histones and selected transcription factors. The HF can mediate protein dimerization (Luger et al. 1997; Gangloff et al. 2001). Since the HF of yeast Spt7 forms a heterodimer with Taf10 in SAGA (see below), this loss is expected to have structural consequences. Loss of the bromodomain may affect the interaction of fly SAGA with acetylated proteins, including histones (Hassan et al. 2002). Human Spt7 does not contain a classical bromodomain or HF, but rather a bromo-associated domain that resembles a histone-like fold. Second, Taf5 from yeast contains a LisH domain, which has been implicated in dimerization. This domain is absent from its counterparts in flies and humans, WDA and Taf5L, respectively (Ogryzko et al. 1998; Guelman et al. 2006b). In contrast, human Taf5, which exclusively assembles into the TFIID complex, contains a LisH domain. Third, and similar to this, yeast Taf6 and human Taf6L (Ogryzko et al. 1998) contain the DUF1546 domain of unknown function, which is absent from its fly counterpart, SAF6 (Weake et al. 2009). Fourth, the human ortholog of yeast Sgf11, ATXN7L3, contains an atypical zinc finger, termed a spinocerebellar ataxia type 7 (SCA7) domain, at its C terminus. This is lacking in flies and yeast (Kohler et al. 2008; Weake et al. 2008, 2009; Bonnet et al. 2010). Fifth, yeast Sgf73 and its human ortholog, ATXN7, also contain a SCA7 domain, but again, this is missing in its fly counterpart (Kohler et al. 2008; Weake et al. 2009; Bonnet et al. 2010). Interestingly, the SCA7 domain of ATXN7/hSgf73 but not of ATXN7L3/hSgf11 can interact with nucleosomes (Bonnet et al. 2010). Finally, the SWIRM domain functioning in protein–protein and protein–DNA interactions (see below) is missing in Ada2b from flies (Muratoglu et al. 2003). Taken together, these structural comparisons suggest that the interaction properties of SAGA complexes may differ between species.
During evolution, gene duplications have occurred for TAF5, TAF6, TAF9, TAF10, GCN5, and ADA2. In yeast, the Taf5 and Taf6 proteins are part of SAGA but also of the related TFIID complex (Grant et al. 1998). In mammals, the genes for TAF5 and TAF6 have been duplicated and have diverged into TAF5L and TAF6L. The Taf5 and Taf6 proteins remain specific for TFIID, while Taf5L and Taf6L are specific for SAGA (Ogryzko et al. 1998). Interestingly, flies have diverged in another direction. While the TAF5L and TAF6L genes exist, their products are not part of SAGA in this species (Guelman et al. 2006b; Weake et al. 2009). Instead, SAGA in flies contains Wda rather than Taf5 or Taf5L, and Saf6 rather than Taf6 or Taf6L. It is not known whether fly Taf5L and Taf6L assemble into alternative transcription complexes. Since both proteins are highly expressed in fly testis, it may be that they are involved in the formation of testis-specific TFIID or SAGA-like complexes (Guelman et al. 2006b; Weake et al. 2009). TAF9 has been duplicated in humans into the TAF9 and TAF9B genes. Taf9 and Taf9b have partially overlapping functions (Frontini et al. 2005). Both proteins can incorporate into SAGA and TFIID complexes, further increasing the number of complexes in humans. Duplication of TAF10 in flies but not in humans led to two genes: TAF10, which encodes a TFIID subunit, and TAF10B, which encodes a SAGA subunit.
Only one GCN5 gene is present in yeast, while flies and humans contain a larger homolog that encodes an N-terminal extension harboring a PCAF homology domain (Koutelou et al. 2010; Nagy et al. 2010). In humans, GCN5 has been duplicated and diverged to generate the PCAF gene. The Gcn5 and Pcaf proteins are very similar, and both harbor the PCAF homology domain. Both Gcn5 and Pcaf can assemble into human SAGA complexes in a mutually exclusive manner (Krebs et al. 2010). Pcaf was named according to its original identification as a p300/CBP-associated factor (Yang et al. 1996). In retrospect, this name is confusing, as SAGA-like complexes lack p300 or CBP. ADA2 represents another gene that has been duplicated in evolution. One ADA2 gene exists in the yeast genome encoding a subunit that is part of both the SAGA and the smaller ADA complex (Lee et al. 2011). In many but not all metazoans, this gene has been duplicated into ADA2A and ADA2B. At this point, the separation between SAGA and ATAC begins. The fly and human Ada2b proteins are exclusively present in SAGA, while Ada2a is specific for ATAC. The entire ATAC complex is specific for higher eukaryotes and is absent from yeast (Barlev et al. 2003; Muratoglu et al. 2003; Suganuma et al. 2008; Wang et al. 2008; Orpinell et al. 2010).
Diversification of SAGA into ATAC and links to other protein complexes
ATAC has emerged later during evolution from SAGA and is exclusive to multicellular eukaryotes. This may be related to the increased complexity of metazoans. SAGA and ATAC diversified from a single variant in flies into two human variants through duplication of its HAT subunit. Interesting physical links exist with related chromatin and transcription complexes. Important differences have been noted between the fly and human ATAC complexes with regard to conserved domains and subunit composition (Suganuma et al. 2008; Wang et al. 2008; Guelman et al. 2009; Nagy et al. 2009).
Subunit composition
ATAC shares a core with SAGA that consists of Ada3, Sgf29, and Gcn5 in flies, or Gcn5/Pcaf in human. This results in the occurrence of one ATAC variant in flies, whereas two variants, with Gcn5 or with Pcaf, exist in humans (Fig. 2). In addition to a shared catalytic core, nine to 10 ATAC-specific subunits exist. A few differences with regard to subunit composition exist between the fly and human complexes. Atac3 has been identified as a bona fide component of fly ATAC by reciprocal immunoprecipitation and mass spectrometry analysis (Suganuma et al. 2008). However, this protein seems to be a fly-specific subunit, since no orthologs have been detected in ATAC purifications from human cells (Wang et al. 2008; Nagy et al. 2009). Hcf1, which is also present in other chromatin-modifying complexes (Wysocka et al. 2003), was found as a specific subunit of ATAC in flies (Suganuma et al. 2008). Human Hcf1 has been detected in both ATAC and SAGA purifications (Wang et al. 2008), but other purifications from human cells failed to detect Hcf1 in either SAGA or ATAC (Guelman et al. 2009; Nagy et al. 2010). This suggests that Hcf1 may not be a stable ATAC or SAGA subunit in human cells, but rather interacts in a dynamic manner. Additional proteins have been identified in ATAC purifications from human but not insect cells. These include Pole4, which is also a subunit of the DNA polymerase ɛ complex; hMap3k7, which links ATAC to mitogen-activated protein kinase (MAPK) signaling; and Ubap2l, a protein with unknown function (Wang et al. 2008).
Conservation of domains
Besides differences in subunits, protein domains are conserved in many but not all cases. The subunit dAtac1 (Guelman et al. 2006a) lacks a C-terminal ZnF domain that is present in human Atac1 (Wang et al. 2008; Guelman et al. 2009; Nagy et al. 2010). Atac2 contains an active HAT domain (see below), the fly homolog contains a canonical PHD finger at the N terminus, and the human homolog may harbor a different type of ZnF (Nagy et al. 2010). PHD fingers have been identified as binding modules to methylated histone tails (Baker et al. 2008). Hence, its absence in human Atac2 may affect the chromatin-binding properties of ATAC. The Mbip subunit shows distinct domain organization in flies and humans. Fly Mbip/CG10238 contains two domains: an N-terminal MoaE domain absent from the human protein, and a C-terminal MBIP domain, which comprises the entire human Mbip protein (Suganuma et al. 2010). Since the MoaE domain has been shown to mediate interaction between ATAC and MAPK signaling, its absence from human Mbip may have functional consequences for the interplay between ATAC and signal transduction pathways (Suganuma et al. 2010). Finally, human Yeats2 contains both YEATS and HF domains (Wang et al. 2008). While several programs fail to detect a HF in Drosophila Yeats2, alignments using ClustalW2 show strong conservation of the HF sequence at residues 869–969.
Links with other protein complexes
ATAC has several links with related protein complexes via shared subunits. The WD repeat proteins fly Wds or human Wdr5 link ATAC to the Set1/MLL complexes involved in methylation of histone H3 at Lys 4 (Shilatifard 2008). Inclusion of Hcf1 could also link functions of ATAC to the MLL and/or the Sin3 HDAC complexes (Wysocka et al. 2003). The Chrac14 subunit is a bona fide HF-containing subunit of fly ATAC. Its human ortholog, hChrac17, has been detected in some (Wang et al. 2008), but not all (Guelman et al. 2009; Nagy et al. 2009), human ATAC purifications. Chrac14/Chrac17 is also a subunit of ISWI-containing ATP-dependent chromatin remodeling complexes (Varga-Weisz et al. 1997; Poot et al. 2000), which initiated functional studies on the role of ATAC during chromatin remodeling (see below). An unexpected HF-containing subunit of ATAC in both flies and humans is Nc2β. This protein is conserved from yeast to humans and forms a stable heterodimeric complex with Nc2α. This NC2 complex is a strong repressor of TBP-mediated in vitro transcription (Kaiser and Meisterernst 1996; Thomas and Chiang 2006). In yeast, the NC2 complex associates with TBP, Mot1, and a small piece of internucleosomal DNA and is involved in regulating TBP promoter association (van Werven et al. 2008). In flies, NC2 is involved in core promoter selectivity (Hsu et al. 2008). The ATAC interaction partner for fly Nc2β is Yeats2, and this heterodimer seems to plays a structural role in ATAC (Wang et al. 2008).
Domains and structural comparison between ATAC and SAGA
The domains presently identified in the SAGA and ATAC subunits are indicated in Figure 1. Several known structural features of SAGA are shared with ATAC, but in a variant form. These include HF-mediated heterodimer pairs, a WD repeat protein, a HAT/chromatin interaction module, and proteins required for structural integrity. Strikingly different subunits decorate these structures.
Heterodimers
In addition to the Yeats2/Nc2β interaction, several other subunit interactions depend on HF motifs, which are known to mediate protein dimerization (Gangloff et al. 2001). In SAGA, the following HF dimers are present: between Taf9 (H3-like) and Taf6/Taf6L (H4-like), between Ada1 (H2A-like) and Taf12 (H2B-like), and between Taf10 and Spt7 (Ogryzko et al. 1998; Wu and Winston 2002; Wu et al. 2004; Koutelou et al. 2010). None of these subunits are present in ATAC. However, some of these pairs are (partially) shared with the basal transcription factor TFIID, which harbors the HF pairs Taf6/Taf9, Taf4/Taf12, Taf3/Taf10, Taf8/Taf10, and Taf11/Taf13 (Gangloff et al. 2001; Cler et al. 2009). ATAC contains three HF proteins: Nc2β, Yeats2, and Chrac14. While Chrac14 and Nc2β fail to form heterodimers (Suganuma et al. 2008), HFs are required for Yeats2/Nc2β interaction (Wang et al. 2008).
WD repeats
Another important structural feature of SAGA, ATAC, and many other complexes, including TFIID, is the presence of a WD repeat protein in a structural subunit, which is involved in protein–protein interactions (Xu and Min 2011). In SAGA and similarly in TFIID, this domain is provided by Taf5/Wda/Taf5L. In ATAC, human Wdr5 and its fly ortholog, Wds, contain a WD repeat domain. It is intriguing that exactly Wdr5 is shared with the H3K4 methyltransferase complex MLL (Wysocka et al. 2003), which raises questions regarding the regulation of complex assembly, the interplay between ATAC and MLL, and the interaction with nucleosomes.
HAT/chromatin interaction module
The HAT/chromatin interaction module of SAGA and ATAC is partially shared and specifies incorporation in either complex. In SAGA, it consists of Gcn5/Pcaf, Ada3, Sgf29, and Ada2b, and in ATAC, it consists of Gcn5/Pcaf, Ada3, Sgf29, and Ada2a. Several domains in these proteins are involved in chromatin recognition. For this reason, we propose to name this the HAT/chromatin interaction module. The first chromatin “reader” is provided by the bromodomain of Gcn5/Pcaf, which can bind acetylated lysine residues on histones (Mujtaba et al. 2007). The second chromatin anchor has recently been identified to be the Tudor domain of Sgf29, which can bind trimethylated Lys 4 of histone H3 (H3K4me3) associated with transcription start sites (TSSs) (Barski et al. 2007; Vermeulen et al. 2010; Bian et al. 2011). Interestingly, peptide pull-down experiments using H3K4me3-modified peptides combined with quantitative mass spectrometry resulted in the specific binding of SAGA but not ATAC subunits from HeLa cells, while Sgf29 is present in both complexes (Vermeulen et al. 2010). It may therefore be possible that the Tudor domain of Sgf29 preferentially binds H3K4me3 when incorporated in SAGA but not when in ATAC. Ada2a and Ada2b both contain three domains involved in chromatin regulation: an N-terminal ZnF, followed by a SANT domain, and a C-terminal SWIRM domain (Muratoglu et al. 2003; Gamper et al. 2009). They also contain conserved Ada boxes of unknown function (Muratoglu et al. 2003). In vitro binding studies have indicated that these domains are involved in contacting Ada3 (via SWIRM) and Gcn5 (via ZnF and SANT) (Gamper et al. 2009). The SWIRM domain of Ada2a can bind DNA and potentiate chromatin remodeling induced by ACF (Qian et al. 2005). It remains to be determined how the other domains are involved in chromatin interaction. Ada2b and Ada3 cooperate to stimulate Gcn5-mediated histone acetylation of nucleosomal templates but not of free histones, which argues for a role of these proteins in chromatin interaction and/or allosteric control (Gamper et al. 2009). In addition to the HAT/chromatin interaction module, several other domains are present in both SAGA and ATAC that may mediate interactions with chromatin. For SAGA, these include the bromodomain of yeast Spt7 and the SCA7 domain of Sgf73. In ATAC, additional potential chromatin anchors include the PHD finger of fly Atac2, and the SANT and ZnF domains of Atac1. Atac2 also adds a second HAT activity to the ATAC complex besides Gcn5 or Pcaf (see below).
Structural integrity
This includes Spt7, Spt20, and probably Taf5/Wda/Taf5L in SAGA, and Yeats2 (Wang et al. 2008) and Atac2 (Guelman et al. 2009) in ATAC. As discussed above, the Wdr5/Wds subunit could also serve an important role, but much less is known about the structural core of ATAC at present.
In conclusion, SAGA and ATAC share a HAT/chromatin interaction core with distinct properties dictated by incorporation of Ada2b (stimulation of Gcn5 HAT activity) or Ada2a (no stimulation). Both complexes use a WD repeat protein and distinct HF-mediated heterodimers. Complex-specific subunits provide ample opportunities for distinct ways of transcriptional regulation, including distinct enzymatic activities, additional chromatin interaction motifs, distinct links with other chromatin complexes, and subunits that link them to transcription factors or signaling pathways.
Functional distinctions in chromatin modification
Several differences exist between SAGA and ATAC with regard to enzymatic activities. First, SAGA contains a DUB module specific for H2Bub (Powell et al. 2004; Ingvarsdottir et al. 2005; Lee et al. 2005), which is absent from ATAC. Several recent studies have shown that the DUB module of metazoan SAGA, like in yeast, plays a role in the regulation of a subset of SAGA-dependent genes, including c-Myc-responsive genes (Pijnappel and Timmers 2008; Zhang et al. 2008; Zhao et al. 2008; Rodriguez-Navarro 2009; Weake et al. 2011). Second, ATAC contains two HAT subunits, Gcn5/Pcaf and Atac2, and Ada2a instead of Ada2b that differentially regulate the activity of Gcn5. Results obtained using purified proteins and from in vivo studies are described hereafter.
In vitro activities
Recombinant Atac2 protein displays HAT activity toward both H3 and H4 when present as free histones, but the activity is ∼40-fold reduced compared with recombinant Gcn5, which specifically acetylates H3 but not H4 (Guelman et al. 2009). Purified preparations of ATAC have also been shown to possess HAT activity for both H3 and H4, both as free histones and in nucleosomes (Suganuma et al. 2008; Guelman et al. 2009; Nagy et al. 2010). Whereas SAGA and ATAC from flies both acetylate H3 and H4 as free histones, SAGA showed strong specificity for H3 and ATAC showed strong specificity for H4 when nucleosomal substrates were used (Suganuma et al. 2008). Altered substrate specificities may be dictated by other complex subunits. In particular, Ada subunits can affect the enzymatic activity of Gcn5 when tested on nucleosomal substrates, and this property is conserved from yeast to humans (Gamper et al. 2009). Interestingly, recombinant human Ada2b and Ada3 strongly stimulated the HAT activity of Gcn5 toward mononucleosomes. Replacement of human Ada2b (SAGA-specific) with Ada2a (ATAC-specific) abolished this stimulation. This suggests that besides the presence of an additional HAT provided by Atac2, ATAC may also possess a different HAT activity caused by the diminished capacity of Ada2a to stimulate Gcn5.
In vivo activities
The contributions of ATAC to histone acetylation in vivo have not been clarified yet. Both human and mouse ATACs have been reported to be required for global acetylation of H3K9, H4K5, H4K12, and H4K16, as shown by knockdown of Atac2 in A549, HEK293 cells, and mouse embryos (Guelman et al. 2009). Atac2 also appeared to be required for complex integrity. No effects were observed for acetylation of H3K14 and H4K8. Another study reported that acetylation of H3K9 and H3K14 was reduced by knockdown of the ATAC-specific subunit Ada2a in HeLa cells and that H4K5, H4K12, and H4K16 acetylation levels were unchanged (Nagy et al. 2010). In this study, the lack of effects on H4 acetylation could be caused by the remaining Atac2 enzyme, which would act either alone or in an ATAC-like subcomplex lacking Ada2a. The consensus between these two studies is represented only by H3K9. In flies, mutation of ATAC2 results in reduced global levels of H4K16ac. The unaffected residues include H3K9, H3K14, H4K5, and H4K8 (Suganuma et al. 2008). Another study reports decreased global levels of H4K12ac in ADA2A mutant flies (Ciurciu et al. 2008). This indicates partially similar roles for Atac2 in the regulation of global acetylation levels in flies and mammals: In both organisms, this HAT is involved in the acetylation of H4K12 and H4K16. It is important to realize that the above studies rely on specificity and sensitivity of antibodies toward acetylated histones. Application of quantitative mass spectrometry to measure histone acetylation could resolve (some of) the discrepancies.
Besides HAT activity toward the histone proteins, a growing number of nonhistone substrates have been identified for Gcn5 and Pcaf. Some of these (cyclin A, ERRα, and Snf2) are discussed in more detail elsewhere in this review. Additional examples include the Pcaf substrates in mammals: p53 (Liu et al. 1999), HIF-1α (Xenaki et al. 2008), β-catenin (Ge et al. 2009), c-myc (Patel et al. 2004), Pcaf autoacetylation (Blanco-Garcia et al. 2009), and Cdk2 (Mateo et al. 2009). Nonhistone substrates for Gcn5 include Cdc6 (Paolinelli et al. 2009), c-Myc (Patel et al. 2004), ISWI in flies (Ferreira et al. 2007), and Rsc4 in yeast (VanDemark et al. 2007). Besides histones, the Trf1 subunit of the shelterin complex is a substrate for the DUB module of SAGA (Atanassov et al. 2009), and more examples are likely to follow. In many cases, acetylation and DUB has been shown to modulate the activity or stability of these nonhistone substrates, and we refer to several reviews for this (Sterner and Berger 2000; Yang 2004; Nagy and Tora 2007).
Interplay with signaling pathways
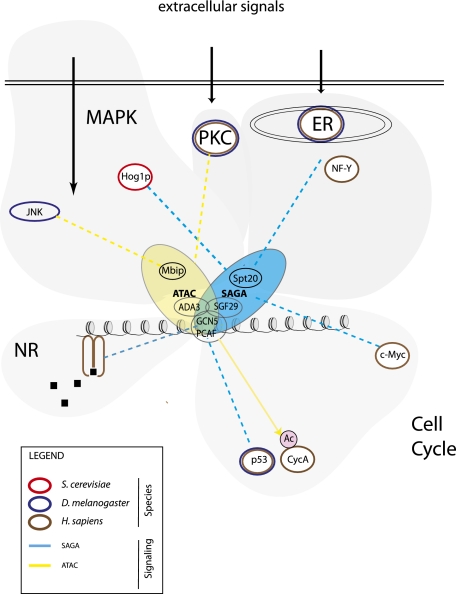
At present, six distinct types of (stress-induced) signaling pathways have been described to involve SAGA-like complexes: phorbol ester-induced (protein kinase C [PKC]), UV-induced (p53), high osmotic stress-induced (MAPK), sodium arsenite-induced (MAPK), thapsigargin-induced (MAPK-ER stress), and nuclear receptor signaling (Fig. 3). This indicates that SAGA and ATAC complexes in metazoans play central roles in gene expression in response to intracellular signaling. This property is conserved in yeast, where SAGA was found to be most relevant for stress-regulated genes (Huisinga and Pugh 2004).
Figure 3.
Interplay between SAGA and ATAC with signaling pathways. See the text for details.
PKC
Activation of the PKC pathway using the phorbol ester TPA leads to induction of many genes, as observed by staining Drosophila polytene chromosomes with antibodies against RNA polymerase II (pol II) (Nagy et al. 2010). Costaining with the ATAC-specific subunit Mbip/CG10238 revealed a similar strong increase of its chromosome association to sites of induced transcription. In contrast, the SAGA-specific subunit Ada2b failed to show TPA-induced chromosome association. As monitored by its Atac1 and Atac2 subunits, human ATAC was recruited to the TPA-induced genes cFOS, EGR1, and FRA1 and was required for TPA-induced expression of these genes. This effect was specific for ATAC, as SAGA was neither recruited nor required for mRNA expression of these genes (Nagy et al. 2010).
The tumor suppressor p53
Upon UV-induced DNA damage, p53 becomes stabilized by phosphorylation and induces a G1/S arrest and a DNA damage response to allow DNA repair. Previous studies have indicated a role for Gcn5 in regulating p53-induced gene expression. More recent work has assigned this role to metazoan SAGA, rather than ATAC. In Drosophila cells, Ada2b but not Ada2a protein interacts with p53. ADA2B but not ADA2A showed genetic interaction with p53 in affecting the pigment contents of adult eyes (Pankotai et al. 2005). In human cells, the SAGA-specific subunit Ada2b but not the ATAC-specific subunit Ada2a was recruited to the p53 response genes CDKN1A/p21, GADD45, and PUMA (Gamper et al. 2009). These findings show that the preferential interaction between p53 and SAGA, rather than ATAC, has been conserved from flies to humans.
MAPK
Distinct MAPK pathways exist in yeast, flies, and humans (Chen and Thorner 2007; Cuadrado and Nebreda 2010). These kinase cascades can be induced by signals including peptide growth factors, hormones, and various stresses, including starvation, oxidative stress, and high osmolarity stress. Pathways include multiple MAPK subfamilies, of which mammalian ERK, JNK, and p38 (Hog1 in yeast) have been most studied. Depending on the stress signal and the specific MAPK pathway involved, either SAGA or ATAC was shown to play an important role in regulating stress-induced gene expression.
Osmotic stress
Stress induced by high osmolarity can be achieved using high concentrations of sorbitol or NaCl in the medium and involves SAGA in yeast and ATAC in flies. In yeast, osmotic stress activates the Hog1 protein kinase, which has a number of functions, including phosphorylation of transcription factors and promoter recruitment of pol II. For example, Hog1 phosphorylation switches Sko1 from a repressor to an activator, which recruits yeast SAGA and SWI–SNF chromatin remodeling complexes to osmotic stress response genes (Proft and Struhl 2002). Nadal and coworkers (Zapater et al. 2007) used the osmotic stress response gene STL1 as a model showing activation by sequential promoter recruitment of Hog1, SAGA, and the Mediator coactivator complex, respectively. Both SAGA and Mediator appeared to be required for STL1 induction. In Drosophila cells, the first evidence for a role of ATAC in MAPK signaling was obtained by proteomic analysis: The ATAC subunit Mbip/CG10238 is known as MUK (MAPK upstream kinase)-binding inhibitory protein, and several MAPK pathway components were identified as substoichiometric ATAC interactors (Suganuma et al. 2010). Using activation of JNK (Jun kinase) by phosphorylation in response to osmotic stress as a readout, Workman and coworkers (Suganuma et al. 2010) found that overexpression of the Mbip/CG10238 subunit of ATAC inhibited, while knockdown of ATAC subunits (Atac2 or Nc2β) potentiated, JNK activation. Detailed analysis of two JNK target genes, Jra/JNK itself (via autoregulation) and Chickadee, revealed differential regulation by ATAC under basal and activated conditions. Without osmotic stress induction, expression of these genes was positively regulated by ATAC, which occurred mostly via Atac2. In contrast, during osmotic stress induction, ATAC functions to inhibit expression of these genes, mostly via Mbip/CG10238. These results show that the role of ATAC in osmotic stress gene regulation is more complex than just the analogous SAGA function in yeast. It would be interesting to know which factors positively regulate osmotic stress-induced gene regulation in Drosophila cells. Could this involve the SAGA complex?
Sodium arsenite-induced signaling
Proteomic analysis also suggested a link between MAPK signaling and human SAGA. Identification of human Spt20 (Nagy et al. 2009) showed that this protein is identical to p38IP, which was previously described as an interactor of p38 MAPK (Zohn et al. 2006). The p38 MAPK pathway can be induced by sodium arsenite and is distinct from the JNK pathway. Activation of p38 MAPK failed to induce recruitment of SAGA to the early response gene EGR1, suggesting that SAGA may not play a role during this type of stress (Nagy et al. 2009). EGR1 is also induced upon PKC activation (see above) and requires ATAC for induction. It would be interesting to determine the involvement of SAGA and ATAC in stress-induced gene expression on a genome-wide scale.
Endoplasmic reticulum (ER) stress pathways
Early studies from yeast have shown that SAGA is involved in the unfolded protein response (UPR) following ER stress (Welihinda et al. 2000). Deletion of the yeast SAGA subunits GCN5, ADA2, or ADA3 prevented a proper UPR. The requirement for SAGA during the ER stress response has been conserved during evolution. In human cells, induction of ER stress using thapsigargin leads to the recruitment of the SAGA subunits Spt20, Spt3, and Sgf11 to the ER stress-induced genes CHOP, ERP70, HERPUD, and GRP78 (Nagy et al. 2009). Knockdown of Spt20 and several DUB module subunits inhibits ER stress induction of these genes, indicating that SAGA recruitment is critical for gene induction (Nagy et al. 2009; Lang et al. 2011). In agreement, it has been reported that human SAGA interacts with the NF-Y transcription factor, which is involved in the ER response (Schroder and Kaufman 2005). These findings indicate that SAGA interacts with the ER stress pathway in both yeast and humans. It is currently unknown whether ATAC can also play a role during ER stress-induced gene expression.
Nuclear receptor (NR) signaling
Several NRs have been shown to use SAGA as a cofactor to regulate transcription of their target genes. These include RARα, RXR, ERα, PR, androgen receptor (AR), and the orphan receptor ERRα. Several direct protein–protein interactions between SAGA and NRs have been reported. These include the Tra1 subunit that contains three LXXLL motifs known to mediate binding to NRs (Yanagisawa et al. 2002). Contacts between Ada3 and RARα, RXR, and ERα have also been observed, which may be mediated by the LXXLL motif of Ada3 (Zeng et al. 2002; Meng et al. 2004; Li et al. 2010). Besides promoting gene expression via histone acetylation, SAGA also may acetylate the NR itself, as is the case for ERRα (Wilson et al. 2010). Acetylation of this NR by Pcaf reduces its DNA-binding activity, resulting in repression of gene expression. Interestingly, evidence indicates that the DUB module of SAGA plays a role in AR-mediated transactivation in both Drosophila embryos and human cells (Zhao et al. 2008).
An important aspect that has been mostly unexplored is whether recruitment of SAGA-like complexes to stress-regulated genes also results in direct regulation of their activity by stress-regulated kinases. Upon activation, several kinases become recruited to target genes in both yeast and human cells (Alepuz et al. 2001; Hu et al. 2009; Lawrence et al. 2009). We hypothesize that these kinases could directly modify the in situ binding and/or different biochemical activities of SAGA-like complexes at target promoters. Large-scale proteomic surveys revealed a large number of acetylations and phosphorylations of subunits of SAGA-like complexes (Supplemental Table S1). It would be interesting to determine whether these modifications are dynamic and/or alter the properties of SAGA-like complexes. Direct kinase-mediated regulation of coactivator complexes would allow for very dynamic and localized events during the activation or desensitization of transcription.
Taken together, SAGA functions during p53 induction, ER stress, and nuclear receptor signaling, while ATAC plays a role during signaling by PKC and osmotic stress-induced MAPK signaling. Some of these pathways have early eukaryotic ancestors such as osmotic stress induction via Hog1 and ER stress, both of which involve regulation by yeast SAGA. This shows that SAGA-like complexes have diversified during evolution to regulate distinct signaling pathways.
Interplay with chromatin remodelers and transcription coactivators
Previous work has suggested a connection between the ATAC-specific subunit Ada2a and chromatin remodeling. These include a physical interaction between human Ada2a and SWI/SNF (Barlev et al. 2003), and the above-mentioned potentiation of ACF-induced chromatin remodeling by the SWIRM domain of human Ada2a (Qian et al. 2005). Recent proteomic links also provided evidence for involvement of ATAC in chromatin remodeling. These include the stable presence of Chrac14/Chrac17 and Nc2β in ATAC (see above) (Suganuma et al. 2008; Wang et al. 2008). These findings initiated the studies described below.
ATAC purified from insect cells stimulated nucleosome sliding induced by several chromatin remodeling complexes, including yeast ISWI, SWI/SNF, and RSC (Suganuma et al. 2008). This effect appeared to be mediated by the action of Chrac14. Inclusion of acetyl-CoA in the reaction further potentiated remodeling, which suggests an involvement of acetylation. How acetylation may influence remodeling is still unclear. Histone acetylation could expose DNA for binding the remodeling machines. However, a recent report describes an inhibitory role for Gcn5-mediated acetylation of the catalytic Snf2 subunit of the SWI/SNF complex, pointing to a more complex mechanism (Kim et al. 2010). In vivo evidence for interplay between ATAC and chromatin remodeling was obtained from Drosophila. Overlapping sets of genes were down-regulated in ADA2A mutant flies compared with mutants for the NURF chromatin remodeling complex (Carre et al. 2008). Mutants of the catalytic subunit of NURF, ISWI, showed strongly reduced Ada2a binding to polytene chromosomes, suggesting that ISWI-mediated remodeling is required for ATAC chromatin binding. Thus, both in vitro and in vivo data point to interplay between ATAC and chromatin remodeling machines. Data so far suggest a mechanism in which chromatin binding of ATAC depends on chromatin remodeling and that the remodeling process can then be further modulated by the action of the Chrac14 subunit of ATAC. How this occurs at the molecular level should be addressed in future experiments.
The Mediator complex represents an important intermediary between gene-specific activators and pol II (Thomas and Chiang 2006). Interestingly, ATAC but not SAGA has been found to form stable interactions with the Mediator complex in mammalian cells (Krebs et al. 2010). The interaction was observed only with the Gcn5 form of ATAC and was most strongly observed in pluripotent embryonic stem cells. The Luzp1 protein is responsible for bridging Gcn5–ATAC with Mediator, and these complexes are particularly important for transcription of several noncoding RNA genes (Krebs et al. 2010). Although several issues (e.g., Luzp1–Gcn5 interactions and Luzp1 interactions with SAGA) require further analyses, these observations reveal another level of functional specialization between SAGA-like complexes in mammals.
Regulation of target gene expression during development
Several target genes have been described that are regulated by either SAGA or ATAC. To investigate this on a genome-wide scale, several studies now report both overlapping and distinct sets of target genes in Drosophila embryos and tissues and in human cells in culture. These studies are described below.
Gene regulation in Drosophila
Larvae mutant for ADA2A (ATAC-specific), ADA2B (SAGA-specific), or ADA3 (shared between ATAC and SAGA) were analyzed using DNA microarrays (Carre et al. 2008; Pankotai et al. 2010). Both SAGA and ATAC appeared essential for embryonic development, but they regulate different sets of genes, resulting in distinct phenotypes. Ablation of ATAC affected an order of a magnitude more genes compared with SAGA. These results are consistent with the early lethal phenotype of ATAC ablation occurring just after this stage and during metamorphosis. A master regulator of metamorphosis is the ecdysone receptor. Gene expression profiles of ADA2A mutants showed that 40% of all transcripts have changed expression levels, many of which are part of the ecdysone transcription pathway. In contrast, ADA2B mutants died much later during development and affected only 3% of transcripts when assayed at the larval stage, of which many overlapped with ADA2A-regulated genes. Genes affected by ADA2B mutations include those involved in antimicrobial defense mechanisms (Zsindely et al. 2009). However, mutations in the SAGA subunits ADA2B, SGF11, and NONSTOP also affect genes in the ecdysone pathway, suggesting that SAGA and ATAC have partially additive or overlapping roles in this process (Weake et al. 2008). A function for the DUB module of SAGA was found to regulate genes involved in photoreceptor axon targeting (Weake et al. 2008). Direct target genes of SAGA in Drosophila tissues have been identified using chromatin immunoprecipitation (ChIP) and sequencing (ChIP-seq) (Weake et al. 2011). Muscle cells or neuronal cells were obtained from late stage Drosophila embryos by GFP labeling and FACS sorting. ChIP-seq of the SAGA-specific subunit Ada2b from these cells identified 1470 target genes in muscle, 59 genes in neurons, and 527 genes in both tissues. This suggests that SAGA regulates many more genes in muscle compared with neuronal cells. A subset of SAGA-binding sites in muscle colocalized with known binding sites for the muscle-specific transcription factor Mef2, suggesting that SAGA may function as its coactivator. These intriguing results may mean that either SAGA preferentially acts in certain tissues or regulation by SAGA is linked to the proliferation status of the cells. The binding location of SAGA in the Drosophila genome strongly correlated with that of pol II. Both were mostly present at TSSs, but in 14% of cases, they resided also within gene bodies. The location of SAGA and pol II in gene bodies is in agreement with previous suggestions that SAGA is involved in transcriptional elongation (Henry et al. 2003). Current models indicate that an elongation function for SAGA may be mediated by the DUB module. Since this module is not present in ATAC, the options for regulating gene expression differ significantly between SAGA and ATAC.
Gene regulation in mammals
In mice, both SAGA and ATAC are required for normal embryonic development. Disruption of SAGA by knockout of p38IP (SPT20) resulted in gastrulation defects due to a failure to down-regulate E-cadherin (Zohn et al. 2006). ATAC2 knockout mice display early embryonic lethality due to cell cycle defects (Guelman et al. 2009). The shared subunits GCN5 and PCAF showed differential requirements for mouse development (Xu et al. 2000; Yamauchi et al. 2000). Whereas PCAF disruption showed no effect, it acted synergistically with GCN5 to induce an earlier and more severe phenotype compared with loss of GCN5 alone. This is explained by redundancy of PCAF and GCN5 and the later developmental expression of PCAF compared with GCN5. Interestingly, mice homozygous for catalytically dead Gcn5 showed a less severe phenotype with longer survival compared with a GCN5 deletion (Bu et al. 2007). This indicates that the HAT activity does not account for all Gcn5/Pcaf functions, which is concordant with studies in yeast and mammals (Candau et al. 1997; Wang et al. 1997; Jiang et al. 1999). Possibly, the Ada2 interaction domains and/or bromodomains of Gcn5/Pcaf are relevant in this. The SAGA subunit hAtxn7 (Sgf73) has been linked to the polyglutamine expansion disease SCA7, which is characterized by cerebellum and brainstem neurodegeneration for poorly understood reasons (David et al. 1997; McCullough and Grant 2010). The expanded glutamine repeat is located prior to the first ZnF domain (Fig. 1) and changes protein conformation, leading to aggregation and cellular toxicity. Interestingly, the expanded protein still incorporates into the SAGA complex, which displays a reduced HAT activity toward nucleosomes (McMahon et al. 2005; Palhan et al. 2005). These results indicate that both SAGA and ATAC play crucial roles during mammalian development and cellular physiology.
Direct target genes of human ATAC have been compared with those of SAGA by ChIP-seq analysis using antibodies to the ATAC-specific subunit ZZZ3/Atac1 and the SAGA-specific subunit Spt20 (Krebs et al. 2011). The results identified three gene categories in both human B-lymphoblasts and HeLa cells: those that bound only SAGA, those that bound only ATAC, or those that bound both. Binding profiles were then compared with published profiles for marks of TSSs (H3K4me3 and pol II) and enhancers (H3K4me1). The analysis indicated that ATAC binds to three types of locations: at known enhancers, at TSSs, and at regions lacking either mark. SAGA bound to two types of locations: known enhancers (but at a lower amount of locations compared with ATAC) and known TSSs. Gene ontology (GO) for both ATAC- and SAGA-bound regions failed to show a preference for binding either housekeeping or tissue-specific genes. This contrasts with the situation in yeast, where SAGA preferentially regulates stress-responsive genes, while TFIID is important for housekeeping genes (Huisinga and Pugh 2004). In metazoans, the role of SAGA-like complexes has therefore expanded to regulate both classes of genes. Interestingly, a subset of the enhancers bound by ATAC (but not by SAGA) lacked p300 binding. Since p300 has been a canonical marker for enhancers, this finding suggests the presence of a novel class of enhancers marked by ATAC binding and lacking p300. It would be interesting to investigate in what functional aspects ATAC-specific enhancers differ from p300-marked enhancers; e.g., do they have differentially acetylated nucleosomes, and do they require specific interplay with chromatin remodelers?
Taken together, these results show that SAGA and ATAC regulate partially overlapping and partially distinct sets of target genes. Remaining questions include the following: How do the observed binding profiles change upon distinct types of cellular stress, and for which of the target genes is binding required for gene expression?
Regulation of cell cycle progression
Previous work has implicated yeast Gcn5 in progression through the G2/M phase of the cell cycle (Howe et al. 2001). Recent work now shows that in metazoans, this function has been conserved in ATAC, rather than in SAGA. Knockdown of human Atac2 results in G2/M accumulation (Guelman et al. 2009). ATAC2-null mice die between embryonic days 8.5 and 11 due to a general increase in apoptotic cells and G2/M arrest. Also, in mouse NIH3T3 cells, knockdown of ATAC subunits results in G2/M arrest, followed by mitotic abnormalities (Orpinell et al. 2010). This effect is specific for ATAC, since SAGA knockdown did not result in cell cycle defects. The ATAC complex remains intact during mitosis and localizes to the mitotic spindle. A model for the mechanism by which ATAC regulates cell cycle progression has been proposed (Orpinell et al. 2010). Via one of its HATs, ATAC directly acetylates cyclin A, which forms a complex with the Cdk2 kinase to regulate early mitotic processes. Progression through mitosis requires degradation of cyclin A, and this is promoted by ATAC-mediated acetylation. A phosphorylation target of cyclin A/Cdk2 is the Sirt2 HDAC, which is also required for mitotic progression. Degradation of cyclin A leads to an activating dephosphorylation of Sirt2. As a result, the Sirt2 targets H4K16 and α-tubulin become deacetylated, which triggers mitotic progression. This shows that ATAC, while regulating transcription in interphase cells, has an important, different role unrelated to transcription during mitosis to modulate post-translational modification of components of the cell cycle machinery. It would be interesting to determine how the relocalization of ATAC from chromatin to the mitotic spindle and back is regulated and whether ATAC also regulates transcription of cell cycle proteins.
Concluding remarks
Current knowledge establishes the multiplicity of SAGA-like complexes in metazoans and defines distinct roles for SAGA and ATAC. These are obvious at the level of subunit composition, structural organization, enzymatic activities, interplay with signaling pathways, regulation of target gene expression, and cell cycle regulation. Although much insight has been obtained, it is clear that regulation by SAGA-like complexes is complex and that many questions remain unanswered. What is the molecular basis for the different activities of Gcn5 within SAGA and ATAC? Why are certain complex subunits shared with other protein complexes? What is the architectural organization of ATAC, and how is it recruited to its genomic locations? What are the assembly routes for SAGA and ATAC in cells? How do these complexes interact with chromatin remodeling machines? Are the activities of the SAGA-like complex subject to regulation by post-translational modifications? Are there fundamental differences between SAGA and ATAC with respect to the mechanism of gene regulation and with respect to their specific chromatin writing and reading capacities? We anticipate that in a few years time, significant progress will be made in addressing these and related questions.
Acknowledgments
We thank Jacques Bonnet, Laszlo Tora, Andrée Schram, Maria Koster, and members of the Timmers and Holstege groups for discussion and suggestions. This work has been supported by the Netherlands Genomics Initiative (Horizon Program no. 93516050), the Netherlands Organization for Scientific Research (NWO-CW, TOP no. 700.57.302), and the Netherlands Proteomics Centre.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.184705.111.
References
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G 2001. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 7: 767–777 [DOI] [PubMed] [Google Scholar]
- Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LA, Allis CD, Wang GG 2008. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat Res 647: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Emelyanov AV, Castagnino P, Zegerman P, Bannister AJ, Sepulveda MA, Robert F, Tora L, Kouzarides T, Birshtein BK, et al. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol 23: 6944–6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol 20: 634–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL 2007. The complex language of chromatin regulation during transcription. Nature 447: 407–412 [DOI] [PubMed] [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L 1992. Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70: 251–265 [DOI] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 30: 2829–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Garcia N, Asensio-Juan E, de la Cruz X, Martinez-Balbas MA 2009. Autoacetylation regulates P/CAF nuclear localization. J Biol Chem 284: 1343–1352 [DOI] [PubMed] [Google Scholar]
- Bonnet J, Wang YH, Spedale G, Atkinson RA, Romier C, Hamiche A, Pijnappel WW, Timmers HT, Tora L, Devys D, et al. 2010. The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep 11: 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Allis CD 1995. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci 92: 6364–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD 1996. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY 2007. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol 27: 3405–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Zhou JX, Allis CD, Berger SL 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J 16: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre C, Ciurciu A, Komonyi O, Jacquier C, Fagegaltier D, Pidoux J, Tricoire H, Tora L, Boros IM, Antoniewski C 2008. The Drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep 9: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RE, Thorner J 2007. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1773: 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurciu A, Komonyi O, Boros IM 2008. Loss of ATAC-specific acetylation of histone H4 at Lys12 reduces binding of JIL-1 to chromatin and phosphorylation of histone H3 at Ser10. J Cell Sci 121: 3366–3372 [DOI] [PubMed] [Google Scholar]
- Cler E, Papai G, Schultz P, Davidson I 2009. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci 66: 2123–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Nebreda AR 2010. Mechanisms and functions of p38 MAPK signalling. Biochem J 429: 403–417 [DOI] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, Weber C, Imbert G, Saudou F, Antoniou E, et al. 1997. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat Genet 17: 65–70 [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6: 1319–1331 [DOI] [PubMed] [Google Scholar]
- Ferreira R, Eberharter A, Bonaldi T, Chioda M, Imhof A, Becker PB 2007. Site-specific acetylation of ISWI by GCN5. BMC Mol Biol 8: 73 doi: 10.1186/1471-2199-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontini M, Soutoglou E, Argentini M, Bole-Feysot C, Jost B, Scheer E, Tora L 2005. TAF9b (formerly TAF9L) is a bona fide TAF that has unique and overlapping roles with TAF9. Mol Cell Biol 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Kim J, Roeder RG 2009. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol Cell Biol 29: 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Romier C, Thuault S, Werten S, Davidson I 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci 26: 250–257 [DOI] [PubMed] [Google Scholar]
- Garcia-Oliver E, Garcia-Molinero V, Rodriguez-Navarro S 2011. mRNA export and gene expression: The SAGA–TREX-2 connection. Biochim Biophys Acta doi: 10.1016/j.bbagrm.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Ge X, Jin Q, Zhang F, Yan T, Zhai Q 2009. PCAF acetylates β-catenin and improves its stability. Mol Biol Cell 20: 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR III, Workman JL 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53 [DOI] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR III, Washburn MP, Abmayr SM, et al. 2006a. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol 26: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Weake V, Swanson SK, Washburn MP, Abmayr SM, Workman JL 2006b. The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol Cell Biol 26: 7178–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, Zha J 2009. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol 29: 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Chen J, Mitchell DL, Johnson DG 2011. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res 39: 1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Nakayama J 2011. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol 2011: 129383 doi: 10.1155/2011/129383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev 15: 3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Juven-Gershon T, Marr MT II, Wright KJ, Tjian R, Kadonaga JT 2008. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev 22: 2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. 2009. Profiling the human protein–DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139: 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol 25: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lu H, Schiltz RL, Pise-Masison CA, Ogryzko VV, Nakatani Y, Brady JN 1999. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol 19: 8136–8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K, Meisterernst M 1996. The human general co-factors. Trends Biochem Sci 21: 342–345 [PubMed] [Google Scholar]
- Kim JH, Saraf A, Florens L, Washburn M, Workman JL 2010. Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes Dev 24: 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E 2008. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10: 707–715 [DOI] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY 2010. Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Demmers J, Karmodiya K, Chang NC, Chang AC, Tora L 2010. ATAC and Mediator coactivators form a stable complex and regulate a set of non-coding RNA genes. EMBO Rep 11: 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L 2011. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell 44: 410–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG 2007. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 26: 4956–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31: 3734–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade L, Rose D, Winston F 2007. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics 177: 2007–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Shao C, McGlynn K, Naziruddin B, Levy MF, Cobb MH 2009. Multiple chromatin-bound protein kinases assemble factors that regulate insulin gene transcription. Proc Natl Acad Sci 106: 22181–22186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol 25: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP 2011. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol 7: 503 doi: 10.1038/msb.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Ai N, Dinh GK, Welsh WJ, Chen JD 2010. Human ADA3 regulates RARα transcriptional activity through direct contact between LxxLL motifs and the receptor coactivator pocket. Nucleic Acids Res 38: 5291–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol 19: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L 1994. Functional similarity and physical association between GCN5 and ADA2: Putative transcriptional adaptors. EMBO J 13: 4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo F, Vidal-Laliena M, Canela N, Zecchin A, Martinez-Balbas M, Agell N, Giacca M, Pujol MJ, Bachs O 2009. The transcriptional co-activator PCAF regulates cdk2 activity. Nucleic Acids Res 37: 7072–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough SD, Grant PA 2010. Histone acetylation, acetyltransferases, and ataxia–alteration of histone acetylation and chromatin dynamics is implicated in the pathogenesis of polyglutamine-expansion disorders. Adv Protein Chem Struct Biol 79: 165–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR III, Grant PA 2005. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci 102: 8478–8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Zhao Y, Nag A, Zeng M, Dimri G, Gao Q, Wazer DE, Kumar R, Band H, Band V 2004. Human ADA3 binds to estrogen receptor (ER) and functions as a coactivator for ER-mediated transactivation. J Biol Chem 279: 54230–54240 [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischerikow N, Spedale G, Altelaar AF, Timmers HT, Pijnappel WW, Heck AJ 2009. In-depth profiling of post-translational modifications on the related transcription factor complexes TFIID and SAGA. J Proteome Res 8: 5020–5030 [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM 2007. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26: 5521–5527 [DOI] [PubMed] [Google Scholar]
- Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, Cserpan I, Lebedeva L, Nabirochkina E, Udvardy A, et al. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol 23: 306–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Tora L 2007. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26: 5341–5357 [DOI] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L 2009. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol 29: 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L 2010. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci 67: 611–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94: 35–44 [DOI] [PubMed] [Google Scholar]
- Orpinell M, Fournier M, Riss A, Nagy Z, Krebs AR, Frontini M, Tora L 2010. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J 29: 2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci 102: 8472–8477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, Tora L, Szabad J, Boros I 2005. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol 25: 8215–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Popescu C, Martin D, Grau B, Zsindely N, Bodai L, Tora L, Ferrus A, Boros I 2010. Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila. Mol Cell Biol 30: 4254–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M 2009. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat Struct Mol Biol 16: 412–420 [DOI] [PubMed] [Google Scholar]
- Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, et al. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol 24: 10826–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Timmers HT 2008. Dubbing SAGA unveils new epigenetic crosstalk. Mol Cell 29: 152–154 [DOI] [PubMed] [Google Scholar]
- Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD 2000. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J 19: 3377–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol 24: 7249–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR III, Grant PA 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol 22: 8774–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K 2002. Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM 2005. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol 12: 1078–1085 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S 2009. Insights into SAGA function during gene expression. EMBO Rep 10: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Hurt E 2011. Linking gene regulation to mRNA production and export. Curr Opin Cell Biol 23: 302–309 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M 2011. Cancer epigenetics reaches mainstream oncology. Nat Med 17: 330–339 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ 2005. ER stress and the unfolded protein response. Mutat Res 569: 29–63 [DOI] [PubMed] [Google Scholar]
- Sermwittayawong D, Tan S 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: Implications for TBP recruitment. EMBO J 25: 3791–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M, Tawfik DS 2010. Mutational effects and the evolution of new protein functions. Nat Rev Genet 11: 572–582 [DOI] [PubMed] [Google Scholar]
- Spedale G, Mischerikow N, Heck AJ, Timmers HT, Pijnappel WW 2010. Identification of Pep4p as the protease responsible for formation of the SAGA-related SLIK protein complex. J Biol Chem 285: 22793–22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL 2000. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL 1999. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Belotserkovskaya R, Berger SL 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci 99: 11622–11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL 2008. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol 15: 364–372 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Mushegian A, Swanson SK, Abmayr SM, Florens L, Washburn MP, Workman JL 2010. The ATAC acetyltransferase complex coordinates MAP kinases to regulate JNK target genes. Cell 142: 726–736 [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178 [DOI] [PubMed] [Google Scholar]
- Timmers HT, Tora L 2005. SAGA unveiled. Trends Biochem Sci 30: 7–10 [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR 2007. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell 27: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT 2008. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev 22: 2359–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598–602 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis CD, Berger SL 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol 17: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E 2008. Human ATAC is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem 283: 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL 2011. SAGA function in tissue-specific gene expression. Trends Cell Biol doi: 10.1016/j.tcb.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL 2008. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J 27: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, Workman JL 2009. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev 23: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, Florens L, Washburn MP, Abmayr SM, Workman JL 2011. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev 25: 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Kaufman RJ 2000. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J Biol Chem 275: 3377–3381 [DOI] [PubMed] [Google Scholar]
- Wieczorek E, Brand M, Jacq X, Tora L 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393: 187–191 [DOI] [PubMed] [Google Scholar]
- Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V 2010. An acetylation switch modulates the transcriptional activity of estrogen-related receptor α. Mol Endocrinol 24: 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Winston F 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol 22: 5367–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15: 199–208 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17: 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenaki G, Ontikatze T, Rajendran R, Stratford IJ, Dive C, Krstic-Demonacos M, Demonacos C 2008. PCAF is an HIF-1α cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene 27: 5785–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J 2011. Structure and function of WD40 domain proteins. Protein Cell 2: 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 26: 229–232 [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci 97: 11303–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, Oishi H, Yamamoto Y, Nagasawa H, McMahon SB, et al. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol Cell 9: 553–562 [DOI] [PubMed] [Google Scholar]
- Yang XJ 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res 32: 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382: 319–324 [DOI] [PubMed] [Google Scholar]
- Zapater M, Sohrmann M, Peter M, Posas F, de Nadal E 2007. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27: 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Kumar A, Meng G, Gao Q, Dimri G, Wazer D, Band H, Band V 2002. Human papilloma virus 16 E6 oncoprotein inhibits retinoic X receptor-mediated transactivation by targeting human ADA3 coactivator. J Biol Chem 277: 45611–45618 [DOI] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29: 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29: 92–101 [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L 2006. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125: 957–969 [DOI] [PubMed] [Google Scholar]
- Zsindely N, Pankotai T, Ujfaludi Z, Lakatos D, Komonyi O, Bodai L, Tora L, Boros IM 2009. The loss of histone H3 lysine 9 acetylation due to dSAGA-specific dAda2b mutation influences the expression of only a small subset of genes. Nucleic Acids Res 37: 6665–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]