Abstract
Bupropion, a clinically-used antidepressant and smoking-cessation drug, acts as a noncompetitive antagonist of nicotinic acetylcholine receptors (nAChRs). To identify its binding site(s) in nAChRs, we developed a photoreactive bupropion analog, (±)-2-(N-tert-butylamino)-3′-[125I]-iodo-4′-azidopropiophenone (SADU-3-72). Based upon inhibition of [125I]SADU-3-72 binding, SADU-3-72 binds with high affinity (IC50 = 0.8 μM) to the Torpedo nAChR in the resting (closed channel) state and in the agonist-induced desensitized state, and bupropion binds to that site with three-fold higher affinity in the desensitized (IC50 = 1.2 μM) than in the resting state. Photolabeling of Torpedo nAChRs with [125I]SADU-3-72 followed by limited in-gel digestion of nAChR subunits with endoproteinase Glu-C established the presence of [125I]SADU-3-72 photoincorporation within nAChR subunit fragments containing M1-M2-M3 helices (αV8-20K, βV8-22/23K and γV8-24K) or M1-M2 helices (δV8-14). Photolabeling within βV8-22/23K, γV8-24K and δV8-14 was reduced in the desensitized state and inhibited by ion channel blockers selective for the resting (tetracaine) or desensitized (thienycyclohexylpiperidine (TCP)) state, and this pharmacologically specific photolabeling was localized to the M2-9 leucine ring (δLeu265, βLeu257) within the ion channel. In contrast, photolabeling within the αV8-20K was enhanced in the desensitized state and not inhibited by TCP, but was inhibited by bupropion. This agonist-enhanced photolabeling was localized to αTyr213 in αM1. These results establish the presence of two distinct bupropion binding sites within the Torpedo nAChR transmembrane domain: a high affinity site at the middle (M2-9) of the ion channel and a second site near the extracellular end of αM1 within a previously described halothane (general anesthetic) binding pocket.
(±)-Bupropion [(±)-2-(tert-butylamino)-1-(3-chlorophenyl) propan-1-one] (Figure 1) is an antidepressant agent (Wellbutrin) that is also effective in treating nicotine dependence (Zyban; (1)). While the consensus view is that bupropion’s therapeutic efficacy as an antidepressant and a smoking cessation agent is attributable to its dual inhibition of dopamine and norepinephrine reuptake transporters, bupropion is also a noncompetitive antagonist (NCA) of several nicotinic acetylcholine receptors (nAChRs, (2–4)). There is emerging evidence that inhibition of neuronal nAChRs, in particular α4β2 and α3β4 subtypes, may contribute to the therapeutic benefit of bupropion as a smoking cessation agent (reviewed in (5)).
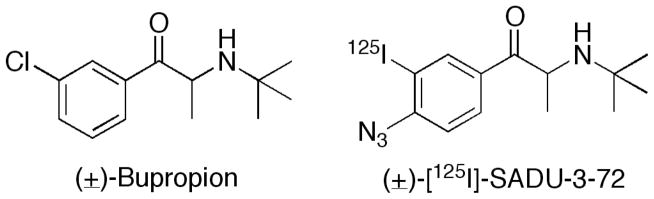
Figure 1.
Chemical structures of bupropion and [125I]-SADU-3-72.
nAChRs are members of the Cys-loop ligand-gated ion channel superfamily, which also includes γ-aminobutyric acid type A (GABAA) receptors, 5-hydroxytryptamine type 3 (5-HT3) receptors), and glycine receptors found in vertebrates, and additional ligand-gated receptors found in invertebrates (reviewed in (6, 7)). Based on the three-dimensional structure of the Torpedo nAChR (8) and the available structural information regarding neuronal nAChRs (reviewed in (9, 10)), nAChRs are pentameric membrane proteins formed by the assembly of homologous subunits, which for the Torpedo nAChR has a subunit stoichiometry of 2α1β1γδ. Each nAChR subunit contains a large extracellular N-terminus and a bundle of four transmembrane α helices (M1–M4). The five M2 helices are arranged about a central axis orthogonal to the membrane forming the channel lumen and the M1, M3, and M4 helices form an outer ring that shield M2 from the lipid bilayer.
Pharmacological studies have shown that bupropion and its analogs noncompetitively inhibit both muscle-type (fetal human α1β1γδ and Torpedo) and neuronal (α4β2, α3β4, α7) nAChRs in the low to intermediate micromolar range (IC50 values range from 0.4–60 μM), with a rank order of potency: α3->α1-~α4->α7- containing nAChRs (4, 5). From an analysis of bupropion effects on agonist-induced macroscopic currents for α1β1εδ (adult mouse muscle-type) nAChRs expressed in HEK-293 cells, it was concluded that bupropion inhibits the receptor via two mechanisms mediated by binding to specific conformational states. Bupropion binding to the resting nAChR results in impaired channel opening (IC50 0.4 μM), while binding to the open state results in either slow channel block or an increase in the rate of desensitization (11). Radioligand competition binding experiments to Torpedo nAChRs further established that bupropion binds to the desensitized state with ~2-fold greater affinity than to the resting (closed) state (11). While molecular docking and dynamics simulations predict that bupropion binds near the middle of the nAChR ion channel (between M2-6 and M2-13; (11)), there has been no direct experimental identification of bupropion binding sites in any nAChR subtype.
To this end, we recently developed a photoreactive analog of bupropion ([125I]-SADU-3-72, Figure 1 (12)). that we use to identify and characterize the binding site(s) for bupropion in the Torpedo nAChR. Having established that [125I]-SADU-3-72 photolabels the Torpedo nAChR in a ‘specific’ manner, that is there is a component of labeling affected by the addition of agonist and inhibitable by bupropion or nAChR NCAs, we employed biochemical and protein chemistry approaches to identify the [125I]-SADU-3-72 -labeled amino acid residues. Our results establish that bupropion does bind in the middle (M2-9) of the Torpedo nAChR channel in the resting and desensitized states. Further, in the desensitized state [125I]-SADU-3-72 and bupropion also bind in proximity to αTyr213 in αM1, a residue that resides within a halothane (general anesthetic) binding pocket (13).
EXPERIMENTAL PROCEDURES
Materials
[125I]-SADU-3-72 (2057 Ci/mmol; Figure 1) was synthesized and radioiodinated according to the procedures outlined in (12). Torpedo californica electric organ was obtained from Aquatic Research Consultants (San Pedro, CA). Bupropion, diisopropylfluorophosphate, bromoacetylcholine, and thienylcyclohexylpiperidine (TCP) were obtained from Sigma-Aldrich (St. Louis, MO). Trypsin (TPCK-treated) was obtained from Worthington, Staphylococcus aureus glutamyl endoproteinase (Glu-C or V8 protease) was obtained from MP Biochemicals (Solon, OH), trifluoroacetic acid (TFA) was obtained from Thermo Fisher Scientific Inc (Rockford, IL), and sodium cholate was obtained from USB Corporation (Cleveland, OH). Protease inhibitor cocktail set III, and Genapol C-100 were obtained from EMD (Calbiochem; Darmstadt, Germany). Prestained low range molecular weight standards and Affigel-10 were obtained from Bio-Rad Laboratories (Hercules, CA). Synthetic lipids were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL) and nonradioactive α–BgTx was obtained from Biotium, Inc. (Hayward, CA). Reversed-phase HPLC columns (Brownlee Aquapore C4 column, 100 × 2.1 mm, BU-300) were obtained from Perkin Elmer Life Sciences Inc. (Boston, MA) and Centriprep-10 concentrators were obtained from Amicon Inc. (Beverly, MA).
Preparation of Affinity-Purified Torpedo nAChR membrane vesicles
Torpedo californica nAChR-rich membranes for radioligand binding studies and for affinity-purification were isolated from frozen electric organs as described previously (14). Torpedo nAChR-rich membranes at 1 mg protein/mL were solubilized in 1% sodium cholate in vesicle dialysis buffer (VDB, 100 mM NaCl, 0.1 mM EDTA, 0.02% NaN3, 10 mM MOPS, pH 7.5) and treated with 0.1 mM diisopropylfluorophosphate after insoluble material was pelleted by centrifugation (91,000g for 1 h). The nAChR was affinity-purified on a bromoacetylcholine bromide-derivatized Affi-Gel 10 column and then reconstituted into lipid vesicles composed of dioleoyl phosphatidylcholine, dioleoyl phosphatidic acid, and cholesterol (DOPC:DOPA:CH at a molar ratio of 3:1:1), as described (15, 16). The lipid to nAChR ratio was adjusted to a molar ratio of 400:1. After purification, the nAChR comprised more than 90% of the protein in the preparation based upon SDS-PAGE. Both the nAChR-rich membranes and purified nAChRs were stored at −80°C.
Electrophysiological Recordings
Voltage clamp electrophysiological studies were conducted using preassembled Torpedo nAChRs incorporated into the plasma membrane of Xenopus oocytes (17, 18). Affinity-purified Torpedo nAChRs reconstituted in DOPC/DOPA/CH (3:1:1 molar ratio) lipid vesicles were microinjected into oocytes (50 nL at 2.5 mg/mL protein). Following an incubation period (48 h) to allow nAChR vesicles to fuse with the plasma membrane, current recordings were performed at room temperature. Currents were recorded from individual oocytes under two-electrode voltage clamp conditions at a holding potential of −60 mV. The ground electrode was connected to the bath via a 3M KCl/agar bridge. Glass microelectrodes had a resistance of <2 megaohms when filled with 3M KCl. Data were acquired and analyzed using a TEV-200 amplifier (Dagan Instruments, Minneapolis, MN), a Digidata 1440A data interface (Molecular Devices, Sunnyvale, CA), and pClamp 10.2 software (Molecular Devices). The concentatration dependence of bupropion inhibition was fit using a variable-slope sigmoidal dose response curve in Prism v5.00 (GraphPad Software, La Jolla, CA). Error bars indicate S.E.M. Acetylcholine (ACh) and bupropion were prepared as 1M and 20 mM stock solutions respectively in distilled water. All solutions were made fresh from stock on the day of the experiment.
[125I]-SADU-3-72 competition binding assays
For competition binding, Torpedo nAChR-rich membranes at 0.11 mg protein/mL or affinity-purified, lipid-reincorporated Torpedo nAChRs at 0.06 mg/mL were incubated for 5 h at room temperature with [125I]-SADU-3-72 (0.2 nM) in the presence of α-bungarotoxin (α-BgTx; 1.4 μM, 30 min preincubation) or carbamylcholine (Carb; 400 μM) and in the presence of increasing concentrations of non-radioactive SADU-3-72, bupropion, or the NCA tetracaine (final concentrations 12.5 nM – 200 μM). α-BgTx, a competitive antagonist, maintains the Torpedo nAChR in the closed (resting) state (19) while Carb stabilizes Torpedo nAChR in the desensitized state. The nonspecific binding was determined in the presence of 200 μM bupropion. Bound and free [125I]-SADU-3-72 were separated by centrifugation (39,000 g for 1 h), then quantified by γ-counting. Bound [125I]-SADU-3-72 was normalized as a percentage of specific binding (total – nonspecific binding) and fit to a simple one-site competition model using Prism v5.00 software. At the [125I]-SADU-3-72 ligand concentration used in these experiments (0.2 nM), for calculated IC50 values of ~ 1μM, the reported IC50 ~Kd.
[125I]-SADU-3-72 photolabeling
For analytical labelings with [125I]-SADU-3-72, 60 μg of affinity-purified nAChR or 125 μg of native nAChR-rich membranes were incubated with ~1.5 nM [125I]-SADU-3-72 (~2100 Ci/mmol) in the presence of 5 μM α-BgTx or 400 μM Carb for 1 h at room temperature under reduced light conditions. The samples were irradiated with a 365 nm hand-held UV lamp (Spectroline EN-280L) for 10 minutes at a distance of less than 1 cm and then pelleted by centrifugation (39,000g for 1 h at 4°C). Pellets were resuspended in electrophoresis sample buffer (12.5 mM Tris-HCl, 2% SDS, 8% sucrose, 1% glycerol, 0.01% bromophenol blue, pH 6.8) and the polypeptides were resolved by SDS-PAGE.
In some experiments, the NCAs tetracaine (130 μM) or thienylcyclohexylpiperidine (TCP; 130 μM) were added to Torpedo nAChR samples incubated with [125I]-SADU-3-72 in the presence of α-BgTx or Carb, respectively. Tetracaine binds in the Torpedo nAChR channel with 30-fold higher affinity in the resting than in the desensitized state, with an established binding locus in the closed channel (20). Reciprocally, TCP, a close structural analog of phencyclidine (PCP), binds to desensitized Torpedo AChR with higher affinity and with nearly identical affinity as PCP, which has an established binding locus in the ion channel in the desensitized state (21). Torpedo nAChR samples were incubated with [125I]-SADU-3-72 in presence of α-BgTx (1.4 μM) or 400 μM Carb and increasing concentrations of bupropion (12.5 nM – 200 μM).
For preparative photolabelings, 3 mg of affinity-purified, lipid-reincorporated (DOPC/DOPA/CH 3:1:1) Torpedo nAChRs (0.2 mg/mL) or 20 mg of native Torpedo nAChR-rich membranes were incubated with 7 or 15 nM [125I]-SADU-3-72 (~240–700 μCi) under two different sets of conditions: a) in the presence of 5 μM α-BgTx or 400 μM Carb; b) in the presence of 400 μM Carb and in the absence or presence of 130 μM TCP. These samples were then photolyzed and processed for SDS-PAGE under the same conditions as analytical labeling experiments.
SDS-Polyacrylamide Gel Electrophoresis
SDS-PAGE was performed according to (22) with separating gels comprised of 8% polyacrylamide/0.33% bisacrylamide (1 mm thick gels for analytical labelings; 1.5 mm thick gels for preparative labeling experiments). Following electrophoresis, gels were stained for 1 h with Coomassie Blue R-250 (0.25% (w/v) in 45% methanol, 10% acetic acid, 45% H2O), and destained (25% methanol, 10% acetic acid, 65% H2O) to visualize bands. Gels were then dried and exposed to Kodak X-OMAT LS film with an intensifying screen at −80°C (5–24 h exposure). After autoradiography, the bands corresponding to the [125I]-SADU-3-72-labeled nAChR subunits were excised from each condition, soaked in overlay buffer (5% sucrose, 125 mM Tris-HCl, 0.1% SDS, pH 6.8) for 30 min, and transferred to the wells of a 15% acrylamide “mapping” gel (23). Each gel slice was overlaid with 5 μg (analytical labeling) or 100 μg (preparative labeling) of S. aureus V8 protease in overlay buffer. After electrophoresis, the gels were stained for 1 h with Coomassie Blue R-250, destained, and either prepared for autoradiography (analytical labeling) or soaked in distilled water overnight (preparative labeling). The 125I-containing bands were excised from the preparative gels and the peptides were retrieved by passive diffusion into 25 mL of elution buffer (0.1M NH4HCO3, 0.1% (w/v) SDS, 1% β-mercaptoethanol, pH 7.8) for 4 days at room temperature with gentle mixing. The eluates were filtered to remove gel pieces and then concentrated using Centriprep-10 concentrators (10 kDa cutoff, Amicon, final volume <150 μL). Samples were then either directly purified by reversed-phase HPLC or acetone precipitated (>85% acetone at −20°C overnight) to remove excess SDS and then subjected to further proteolytic digestion.
Proteolytic Digestions and Tricine SDS-PAGE
For digestion with trypsin, acetone-precipitated subunit fragments were suspended in 30 μL 0.1M NH4HCO3, 0.1% SDS, pH 7.8, and then the SDS content was diluted by addition of 113 μL 0.1M NH4HCO3 and 7.5 μL Genapol C-100 (final concentrations: 0.02% (w/v) SDS, 0.5% Genapol C-100, pH 7.8). Trypsin (60 μg) was added and the digestion was allowed to proceed for 5 days at room temperature.
Material from each digest was then resolved on 1.0 mm thick small pore (16.5%T/6%C) Tricine SDS-PAGE gels (24, 25). After Coomassie Blue R-250 staining (1 h) and destaining (3–4 h), Tricine gels were dried and exposed to film (8–12 h). The 125I-containing bands were excised from the Tricine gels and subjected to reversed-phase HPLC purification as indicated in the previous section.
Reversed-Phase HPLC Purification
HPLC was performed on a Shimadzu LC-10A binary HPLC system using a Brownlee Aquapore C4 column (100 × 2.1mm). Solvent A was comprised of 0.08% trifluoroacetic acid (TFA) in water and Solvent B was comprised of 0.05% TFA in 60% acetonitrile/40% 2-propanol. A nonlinear elution gradient at 0.2 mL/min was employed (25% to 100% Solvent B in 100 min, shown as a dotted line in the figures) and fractions were collected every 2.5 min (42 fractions/run). The elution of peptides was monitored by the absorbance at 210 nm and the amount of 125I associated with each fraction was determined by γ-counting (5 minute/fraction) in a Packard Cobra II γ-counter.
Sequence Analysis
Amino terminal sequence analysis of Torpedo nAChR subunit fragments was performed on a Applied Biosystems PROCISE 492 protein sequencer configured to utilize 1/6 of each cycle of Edman degradation for amino acid identification/quantification and collect the other 5/6 for 125I counting. The pooled HPLC fractions were diluted 3-fold with 0.1% TFA in distilled water (to reduce organic concentration) and loaded onto PVDF disks using Prosorb sample preparation cartridges (Applied Biosystems No. 401950). Before sequencing, filters were processed as recommended by the manufacturer. To determine the amount of the sequenced peptide, the pmol of each amino acid in a detected sequence was quantified by peak height and fit to the equation f(x) = I0Rx, where I0 was the initial amount of the peptide sequenced (in pmol), R was the repetitive yield, and f(x) was the pmol detected in cycle x. Ser, His, Trp, and Cys were not included in the fits due to known problems with their accurate detection/quantification. The fit was calculated in SigmaPlot 11 (SPSS) using a non-linear least-squares method and figures containing 125I release profiles (Figures 6C, 6D, 7B and 8B) include this fit as a dotted line. Quantification of 125I incorporated into a specific residue was calculated by (cpmx−cpm(x−1))/5IoRx.
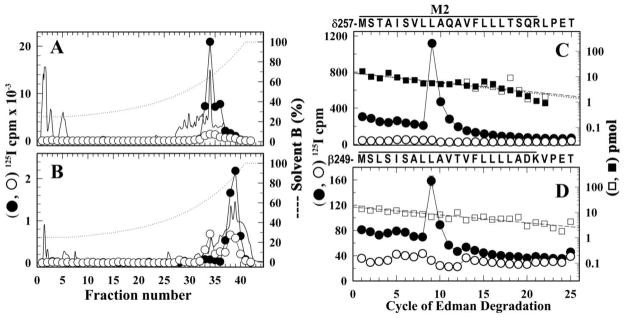
Figure 6. Identification of amino acids photolabeled by [125I]-SADU-3-72 in the δ M2 and βM2 segments in the Torpedo nAChR.
Affinity-purified nAChR was photolabeled with [125I]-SADU-3-72 in the presence of α-BgTx (●) or Carb (○) as detailed in Experimental Procedures. Shown are reversed-phase HPLC fractionation (A, B) and amino acid sequence analysis (C, D) of [125I]-SADU-3-72-labeled proteolytic fragments δT-5K (A, C) and βT-7K (B, D). The elution of peptides during HPLC was monitored by absorbance at 210 nm (solid line) and 125I elution was quantified by γ counting of each fraction (●, ○). C, 125I (●, ○) and PTH- amino acids (■, □) released during amino acid sequence analysis of 125I peak (fractions 33–35; ●, 20,000 cpm; ○, 2,000 cpm) from the HPLC purification of δT-5K. In each sample, the primary peptide detected began at δMet257 (●, I0 = 13.8 ± 2.4 pmol, R = 92%; ○, I0 = 14 ± 1.5 pmol, R = 92%) and for the (●) sample there was a peak of 125I cpm release in cycle 9 corresponding to labeling of δLeu265 (30 cpm/pmol). D, 125I (●, ○) and PTH-amino acids (■, □) released during amino acid sequence analysis of 125I peak (fractions 37–40; ●, 5,000 cpm; ○, 1,700 cpm) from the HPLC purification of βT-7K. In each sample, the primary peptide detected began at βMet249 (●, I0 = 9 ± 1 pmol, R = 92%; ○, I0 = 16 ± 3 pmol, R = 93%) and for the (●) sample there was a peak of 125I cpm release in cycle 9 corresponding to labeling of βLeu257 (5 cpm/pmol).
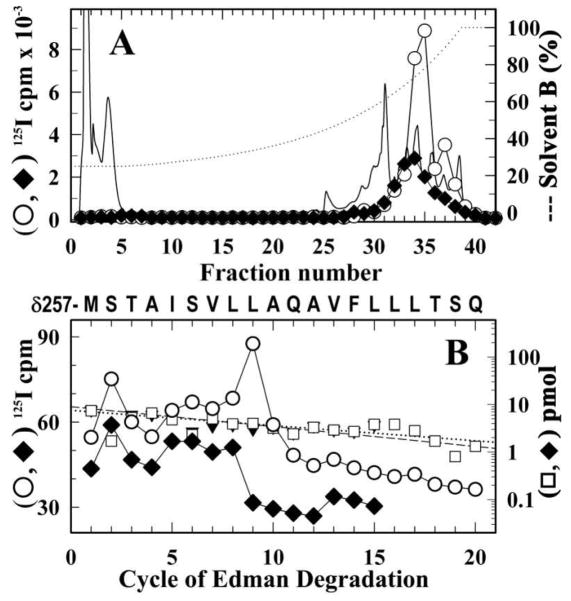
Figure 7. Identification of amino acids photolabeled by [125I]-SADU-3-72 in the δ M2 segment in the desensitized Torpedo nAChR.
nAChR-rich membranes were labeled with [125I]-SADU-3-72 in the presence of 400 μM Carb and in the absence (○, □) or presence 130 μM TCP (◆, ▼). As detailed in Experimental Procedures, the δ subunit was isolated from an 8% SDS-PAGE gel and digested ‘in-gel’ with V8 protease. The labeled fragment δV8-14K was isolated and further digested with trypsin. The fragment δT5K was then isolated from a small-pore Tricine SDS-PAGE gel. Shown are reversed-phase HPLC fractionation (A) and amino acid sequence analysis (B) of [125I]-SADU-3-72-labeled proteolytic fragments δT-5K (A). The elution of peptides during HPLC was monitored by absorbance at 210 nm (solid line) and 125I elution was quantified by γ counting of each fraction (○, ◆). B, 125I (○, ◆ ) and PTH-amino acids (□, ▼) released during amino acid sequence analysis of 125I peak (fractions 33–35; ○, 7,000 cpm; ◆, 3,000 cpm) from the HPLC purification of δT-5K. In each sample, the primary peptide detected began at δMet257 (□, I0 = 9 ± 1 pmol, R = 91%; ▼, I0 = 8 ± 1 pmol, R = 93%) and for the (◆) sample there were peaks of 125I cpm release in cycles 2 and 9 corresponding to labeling of δSer258 (3 cpm/pmol) and δLeu265 (5 cpm/pmol).
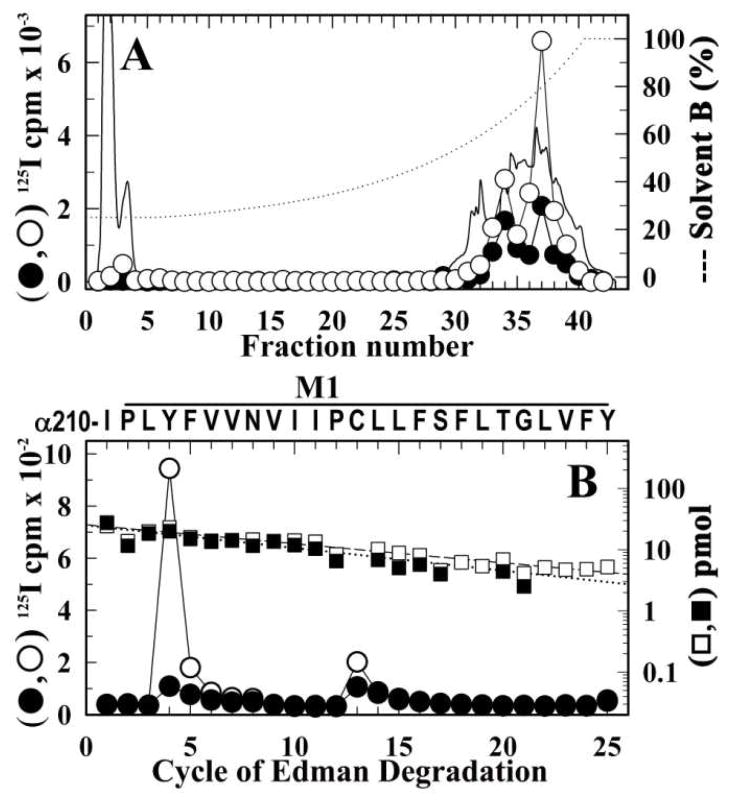
Figure 8. Identification of amino acids photolabeled by [125I]-SADU-3-72 in the α M1 segment of the resting Torpedo nAChR.
From the photoabeling experiment of Figure 6, α subunits from affinity-purified nAChR photolabeled in the presence of α-BgTx (●) or Carb (○) were digested ‘in-gel’ with V8 protease. The labeled fragment αV8-20K was then isolated and further digested with trypsin. The tryptic fragment αT5K was then isolated from a small-pore Tricine SDS-PAGE gel. Shown are reversed-phase HPLC fractionation (A) and amino acid sequence analysis (B) of the [125I]-SADU-3-72-labeled αT5K. A, The elution of peptides during HPLC was monitored by absorbance at 210 nm (solid line) and 125I elution was quantified by γ counting of each fraction (●, ○). B, 125I (●, ○) and PTH-amino acids (■, □) released during amino acid sequence analysis of 125I peak (fractions 36–38; ●, 3,000 cpm; ○, 10,000 cpm) from the HPLC purification of αT5K. In each sample, the primary peptide detected began at αIle210 (●, I0 = 25 ± 2 pmol, R = 92%; ○, I0 = 26 ± 2 pmol, R = 93%) and for both samples there were peaks of 125I cpm release in cycles 4 and 13 corresponding to labeling of αTyr213 (●, 1 cpm/pmol; ○, 10 cpm/pmol) and αCys222 (●, 2 cpm/pmol; ○, 3 cpm/pmol).
RESULTS
Functional inhibition of Torpedo nAChRs by Bupropion
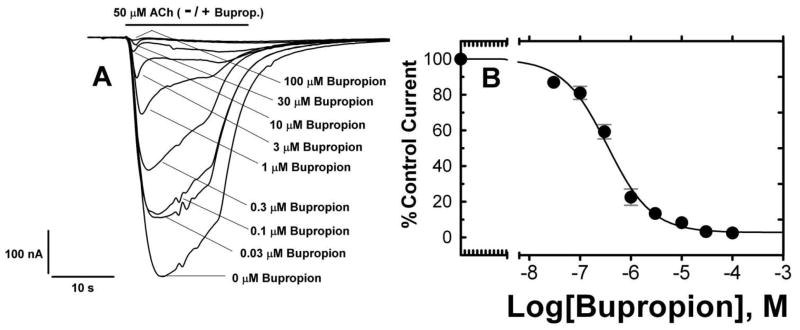
To characterize bupropion as a Torpedo nAChR inhibitor, we employed a rather unique variant of the Xenopus oocyte/electrophysiology expression system introduced by the Ricardo Miledi group (17, 18). In this system, pre-assembled Torpedo nAChRs are microinjected into oocytes, and following fusion of receptor containing lipid vesicles with the oocyte plasma membrane, whole-cell two-electrode voltage-clamp recordings are used to monitor receptor functionality. This system allowed us to monitor receptor functionality and bupropion effects with the same protein/receptor material that we used subsequently for bupropion binding and photoaffinity labeling experiments. In preliminary experiments, we measured ACh responses as a function of time after microinjection (50 nL; 2.5 mg/mL protein) of Torpedo nAChR-rich membranes (native nAChR) or affinity-purified and lipid reconstituted (DOPC/DOPA/CH 3:1:1) Torpedo nAChR (purified nAChR) that are structurally and functionally nearly indistinguishable from Torpedo nAChR-rich membranes (16, 21, 26). Maximum acetylcholine (ACh) induced currents were obtained approximately 48 h after nAChR microinjection, and the ACh dose-response curve yielded values (EC50 = 50 μM; Hill coefficient, nH= 1.5; n =1) consistent with previously published data (18). Co-application of bupropion resulted in a dose-dependent and reversible reduction in ACh-induced currents (Figure 2) with an IC50 = 0.34 ± 0.07 μM, nH = 0.98 ± 0.14 (n =7). Thus bupropion inhibits Torpedo nAChR function with similar potency as observed for mouse α1βεδ nAChRs (11).
Figure 2. Bupropion Inhibition of ACh-evoked currents of Torpedo AChRs microinjected into Xenopus oocytes.
Currents elicited by ACh from affinity-purified and lipid-reconstituted (DOPC/DOPA/CH 3:1:1) Torpedo nAChR vesicles microinjected into Xenopus oocytes were measured using a standard two-electrode voltage clamp at a holding potential of −60 mV. (A) Once a stable response was observed for an EC50 dose (50 μM) of ACh, responses were measured when ACh was applied simultaneously for 20 seconds with increasing concentrations of bupropion (0.03–100 μM), and in each case a representative current trace is displayed. Inhibition by bupropion was reversible since peak responses to ACh returned to control levels after exposure to 3 μM bupropion followed by a six minute wash (not shown). (B) Nonlinear least-squares analysis of the curves yielded an IC50 = 0.34 ± 0.07 μM, nH = 0.98 ± 0.14 (7 oocytes). Currents were normalized to the 50 μM ACh response.
Pharmacological characterization of [125I]SADU-3-72 binding to Torpedo nAChRs
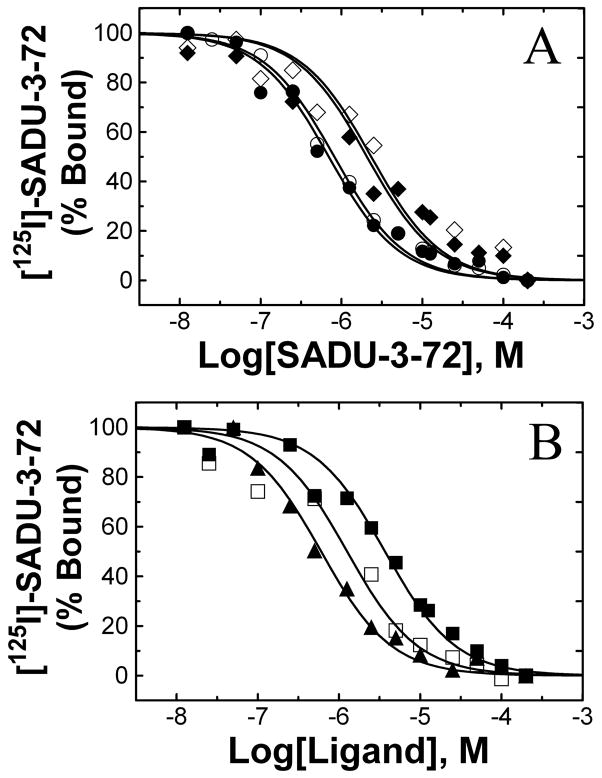
The effect of non-radioactive SADU-3-72 and bupropion on the equilibrium binding of [125I]SADU-3-72 to the Torpedo nAChR in the resting and Carb-induced desensitized states was assessed (Figure 3). SADU-3-72 inhibited [125I]SADU-3-72 binding to native nAChR (IC50,0.8 μM) and to purified Torpedo nAChRs (IC50, 2 μM) in the resting or desensitized state. Bupropion had the same potencies as an inhibitor of [125I]SADU-3-72 binding to the native Torpedo nAChR in the desensitized state (IC50, 1.2 ± 0.34 μM) or resting state (IC50, 3.6 ± 0.46 μM) (Figure 3B) as reported previously for its inhibition of [3H]TCP binding (11). Tetracaine, which binds in the resting state ion channel at the level of M2-9 (20), also fully inhibited [125I]SADU-3-72 binding (Figure 3B; IC50, 0.42 ± 0.05 μM). Competition binding data were in each case well fit to a simple one-site competition model, suggesting that tetracaine, bupropion, and [125I]SADU-3-72 bind at a common site within the resting receptor.
Figure 3. Inhibition of [125I]-SADU-3-72 binding to Torpedo nAChRs by (A) SADU-3-72 and (B), bupropion or tetracaine.
The equilibrium binding of [125I]-SADU-3-72 (0.22 nM) to Torpedo nAChRs in the absence (closed symbols) or presence (open symbols) of 400 μM Carb was determined by centrifugation. A, SADU-3-72 inhibited binding to native nAChR-rich membranes (●,○) with IC50s of 0.7± 0.07 (●) and 0.8 ± 0.08 (○) μM and to affinity-purified, lipid-reconstituted nAChR with IC50s of 2.3 ± 0.7 (◆) and 2.0 ± 0.7 (◇) μM. For nAChR-rich membranes, the total binding of [125I]-SADU-3-72 was 29,078 and 26,301 cpm in the absence and presence of Carb, with 10,305 and 11,215 cpm bound non-specifically in the presence of 200 μM SADU-3-72. For the purified nAChR, the total binding of [125I]-SADU-3-72 was 71,980 and 83,161 cpm in the absence and presence of Carb, with 12,211 and 12,183 cpm bound non-specifcally in the presence of 200 μM SADU-3-72. B, Bupropion (■,□) and tetracaine (▲) inhibited binding to Torpedo nAChR-rich membranes with IC50s of, 3.6 ± 0.5 (■),1.2 ± 0.3 (□), and 0.42 ± 0.05 (▲) μM. Nonspecific binding was determined in the presence of 200 μM bupropion. In each case values of IC50 were determined by fitting the normalized specific binding to a one-site model.
[125I]SADU-3-72 photolabeling of the Torpedo nAChR
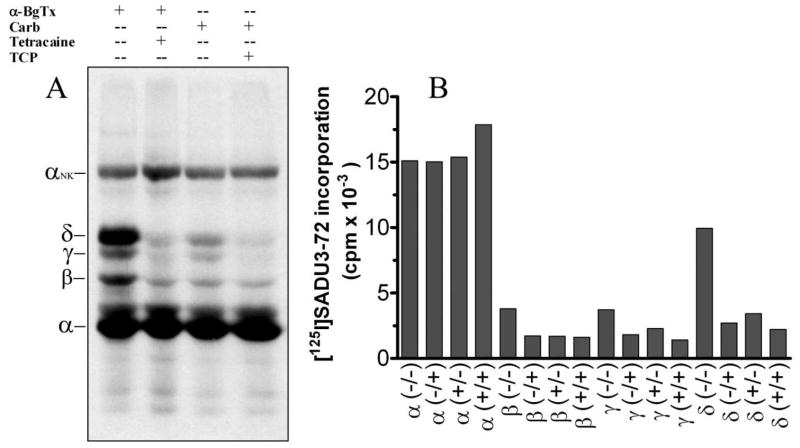
In initial photolabeling experiments we characterized photoincorporation into the Torpedo nAChR in the resting state (stabilized by α-BgTx (19)) and in the desensitized state (stabilized by Carb), and we examined the effect of tetracaine or TCP on the extent of photolabeling. Torpedo nAChR-rich membranes or purified nAChR in lipid vesicles were photolabeled with [125I]SADU-3-72 (1.5 nM), and after UV-irradiation, nAChR subunits were separated by SDS-PAGE, and photolabeling was monitored by autoradiography (Figure 4A) and by γ counting of excised subunit bands (Figure 4B). A representative autoradiograph of an SDS-PAGE gel of [125I]SADU-3-72 labeled native nAChR (Figure 4A) demonstrates photoincorporation into each nAChR subunit. In the nAChR resting state (+α-BgTx), tetracaine reduced photolabeling in the β–, γ- and δ-subunits by 55, 51, and 73% respectively, but by <5% in the α-subunit. Compared to the nAChR resting state, [125I]SADU-3-72 photolabeling in the desensitized state (Carb +) was unchanged in the α subunit, but was reduced by 50–70% in the β-, γ-, and δ-subunits. In the nAChR desensitized state, TCP inhibited [125I]SADU-3-72 photolabeling in the γ– and δ-subunits by ~35%, while photolabeling in the α-subunit was increased by ~15%, and β subunit labeling was unchanged. As a control, no reproducible effect on the extent of [125I]SADU-3-72 photolabeling of the α-subunit of the Na,K-ATPase (αNK band) was observed between any of the different labeling conditions (data not shown). Qualitatively and quantitatively similar results for nAChR labeling were observed for [125I]SADU-3-72 photolabeling experiments conducted with purified Torpedo nAChRs (data not shown).
Figure 4. Photoincorporation of [125I]-SADU-3-72 into the Torpedo nAChR in the absence and presence of Carb.
A, An autoradiograph (12–24 h exposure with intensifying screen) of an 8% SDS-PAGE gel containing native Torpedo nAChR-rich membranes photolabeled with [125I]-SADU-3-72 in the absence (−) and/or the presence (+) of the agonist Carbamylcholine (Carb), the competitive antagonist α-bungarotoxin (αBgTx), the resting state-selective channel blocker tetracaine, or the desensitized state-selective channel blocker thienycyclohexylpiperidine (TCP). The migration of individual nAChR subunits and the alpha subunit of Na/K ATPase (αNK) is indicated on the left. B, 125I cpm incorporation into each nAChR subunit for each of the labeling conditions of Panel A, as determined by γ-counting of individual nAChR subunit bands excised from the dried gel after autoradiography (n =1). The notations −/−, +/−, −/+ and +/+ indicate [125I]SADU-3-72 photolabelings in the presence of αBgTx (resting state), Carb (desensitized sate), αBgTx and tetracaine or Carb and TCP, respectively.
Bupropion also inhibited [125I]SADU-3-72 photolabeling in a concentration-dependent manner, as determined by the level of subunit photolabeling determined by SDS-PAGE. The concentration dependence of bupropion inhibition of δ subunit photolabeling in the absence (IC50, 5.5 ± 0.7 μM) and presence of Carb (IC50, 2.1 ± 0.7 μM) (Supplementary Figure S1) was similar to that seen for the inhibition of reversible [125I]SADU-3-72 binding (Figure 3B).
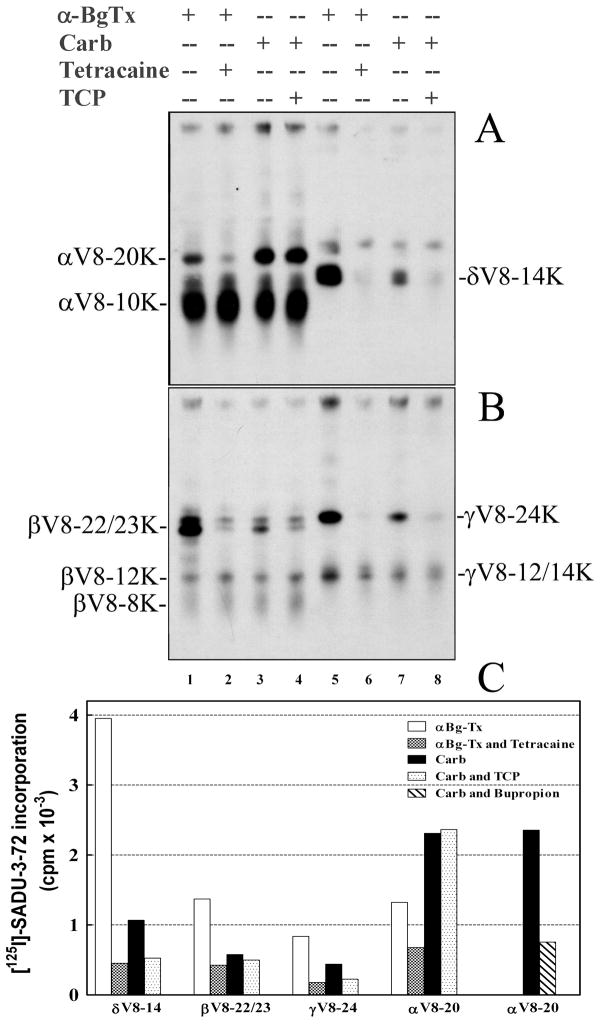
To further localize the site(s) of [125I]SADU-3-72 labeling within each nAChR subunit, we employed limited digestion with S. aureus V8 protease in a 15% acrylamide mapping gel (23). Limited V8 digestion reproducibly generates a set of non-overlapping fragments for each receptor subunit (25, 27–29) including fragments of ~20kDa containing the M1, M2, and M3 helices (αV8-20K, βV8-22/23K, γV8-24K) or of 14 kDa containing the M1 and M2 helices (δV8-14K), and fragments of 10–14 kDa containing the M4 helix (αV8-10K, βV8-12K, and γV8-14K).
Inspection of the autoradiograph of the V8 protease digests (Figure 5A and B) and γ counting of the excised gel bands (Figure 5C) revealed that for nAChR in the resting state, tetracaine inhibited [125I]SADU-3-72 labeling within the αV8-20K, βV8-22/23K, γV8-24K, and δV8-14K fragments by 49, 76, 79, and 88% respectively, which is likely to result from photolabeling in each subunit in the M2 ion channel domain. Compared to the resting state, for the nAChR desensitized state photolabeling in the βV8-22/23K, γV8-24K, and δV8-14K fragments was reduced by 60, 45, and 75%, and TCP further inhibited labeling in each of these fragments. The TCP-inhibitable [125I]SADU-3-72 labeling within βV8-22/23K, γV8-24K, and δV8-14K predicts photolabeling of amino acids within these M2 segments, In contrast, there was an approximately 2-fold increase in photolabeling within αV8-20K in the desensitized compared to resting state. This agonist-enhanced photolabeling within αV8-20K was not inhibited by TCP, but bupropion did reduce the photolabeling by 70% (Figure 5C). In each subunit there was also [125I]SADU-3-72 labeling that was neither affected by agonist (Carb) nor inclusion of tetracaine or TCP and that mapped to the αV8-10K, βV8-12K, βV-8K, and γV8-14K subunit fragments that contain the M4 helices.
Figure 5. Proteolytic mapping of the sites of [125I]-SADU-3-72 incorporation into AChR subunits by autoradiography (A and B) and γ-counting of subunit fragments (C).
Native Torpedo nAChR-rich membranes were photolabeled with [125I]-SADU-3-72 (1.5 nM) in the presence of α-BgTx (resting state), − or + tetracaine (130 μM), or Carb (desensitized state), − or + TCP (140 μM). Subunits were resolved by SDS-PAGE on an 8% acrylamide gel, and the nAChR subunit bands were excised, transferred to the wells of a 15% mapping gel for “in gel” digestion with S. aureus V8 protease (see Experimental Procedures). [125I]-SADU-3-72 incorporation into nAChR subunit fragments was determined by autoradiography (A, α and δ subunits; B, β and γ subunits (24 h exposure with intensifying screen)) and by γ - counting of excised gel bands (C). The principal [125I]-SADU-3-72 labeled proteolytic fragments are shown using the nomenclature of Blanton and Cohen (25) (see text). C, 125I cpm incorporated in δV8-14, βV8-22/23K, V8-24K, and αV8-20K from the experiment of Panels A and B, and in αV8-20K from a separate experiment for nAChRs in the presence of Carb (desensitized state) photolabeled with [125I]-SADU-3-72 in the absence or presence of 130 μM bupropion. Not included in C is the quantification by γ-counting of 125I incorporation (mean ± SEM for the 4 conditions) in the other excised gel bands of Panels A and B: aV8-10K (4,630 ± 290 cpm), βV8-12K (313 ± 12 cpm), βV-8K ( 326 ± 18 cpm), γV8-12/14K (560 ± 31 cpm).
[125I]SADU-3-72 photolabeling in the M2 ion channel domain
For the Torpedo nAChR in the resting state, the principal site of tetracaine-inhibitable labeling resides within δV8-14K and βV8-22/23K (Figure 5). To identify the photolabeled amino acids, δV8-14 and βV8-22/23 fragments, isolated from Torpedo nAChRs labeled with [125I]SADU-3-72 in the presence of α-BgTx or Carb, were further digested with trypsin, and the digests were separated by Tricine SDS-PAGE (Supplementary Figure S2, Panels B, C). The principal labeled fragments (δT-5K and βT-7K) isolated from the gel were further purified by reversed-phase HPLC (Figure 6A, 6B), and peak 125I cpm fractions were subjected to amino acid sequence analysis (Figure 6C, 6D). Sequencing revealed in each case the presence of a primary peptide beginning at the start of the M2 segment (δMet257 and βMet249) and that [125I]SADU-3-72 labeled homologous leucine residues (δLeu265, 30 cpm/pmol; βLeu257, 5 cpm/pmol) located nine residues from the start of the M2 segment (M2-9).
Photolabeling of βM2-9 and δM2-9 was reduced by >90% in the desensitized compared to resting state, despite the fact [125I]SADU-3-72 binds with similar affinity to the nAChR in both states (Figure 3A). There was, however, TCP-inhibitable photolabeling in δV8-14K and βV8-22/23K (Figure 5). To determine whether [125I]SADU-3-72 photolabeled the ion channel in the desensitized state, we carried out a preparative photolabeling of Torpedo nAChR in the desensitized state in the absence and presence of TCP using a higher concentration of [125I]SADU-3-72. As before, the fragment beginning at δMet-257 was isolated from trypsin digests of δV8-14K by Tricine SDS-PAGE (Supplementary Figure S2, Panel D) and reversed-phase HPLC (Figure 7A). Amino acid sequence analysis (Figure 7B) revealed the presence of a primary peptide beginning at the start of the M2 segment (δMet257), with a small peak of 125I release in cycle 9, consistent with photolabeling of δLeu265 (M2-9, 5 cpm/pmol), and an additional peak of 125I release in cycle 2, which indicates photolabeling of δSer258 (M2-2, 3 cpm/pmol). Inclusion of 130 μM TCP eliminated the [125I]SADU-3-72 labeling of δLeu265 (M2-9).
Agonist-dependent [125I]SADU-3-72 photolabeling within the α M1
[125I]SADU-3-72 photolabeling of αV8-20K was 2-fold greater in the nAChR desensitized state than in the resting state, and that photolabeling was inhibited by bupropion but not by the channel blocker TCP (Figure 5C). To further localize this Carb-enhanced, bupropion-inhibitable labeling, the αV8-20K fragments isolated from Torpedo nAChRs labeled with [125I]SADU-3-72 in the absence and presence of Carb, were digested with trypsin, the digests separated by Tricine SDS-PAGE gel (Supplementary Figure S2, Panel A). The principal photolabeled fragment (αT-5K) was isolated and purified by reversed-phase HPLC (Figure 8A). Amino acid sequence analysis (Figure 8B) revealed a primary peptide beginning at αIle210 at the NH2-terminus of the αM1 segment. The major peak of 125I release in cycle 4 indicated photolabeling of αTyr213, with labeling of this residue ~10-fold greater in the desensitized vs. resting Torpedo nAChR (10 and 1 cpm/pmol, respectively). In addition, there was a small peak of 125I release in cycle 13, consistent with [125I]SADU-3-72 labeling of αCys222 at similar efficiency in the resting and desensitized states (2 cpm/pmol).
[125I]SADU-3-72photolabeling in α M4
Approximately 80% of the total [125I]SADU-3-72 photoincorporation into the α-subunit resides within the proteolytic fragment αV8-10 ((Asn339-Gly437) Figure 5; with that photolabeling unaltered by inclusion of agonist or channel blockers (TCP, tetracaine). To identify the photolabeled amino acids, αV8-10K fragments, isolated from Torpedo nAChRs labeled with [125I]SADU-3-72 in the absence and presence of Carb, were digested with trypsin. When the digests were fractionated by reversed-phase HPLC (Supplementary Figure S3A), all 125I was recovered in a broad hydrophobic peak, which amino acid sequence analysis (Supplementary Figure S3B) identified as a fragment beginning at aTyr401 and extending through aM4, with peaks of 125I release indicating [125I]SADU-3-72 labeling of αCys412 and αCys418, residues previously identified as residing at the lipid-exposed helical face of the M4 segment (8, 25, 30).
DISCUSSION
Bupropion became the first FDA approved non-nicotine medication (Zyban®) for smoking cessation in 1997, after many years of clinical use as an antidepressant (Wellbutrin®). While inhibition of presynaptic dopamine and norepinephrine transporters is believed to be the primary mechanism underlying bupropion’s efficacy, there is emerging evidence that inhibition of neuronal nAChRs also contributes to its efficacy in treating nicotine dependence (reviewed in (4, 5). Bupropion inhibits noncompetitively a diverse group of muscle and neuronal nAChR subtypes (4, 5), with a predicted binding site within the receptor’s ion channel (11). A novel photoreactive analogue of bupropion, (±)-2-(N-tert-butylamino)-3′-[125I]-iodo-4′-azidopropiophenone ([125I]-SADU-3-72), has been recently developed that photoincorporates into dopamine transporters and nAChRs (12). In this report we used [125I]-SADU-3-72 to identify the amino acids contributing to two distinct binding sites for SADU-3-72 and bupropion in the Torpedo (muscle-type) nAChR transmembrane domain: the predicted binding site in the ion channel and a binding site near the extracellular end of αM1.
Pharmacological Characterization of Bupropion and [125I]-SADU-3-72 Interactions with the Torpedo nAChR
For Torpedo nAChRs injected into Xenopus oocytes, we found that bupropion inhibits reversibly ACh-induced currents with an IC50 of 0.3 μM, similar to the reported IC50 for bupropion inhibition of mouse muscle-type nAChR (11). We also established that bupropion inhibits the reversible binding and UV-induced photoincorporation of [125I]-SADU-3-72 into the Torpedo nAChR with high affinity in the desensitized state (IC50’s of 1 and 2 μM), as expected for mutually exclusive binding at a common site, and with three-fold lower affinity in the resting state. Furthermore, tetracaine, a resting state selective ion channel blocker, inhibited the reversible binding of [125I]-SADU-3-72 (IC50, 0.4 μM) and [125I]-SADU-3-72 photoincorporation within nAChR subunit fragments containing the M2 segments (50–90% inhibition in αV8-20, βV8-22, γV8-24 and δV8-14), which suggested the presence of a binding site for bupropion/[125I]-SADU-3-72 within the ion channel in the nAChR resting state. For the nAChR in the desensitized state, TCP, which binds with high affinity in the ion channel in the desensitized state, inhibited [125I]-SADU-3-72 photolabeling within γV8-24 and δV8-14, but not within αV8-20, where there was agonist-enhanced photolabeling that was inhibitable by bupropion. These results indicate the presence of at least two binding sites for [125I]-SADU-3-72/bupropion in the nAChR in the desensitized state: a site within the ion channel that is inhibitable by TCP and a second site, inhibitable by bupropion, that is in proximity to amino acid(s) within αM1/α M2/and/or αM3.
[125I]-SADU-3-72/Bupropion Binding Site(s) in the Torpedo nAChR Transmembrane Domain
The nAChR amino acids photolabeled by [125I]-SADU-3-72 are located in three different regions of the nAChR transmembrane domain (Figure 9): (i) within the ion channel, identified by photolabeling in the resting and desensitized states of the homologous residues, δLeu265 and βLeu257, at position M2-9; (ii) at the α subunit interfaces not containing the transmitter binding sites (α−), in proximity to amino acids from the γ and/or β subunits, identified by the photolabeling in the desensitized state of αTyr213 near the extracellular end of αM1; (iii) at the lipid interface, identified by the photolabeling of αCys412 and αCys418 in αM4 at similar efficiency in the resting and desensitized states. αCys412 and αCys418, and others lying on a common strip of the αM4 helix, have been photolabeled by a variety of hydrophobic probes of diverse structure and photoreactivity that partition into lipid (25, 27, 31, 32).
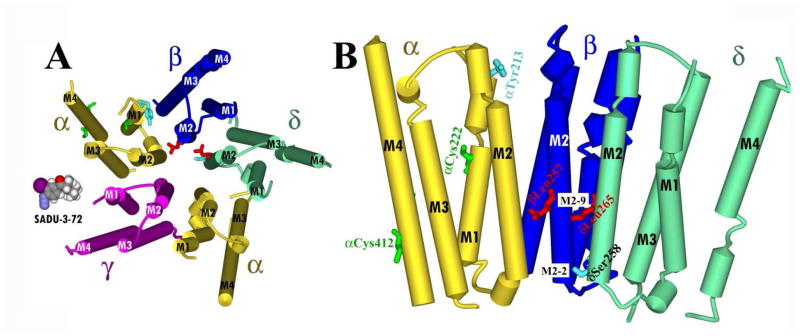
Figure 9. Molecular model of sites of [125I]-SADU-3-72 labeling in the Torpedo nAChR structure (PDB # 2BG9).
Residues photolabeled by [125I]-SADU-3-72 within the transmembrane domain of the Torpedo nAChR. Views of the membrane-spanning helices (shown as cylinders) of the Torpedo nAChR structure (PDB # 2BG9): A, looking down the channel from the base of the extracellular domain; and B, looking parallel to the membrane with 2 subunits removed for clarity, rotated 90° from (A). Subunits are color-coded: α, yellow; β blue; and δ, green. Residues photolabeled by [125I]-SADU-3-72 are included in stick format, color-coded by domain and conformation: ion channel, resting state (red); ion channel, desensitized state (cyan); lipid-protein interface (green). A Connolly surface model of [125I]-SADU-3-72 is included in A for scale.
[125I]-SADU-3-72/Bupropion Binding in the Ion Channel
[125I]-SADU-3-72 photolabeled βM2-9 and δM2-9 within the ion channel at >10-fold higher efficiency in the resting state than in the desensitized state, despite the fact that [125I]-SADU-3-72 binds with similar high affinity (~1 μM) in both states (Figure 3A), as does bupropion (11). In this regard, [125I]-SADU-3-72 mirrors [125I]TID, a NCA that binds in the nAChR ion channel with similar high affinity in the desensitized and resting states but photolabels amino acids in the nAChR ion channel with ~10-fold higher efficiency in the resting state than in the desensitized state (33, 34). By photolabeling nAChRs in the desensitized state at a higher concentration of [125I]SADU-3-72, we also identified photolabeling of δLeu265 (M2-9), inhibitable by TCP, and δSer258 (M2-2). This indicates that SADU3-72 and bupropion bind to the same site (M2-9) within the ion channel in the resting and desensitized states, but with a broader binding locus in the desensitized state.
[125I]-SADU-3-72/Bupropion Binding Site in Proximity to αTyr213
The photolabeling of αTyr213 located near extracellular end of the αM1 helix was enhanced by >10-fold in the desensitized state compared to the resting state, and photolabeling of αTyr213 can account for the agonist-enhanced photolabeling in αV8-20 that was inhibitable by bupropion but not by TCP. TCP binding in the ion channel inhibited channel photolabeling by [125I]SADU-3-72, but not photolabeling of αTyr213. Hence, it is unlikely that bupropion binding in the ion channel would allosterically inhibit αTyr213 photolabeling. Rather bupropion must inhibit competitively [125I]SADU-3-72 binding in proximity to αTyr213.
αTyr213 is photolabeled by the general anesthetic [14C]-halothane (13), and amino acid substitutions at this position perturb gating, with the accessibility of αTyr213 Cys-substituted receptors to chemical modification increased by agonist (35, 36). Thus, αTyr213 contributes to a water-accessible, general anesthetic binding pocket located at the same interface between subunits where uncharged positive and negative nAChR allosteric modulators bind more towards the middle of the transmembrane domain (37, 38). The agonist-enhanced photolabeling of αTyr213 may be explained by the formation of SADU-3-72–accessible pocket in the vicinity of αTyr213 during desensitization, but further studies are required to examine the presence of this pocket and its accessibility to SADU-3-72 in resting and open states. Interestingly, we found no evidence that [125I]SADU-3-72 photolabeled amino acids in the nAChR d subunit helix bundle, where the uncharged inhibitor [125I]TID binds in the open and desensitized states (21, 39, 40).
Conclusions and Implications
In this report, we provide a direct identification, at the amino acid level, of two distinct [125I]SADU-3-72/bupropion binding sites in the Torpedo nAChR. One site is at the middle of the Torpedo nAChR ion channel (M2-9) where SADU3-72/bupropion binds with micromolar affinity in the resting and desensitized states and is likely to contribute to their functional inhibition of the Torpedo nAChR. The second site of [125I]-SADU-3-72/bupropion binding resides within a desensitized state pocket in the proximity of αTyr213 in αM1. Though photolabeling of αTyr213 by [125I]SADU-3-72 provides a positive identification of this site, further studies are required to determine the affinity of SADU3-72/bupropion at this site and the functional contribution of this site to bupropion inhibition of the nAChR. Given that the channel-lining leucine residues labeled by [125I]-SADU-3-72 in the Torpedo δ- and β-subunit are conserved in all nAChRs (as well as across the Cys Loop receptor family), it is tempting to predict a common bupropion binding site at M2-9 in the ion channel in all nAChR subtypes. However, given the range of bupropion potencies for inhibition of diverse nAChR subtypes (5), such an extrapolation could be an oversimplification, and the molecular determinants of bupropion binding to different nAChR ion channels may be subtype-specific. Moreover, it remains to be determined whether or not an equivalent bupropion binding site at αTyr213 is present in neuronal nAChRs.
Supplementary Material
Acknowledgments
This research was supported in part by the South Plains Foundation (M.P.B and M.J.), by the Center for Membrane Protein Research, TTUHSC (M.P.B. and M.J.), by a Hunkele Dreaded Disease Award (D.J.L.), the Mylan School of Pharmacy at Duquesne University (D.J.L.), and by National Institutes of Health grants GM58448 (J.B.C.) and DA27081 (D.J.L.).
We thank Dr. Jose-Luis Redondo (Department of Pharmacology and Neuroscience, TTUHSC) for his cell culture assistance.
Abbreviations
- nAChR
nicotinic acetylcholine receptor
- α–BgTx
α-bungarotoxin
- LGIC
ligand-gated ion channel
- Carb
carbamylcholine
- TCP
1-(1-(2-Thienyl)cyclohexyl) piperidine (tenocyclidine)
- HPLC
high-performance liquid chromatography
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TFA
trifluoroacetic acid
- PTH
phenylthiohydantoin
- [125I]-SADU-3-72
(±)-2-(N-tert-Butylamino)-3′-[125I]-iodo-4′-azidopropiophenone hydrochloride
- Tricine
N-tris(hydroxylmethyl)methylglycine
- VDB
vesicle dialysis buffer
- V8 protease
S. aureus endoproteinase Glu-C
- [125I]-TID
3-trifluoromethyl-3-(m-[125I]-iodophenyl) diazirine
Footnotes
Bupropion inhibition of [125I]-SADU-3-72 photoincorporation into the nAChR δ subunit (Supplementary Figure S1); Autoradiographs of Tricine SDS-PAGE separation of tryptic digests of [125I]-SADU-3-72-labeled fragments αV8-20K, βV8-22K, and δV8-14K isolated from Torpedo nAChR in the resting or desensitized state (Supplementary Figure S2); Identification of amino acids photolabeled by [125I]-SADU-3-72 in the M4 segment of the Torpedo nAChR (Supplementary Figure S3).
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wilkes S. The use of bupropion SR in cigarette smoking cessation. Int J Chron Obstruct Pulmon Dis. 2008;3:45–53. doi: 10.2147/copd.s1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fryer JD, Lukas RJ. Noncompetitive functional inhibition at diverse, human nicotinic acetylcholine receptor subtypes by bupropion, phencyclidine, and ibogaine. J Pharmacol Exp Ther. 1999;288:88–92. [PubMed] [Google Scholar]
- 3.Slemmer JE, Martin BR, Damaj MI. Bupropion is a nicotinic antagonist. J Pharmacol Exp Ther. 2000;295:321–327. [PubMed] [Google Scholar]
- 4.Carroll FI, Blough BE, Mascarella SW, Navarro HA, Eaton JB, Lukas RJ, Damaj MI. Synthesis and biological evaluation of bupropion analogues as potential pharmacotherapies for smoking cessation. J Med Chem. 2010;53:2204–2214. doi: 10.1021/jm9017465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias HR. Molecular interaction of bupropion with nicotine acetylcholine receptors. J Pediatr Biochem. 2010;1:185–197. [Google Scholar]
- 6.Thompson AJ, Lester HA, Lummis SC. The structural basis of function in Cys-loop receptors. Q Rev Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 7.Bouzat C. New insights into the structural bases of activation of Cys-loop receptors. J Physiol Paris. 2011 doi: 10.1016/j.jphysparis.2011.09.012. (EPUB 10/02/2011) [DOI] [PubMed] [Google Scholar]
- 8.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 9.De Biasi M, Dani JA. Reward, addiction, withdrawal to nicotine. Annu Rev Neurosci. 2011;34:105–130. doi: 10.1146/annurev-neuro-061010-113734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu Rev Pharmacol Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- 11.Arias HR, Gumilar F, Rosenberg A, Targowska-Duda KM, Feuerbach D, Jozwiak K, Moaddel R, Wainer IW, Bouzat C. Interaction of bupropion with muscle-type nicotinic acetylcholine receptors in different conformational states. Biochemistry. 2009;48:4506–4518. doi: 10.1021/bi802206k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lapinsky DJ, Aggarwal S, Nolan TL, Surratt CK, Lever JR, Acharya R, Vaughan RA, Pandhare A, Blanton MP. (+/−)-2-(N-tert-Butylamino)-3′-[(125)I]-iodo-4′-azidopropiophenone: A dopamine transporter and nicotinic acetylcholine receptor photoaffinity ligand based on bupropion (Wellbutrin, Zyban) Bioorg Med Chem Lett. 2012;22:523–526. doi: 10.1016/j.bmcl.2011.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiara DC, Dangott LJ, Eckenhoff RG, Cohen JB. Identification of nicotinic acetylcholine receptor amino acids photolabeled by the volatile anesthetic halothane. Biochemistry. 2003;42:13457–13467. doi: 10.1021/bi0351561. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen SE, Dreyer EB, Cohen JB. Location of ligand-binding sites on the nicotinic acetylcholine receptor alpha-subunit. J Biol Chem. 1986;261:13735–13743. [PubMed] [Google Scholar]
- 15.Fong TM, McNamee MG. Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry. 1986;25:830–840. doi: 10.1021/bi00352a015. [DOI] [PubMed] [Google Scholar]
- 16.Hamouda AK, Sanghvi M, Sauls D, Machu TK, Blanton MP. Assessing the lipid requirements of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry. 2006;45:4327–4337. doi: 10.1021/bi052281z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsal J, Tigyi G, Miledi R. Incorporation of acetylcholine receptors and Cl- channels in Xenopus oocytes injected with Torpedo electroplaque membranes. Proc Natl Acad Sci U S A. 1995;92:5224–5228. doi: 10.1073/pnas.92.11.5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morales A, Aleu J, Ivorra I, Ferragut JA, Gonzalez-Ros JM, Miledi R. Incorporation of reconstituted acetylcholine receptors from Torpedo into the Xenopus oocyte membrane. Proc Natl Acad Sci U S A. 1995;92:8468–8472. doi: 10.1073/pnas.92.18.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore MA, McCarthy MP. Snake venom toxins, unlike smaller antagonists, appear to stabilize a resting state conformation of the nicotinic acetylcholine receptor. Biochim Biophys Acta. 1995;1235:336–342. doi: 10.1016/0005-2736(95)80022-8. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher MJ, Cohen JB. Identification of amino acids of the torpedo nicotinic acetylcholine receptor contributing to the binding site for the noncompetitive antagonist [(3)H]tetracaine. Mol Pharmacol. 1999;56:300–307. doi: 10.1124/mol.56.2.300. [DOI] [PubMed] [Google Scholar]
- 21.Hamouda AK, Chiara DC, Blanton MP, Cohen JB. Probing the structure of the affinity-purified and lipid-reconstituted torpedo nicotinic acetylcholine receptor. Biochemistry. 2008;47:12787–12794. doi: 10.1021/bi801476j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 24.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 25.Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- 26.Baenziger JE, Morris ML, Darsaut TE, Ryan SE. Effect of membrane lipid composition on the conformational equilibria of the nicotinic acetylcholine receptor. J Biol Chem. 2000;275:777–784. doi: 10.1074/jbc.275.2.777. [DOI] [PubMed] [Google Scholar]
- 27.Blanton MP, Dangott LJ, Raja SK, Lala AK, Cohen JB. Probing the structure of the nicotinic acetylcholine receptor ion channel with the uncharged photoactivable compound -3H-diazofluorene. J Biol Chem. 1998;273:8659–8668. doi: 10.1074/jbc.273.15.8659. [DOI] [PubMed] [Google Scholar]
- 28.Blanton MP, McCardy EA, Huggins A, Parikh D. Probing the structure of the nicotinic acetylcholine receptor with the hydrophobic photoreactive probes [125I]TID-BE and [125I]TIDPC/16. Biochemistry. 1998;37:14545–14555. doi: 10.1021/bi981435q. [DOI] [PubMed] [Google Scholar]
- 29.Blanton MP, Li YM, Stimson ER, Maggio JE, Cohen JB. Agonist-induced photoincorporation of a p-benzoylphenylalanine derivative of substance P into membrane-spanning region 2 of the Torpedo nicotinic acetylcholine receptor delta subunit. Mol Pharmacol. 1994;46:1048–1055. [PubMed] [Google Scholar]
- 30.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 31.Blanton MP, Xie Y, Dangott LJ, Cohen JB. The steroid promegestone is a noncompetitive antagonist of the Torpedo nicotinic acetylcholine receptor that interacts with the lipid-protein interface. Mol Pharmacol. 1999;55:269–278. doi: 10.1124/mol.55.2.269. [DOI] [PubMed] [Google Scholar]
- 32.Garcia G, 3rd, Chiara DC, Nirthanan S, Hamouda AK, Stewart DS, Cohen JB. [3H]Benzophenone photolabeling identifies state-dependent changes in nicotinic acetylcholine receptor structure. Biochemistry. 2007;46:10296–10307. doi: 10.1021/bi7008163. [DOI] [PubMed] [Google Scholar]
- 33.White BH, Howard S, Cohen SG, Cohen JB. The hydrophobic photoreagent 3-(trifluoromethyl)-3-m-([125I] iodophenyl) diazirine is a novel noncompetitive antagonist of the nicotinic acetylcholine receptor. J Biol Chem. 1991;266:21595–21607. [PubMed] [Google Scholar]
- 34.White BH, Cohen JB. Agonist-induced changes in the structure of the acetylcholine receptor M2 regions revealed by photoincorporation of an uncharged nicotinic noncompetitive antagonist. J Biol Chem. 1992;267:15770–15783. [PubMed] [Google Scholar]
- 35.Akabas MH, Karlin A. Identification of acetylcholine receptor channel-lining residues in the M1 segment of the alpha-subunit. Biochemistry. 1995;34:12496–12500. doi: 10.1021/bi00039a002. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y, Shi L, Karlin A. Structural effects of quinacrine binding in the open channel of the acetylcholine receptor. Proc Natl Acad Sci U S A. 2003;100:3907–3912. doi: 10.1073/pnas.0730718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nirthanan S, Garcia G, 3rd, Chiara DC, Husain SS, Cohen JB. Identification of binding sites in the nicotinic acetylcholine receptor for TDBzl-etomidate, a photoreactive positive allosteric effector. J Biol Chem. 2008;283:22051–22062. doi: 10.1074/jbc.M801332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamouda AK, Stewart DS, Husain SS, Cohen JB. Multiple transmembrane binding sites for p-trifluoromethyldiazirinyl-etomidate, a photoreactive Torpedo nicotinic acetylcholine receptor allosteric inhibitor. J Biol Chem. 2011;286:20466–20477. doi: 10.1074/jbc.M111.219071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor’s {delta}-subunit. A time-resolved photolabeling study. J Biol Chem. 2005;280:13631–13640. doi: 10.1074/jbc.M413911200. [DOI] [PubMed] [Google Scholar]
- 40.Yamodo IH, Chiara DC, Cohen JB, Miller KW. Conformational changes in the nicotinic acetylcholine receptor during gating and desensitization. Biochemistry. 2010;49:156–165. doi: 10.1021/bi901550p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.