This article shows that the phytochrome A photoreceptor promotes reorientation of the hypocotyl toward blue light (phototropism) by regulating the expression of nuclear genes. It also shows that phytochrome A nuclear signaling events still operate in a mutant where phytochrome A does not significantly accumulate in the nucleus.
Abstract
Phototropin photoreceptors (phot1 and phot2 in Arabidopsis thaliana) enable responses to directional light cues (e.g., positive phototropism in the hypocotyl). In Arabidopsis, phot1 is essential for phototropism in response to low light, a response that is also modulated by phytochrome A (phyA), representing a classical example of photoreceptor coaction. The molecular mechanisms underlying promotion of phototropism by phyA remain unclear. Most phyA responses require nuclear accumulation of the photoreceptor, but interestingly, it has been proposed that cytosolic phyA promotes phototropism. By comparing the kinetics of phototropism in seedlings with different subcellular localizations of phyA, we show that nuclear phyA accelerates the phototropic response, whereas in the fhy1 fhl mutant, in which phyA remains in the cytosol, phototropic bending is slower than in the wild type. Consistent with this data, we find that transcription factors needed for full phyA responses are needed for normal phototropism. Moreover, we show that phyA is the primary photoreceptor promoting the expression of phototropism regulators in low light (e.g., PHYTOCHROME KINASE SUBSTRATE1 [PKS1] and ROOT PHOTO TROPISM2 [RPT2]). Although phyA remains cytosolic in fhy1 fhl, induction of PKS1 and RPT2 expression still occurs in fhy1 fhl, indicating that a low level of nuclear phyA signaling is still present in fhy1 fhl.
INTRODUCTION
Plants have advanced light sensing and signaling systems to control their growth and development. Seedling development is strongly influenced by the intensity, wavelength, photoperiod, and direction of the light (Kami et al., 2010). In Arabidopsis thaliana, multiple photoreceptors, including five phytochromes (phyA to phyE), two cryptochromes (cry1 and cry2), two phototropins (phot1 and phot2), and UVB-RESISTANCE8 regulate seed germination, deetiolation, and/or phototropism (Franklin et al., 2005; Christie, 2007; Demarsy and Fankhauser, 2009; Kami et al., 2010; Rizzini et al., 2011). Later in the life cycle, photoreceptors also regulate vegetative development (e.g., shade avoidance) and the transition to reproduction (Kami et al., 2010). Although some light responses are primarily regulated by a single photoreceptor, there are numerous examples of photoreceptor coaction leading to an optimal physiological or developmental response in a changing light environment (Casal, 2000; Sellaro et al., 2009; Kami et al., 2010). Such coaction is very important during seedling establishment, a growth stage when plantlets are particularly vulnerable (Sellaro et al., 2009).
The phototropins regulate several blue light responses, including phototropism, leaf flattening, chloroplast movements, and opening of the stomata (Christie, 2007). phot1 and phot2 regulate many of these responses together; however, phot1 is more sensitive to blue light than is phot2, as exemplified by the essential nature of phot1 for phototropism in response to low blue light (Kagawa and Wada, 2000; Sakai et al., 2001; Christie, 2007). Upon light perception, the phototropins autophosphorylate, a step that is essential for all tested physiological responses (Inoue et al., 2008; Inoue et al., 2011). How this initial step is connected to the subsequent signaling events remains to be determined. In the case of phototropism, a gradient of auxin has been proposed to be a prerequisite for the asymmetric growth response allowing optimal positioning of the leaves/cotyledons (Esmon et al., 2006). Genetic studies have identified a limited number of phototropin signaling components, including NON PHOTOTROPIC HYPOCOTYL3 (NPH3), ROOT PHOTOTROPISM2 (RPT2), ABCB19, PHYTOCHROME KINASE SUBSTRATE1 (PKS1), and PKS2, that interact with the phototropins and act early downstream of phototropin activation (Motchoulski and Liscum, 1999; Sakai et al., 2000; Lariguet et al., 2006; de Carbonnel et al., 2010; Christie et al., 2011). In Oryza sativa (rice), it has been shown that CP1, the ortholog of NPH3, acts upstream of auxin redistribution in the coleoptile (Haga et al., 2005). The importance of auxin transport and signaling for phototropism has been confirmed genetically (Tatematsu et al., 2004; Stone et al., 2008; Möller et al., 2010; Christie et al., 2011; Ding et al., 2011).
Interestingly, phyA, cry1, and cry2 also modulate the phototropic response, and some studies have linked their activity to a modulation of auxin transport (Parks et al., 1996; Janoudi et al., 1997; Whippo and Hangarter, 2003; Lariguet and Fankhauser, 2004; Whippo and Hangarter, 2004; Nagashima et al., 2008; Tsuchida-Mayama et al., 2010). Whereas some studies suggest a direct role of these photoreceptors on phototropism, others propose that they act indirectly by inhibiting the gravitropic response (Whippo and Hangarter, 2003; Lariguet and Fankhauser, 2004; Whippo and Hangarter, 2004; Iino, 2006; Nagashima et al., 2008). In addition to phototropism enhancement, the phytochromes also modulate other phototropin responses, such as the regulation of chloroplast movements and opening of the stomata (DeBlasio et al., 2003; Wang et al., 2010). Despite the well-known importance of photoreceptor coaction, particularly during early seedling establishment (Sellaro et al., 2009), the molecular mechanisms underlying phototropic enhancement by phytochromes and cryptochromes remain poorly understood.
Whereas in angiosperms, phototropism occurs only in response to blue light, in numerous cryptogams, both red and blue light trigger a phototropic response (Suetsugu et al., 2005). The fern Adiantum possesses a chimeric photoreceptor consisting of a phytochrome photosensory domain fused to a phototropin-type photoreceptor (Kawai et al., 2003). When expressed in Arabidopsis, this photoreceptor triggers phototropism toward both red and blue light (Kanegae et al., 2006). This can be used as an evolutionary argument to propose that phytochromes and phototropins presumably act closely together in the regulation of phototropism (Marcotte et al., 1999). However, in angiosperms, the phytochromes and phototropins are mostly present in different subcellular compartments (Christie, 2007; Fankhauser and Chen, 2008). Phototropins are localized at the plasma membrane in the dark. In response to blue light, a fraction of phot1 and phot2 is relocalized to the cytoplasm and Golgi, respectively (Sakamoto and Briggs, 2002; Kong et al., 2006; Wan et al., 2008). Phytochromes are cytosolic in the dark and enter the nucleus upon light activation (Nagatani, 2004; Fankhauser and Chen, 2008). Nuclear import of phyB is triggered by the light-regulated unmasking of a nuclear localization signal (NLS) sequence (Chen et al., 2005). phyA nuclear import depends on its light-regulated interaction with FHY1 and FHL, a pair of related proteins comprising an NLS sequence followed by an extended linker region and a phyA interaction domain (Hiltbrunner et al., 2006; Rösler et al., 2007; Genoud et al., 2008; Rausenberger et al., 2011). Although these two classes of photoreceptors are primarily encountered in different subcellular compartments, they are both present in the cytosol when etiolated seedlings are first exposed to light.
Many findings are consistent with the idea that phytochromes and phototropins act together in the cytosol to regulate phototropism (Rösler et al., 2010). The phyA mutant has rather severe phototropic defects in response to low blue light (Parks et al., 1996; Janoudi et al., 1997; Lariguet and Fankhauser, 2004; Whippo and Hangarter, 2004; Rösler et al., 2007). By contrast, the fhy1 fhl double mutant, in which phyA cannot enter the nucleus, displays a normal phototropic response (Rösler et al., 2007). This is consistent with the idea that cytosolic phyA has a predominant function in promoting phototropism. A subsequent study demonstrated that phyA plays a role in the light-regulated relocalization of phot1 to the cytosol, again suggesting a possible cytosolic role for phyA (Han et al., 2008). Other studies suggest that the mechanism by which cryptochromes and phytochromes, including phyA, promote phototropism is through light-regulated induction of the expression of phototropism-signaling components (Stowe-Evans et al., 2001; Lariguet et al., 2006; Tsuchida-Mayama et al., 2010). The two hypotheses are not mutually exclusive, but it is currently unknown whether phytochromes primarily enhance phototropism by acting with the phototropins in the cytosol or whether phototropism enhancement depends on nuclear entry of the phytochrome.
To address this question, we have studied phyA-mediated enhancement of phototropism. phyA is well suited for this study, because the loss-of-function mutant has severe phototropic defects and because we possess genetic tools to keep phyA either in the nucleus or in the cytosol. fhy1 fhl mutants retain normal levels of phyA, but the photoreceptor is not imported into the nucleus upon light perception (Hiltbrunner et al., 2006; Rösler et al., 2007). In phyA mutants expressing phyA-NLS-green fluorescent protein (GFP) (hereafter called phyA-NLS-GFP), the photoreceptor is constitutively present in the nucleus (Genoud et al., 2008; Toledo-Ortiz et al., 2010). By comparing the wild type, phyA, fhy1 fhl, and phyA-NLS-GFP, we showed that nuclear phyA leads to accelerated phototropic bending, whereas when phyA is present in the cytosol, phototropism proceeds more slowly. Consistent with these data, transcriptional regulators involved in phyA signaling are required for a normal phototropic response. Interestingly, in response to blue light, fhy1 fhl retains low levels of light-induced gene expression correlating with the slow phototropic response of the mutant. Our gene expression analysis is consistent with the notion that fhy1 fhl retains a small degree of nuclear phyA signaling, particularly in response to blue light. Our study shows that phyA enhances phototropism most efficiently when localized in the nucleus; however, it does not exclude a role for phyA in the cytosol.
RESULTS
Both fhy1 fhl and phyA-NLS-GFP Seedlings Show a Robust Phototropic Response
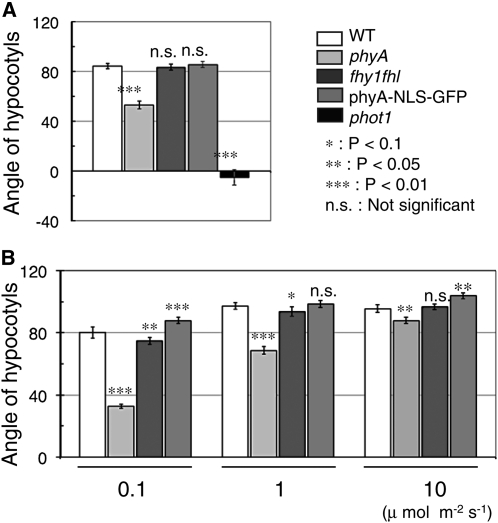
We have previously shown that phyA mutant seedlings grown from the time of germination in unilateral blue light display a clear phototropic phenotype (Lariguet and Fankhauser, 2004). We thus compared the wild type, phyA, fhy1 fhl, and phyA-NLS-GFP using this long-term phototropic protocol. Importantly, phyA levels in the phyA-NLS-GFP and phyA-NLS lines used here were quantified and were not higher than in the wild type (Genoud et al., 2008). Our data confirmed previous observations that showed a reduced response in phyA but a similar phototropic response in fhy1 fhl and the wild type (Rösler et al., 2007). Surprisingly, however, phyA-NLS-GFP seedlings also displayed a normal phototropic response, suggesting that phyA either in the nucleus or in the cytosol is sufficient to promote phototropism (Figure 1A).
Figure 1.
Phototropic Response in the Wild Type, phyA, fhy1 fhl, and phyA-NLS-GFP.
(A) Long-term phototropism experiment. Seedlings were grown for 3 d in unilateral blue light (0.1 μmol m−2 s−1). Final growth direction relative to vertical was measured (0° represents vertical growth). Data are average angles relative to vertical ± 2 × se (n > 120). WT, wild type.
(B) Short-term phototropism experiment. Hypocotyl curvatures of the wild type, phyA, fhy1 fhl, and phyA-NLS-GFP. The 3-d-old etiolated seedlings were exposed to blue light (0.1, 1, or 10 μmol m−2 s−1) for 24 h. Data are average angles relative to vertical ± 2 × se of hypocotyl (n > 180).
Asterisks indicate the P value for statistical difference with the wild type in each condition. *, P < 0.1; **, P < 0.05; ***, P < 0.01; n.s., not significant.
Most phototropism studies are performed with etiolated seedlings treated with a unilateral light source, which prompted us to test this hypothesis further using a more conventional protocol. We used 2-d-old etiolated seedlings (4 to 6 mm) and exposed them for 24 h to different fluence rates of blue light before measuring the deviation from vertical growth. phyA showed a reduced phototropic response at all tested fluence rates, but the phenotype was strongest at the lowest fluence rate (Figure 1B). Interestingly, at low fluence rates, fhy1fhl showed a significantly reduced phototropic response, whereas phyA-NLS-GFP showed enhanced bending (Figure 1B).
Previous studies have shown that phyA-GFP enters the nucleus in response to white, red, blue, and far-red (FR) light (Kircher et al., 1999; Kim et al., 2000). To verify that the same occurs under our experimental conditions, we analyzed the subcellular localization of phyA-GFP and phyA-NLS-GFP in etiolated seedlings treated with low blue light by confocal microscopy. These light conditions triggered entry of phyA-GFP into the nucleus and formation of nuclear bodies (see Supplemental Figures 1A to 1D online). A prolonged light treatment led to a reduced phyA-GFP signal, consistent with the light-regulated degradation of phyA (see Supplemental Figures 1A to 1D online). Consistent with a previous report, phyA-NLS-GFP was constitutively nuclear, the levels of GFP fluorescence decreased upon light treatment, and nuclear bodies only appeared in response to light (see Supplemental Figures 1E and 1F online) (Genoud et al., 2008; Toledo-Ortiz et al., 2010). Collectively, these experiments indicate that phyA entered the nucleus in response to a light treatment that triggered phototropism and that phyA-NLS-GFP was present in the nucleus from the beginning of the experiment.
Nuclear phyA Accelerates Phototropic Bending
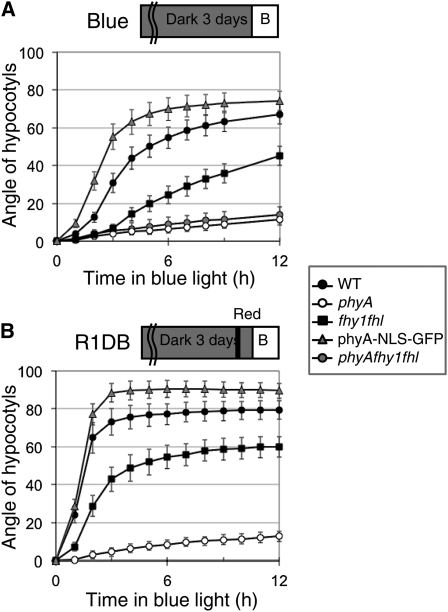
Our experiment indicated that phyA enters the nucleus rapidly in response to a phototropism-stimulating light treatment and that nuclear phyA may be more effective than cytosolic phyA in promoting phototropism (Figure 1; see Supplemental Figure 1 online). To test this hypothesis more carefully, we performed phototropism time-course experiments, because classical experiments have shown that phytochrome accelerates phototropic bending (Parks et al., 1996; Janoudi et al., 1997; Iino, 2006). Our analysis of time-lapse images showed that the kinetics of hypocotyl bending were influenced by the length of the etiolated hypocotyl when the light treatment started and confirmed previous findings showing that the position of the cotyledon relative to the unilateral light source strongly influences the response (see Supplemental Figure 2 online) (Khurana et al., 1989). To account for those developmental effects on phototropic bending, we size-selected seedlings (4- to 5.9-mm–long hypocotyls) and used 20 seedlings with the cotyledons on each side for each time point. By following the phototropic bending response, we observed a strong phenotype in fhy1 fhl that was much more striking than in end-point experiments (cf. Figures 1 and 2A). The bending response was very slow in phyA and much slower in fhy1 fhl than in the wild type (Figure 2A). Interestingly, bending occurred more rapidly in phyA-NLS-GFP than in the wild type (Figure 2A). These results indicate that nuclear phyA is more efficient than cytosolic phyA in promoting phototropism. A classic way to demonstrate the promoting effect of phyA on phototropism is to pretreat etiolated seedlings with a red light pulse prior to subjecting them to unilateral blue light (Parks et al., 1996; Janoudi et al., 1997). Such a light treatment leads to nuclear accumulation of phyA, which may explain why phyA-NLS-GFP reoriented the hypocotyl growth direction faster than the wild type (Figure 2A). To test this idea, we pretreated etiolated seedlings with red light 1 h prior to unilateral blue light irradiation and followed bending of the hypocotyls over time. Interestingly, this treatment enhanced the speed of bending in the wild type, which showed an initial speed of reorientation very similar to that of phyA-NLS-GFP (Figure 2B). phyA-NLS-GFP also responded to the red light pretreatment, indicating that translocation of phyA into the nucleus is not sufficient for phyA activation, which is consistent with previous studies using these lines (Genoud et al., 2008; Toledo-Ortiz et al., 2010). Importantly, however, the red light pulse had a lower effect on phyA-NLS-GFP than the wild type (cf. Figures 2A and 2B). As anticipated, phyA did not respond to this red light treatment, whereas, interestingly, bending in fhy1 fhl reoriented more rapidly after a red light pretreatment (Figure 2B).
Figure 2.
Kinetics of the Phototropic Response in the Wild Type, phyA, fhy1 fhl phyA fhy1 fhl, and phyA-NLS-GFP.
(A) Phototropism kinetics from time-lapse images of seedlings grown under unidirectional blue light (0.1 μmol m−2 s−1).
(B) Phototropism kinetics from time-lapse images under blue light with a red light pretreatment. For the red light pretreatment, etiolated seedlings were exposed with red light (1 μmol m−2 s−1, 10 s) and incubated for 1 h in darkness before exposure to unilateral blue light (0.1 μmol m−2 s−1).
Data show average hypocotyl angles (n = 40, 20 with cotyledons facing blue light, and 20 with cotyledons in the opposite direction; see Methods) ± 2 × se. WT, wild type.
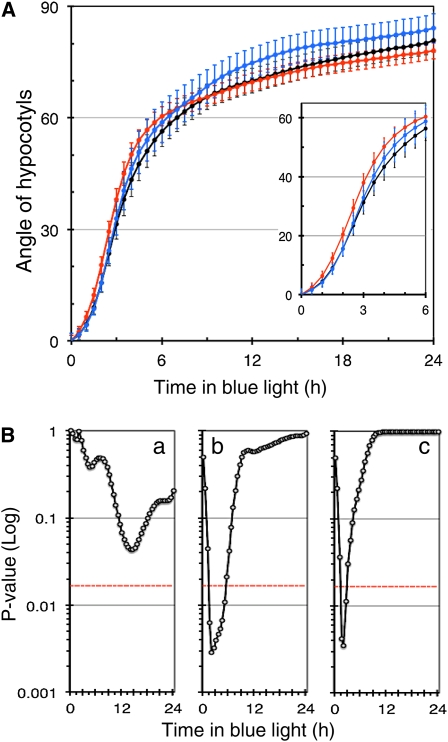
To confirm that plants with constitutively nuclear phyA have a more rapid phototropic response than plants in which phyA enters the nucleus only in response to light, we compared the kinetics of phototropic bending of the wild type, phyA-GFP, and phyA-NLS seedlings (Genoud et al., 2008) (Figure 3). The experiment was performed as in Figure 2, and measurements were either done manually or with semiautomatic analysis software designed to this end (HypoPhen) (Figure 3; see Supplemental Figure 3 online). The HypoPhen software takes as input time-lapsed images of growing hypocotyls and measures bending angles and hypocotyl growth (see Methods). Bending values are then computed as the direction of the upper tip of the hypocotyl. The software is open-source and based on the OpenCV library (Bradski and Kaehler, 2008). It is documented and freely available at http://www.unil.ch/cbg/index.php?title=HypoPhen. HypoPhen allowed us to obtain higher temporal resolution and greater reproducibility (no user-induced bias) than manual measurements. During the first hours of phototropism, phyA-NLS reoriented faster than the wild type and phyA-GFP, whereas in phyA and fhy1 fhl, the response was much slower (Figure 3; see Supplemental Figure 3 online). These data confirm that in plants where phyA is constitutively nuclear, the early bending response is more rapid than in the wild type (Figures 2 and 3).
Figure 3.
phyA-NLS Seedlings Have a Faster Phototropic Response than Wild-Type and phyA-GFP Seedlings.
(A) Phototropism kinetics of seedlings grown as in Figure 2A (black, wild type; blue, phyA-GFP; red, phyA-NLS) analyzed using semiautomatic measurements (HypoPhen). The data are average values of both cotyledon positions ± 2 × se (n > 60) with the same number of cotyledons for each position.
(B) Analysis of a difference in phototropism kinetics for the data shown in (A). Data are represented as pairwise comparisons between the wild type versus phyA-GFP (a), the wild type versus phyA-NLS (b), and phyA-GFP versus phyA-NLS (c) for each time point. The P values resulting from a Student’s t test are represented. The red line represents the 5% threshold corrected for multiple testing.
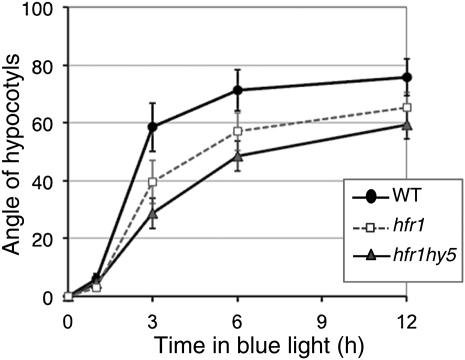
Our data indicate that phyA in the nucleus is very efficient in promoting phototropism (Figures 2 and 3). To test this hypothesis further, we analyzed phototropism using mutants deficient in nuclear phyA signaling events. We selected LONG HYPOCOTYL IN FR LIGHT1 (HFR1) and HYPOCOTYL5 (HY5), which code for a bHLH and a bZIP transcription factor, respectively (Kami et al., 2010). A phototropic time course showed that hfr1 displays a significantly slower phototropic response (Figure 4). Moreover, this phenotype is further enhanced in an hfr1 hy5 double mutant consistent with the enhanced deetiolation phenotype of such a double mutant in FR light conditions (Figure 4) (Kim et al., 2002). These data are consistent with the importance of nuclear phyA events in the promotion of phototropism.
Figure 4.
Nuclear Components of phyA Signaling Are Required for a Fast Phototropic Response.
Phototropism kinetics of seedlings grown under unilateral blue light (0.1 μmol m−2 s−1). Data show average hypocotyl angles (n = 40, 20 with cotyledons facing blue light, and 20 with cotyledons in the opposite direction, except for hfr1 hy5, in which n = 34 with 17 seedlings in each orientation; see Methods) ± 2 × se. WT, wild type.
In Blue Light, FHY1 and FHL Are Required for Nuclear Import of phyA, but phyA Nuclear Signaling Still Works Partially in fhy1 fhl
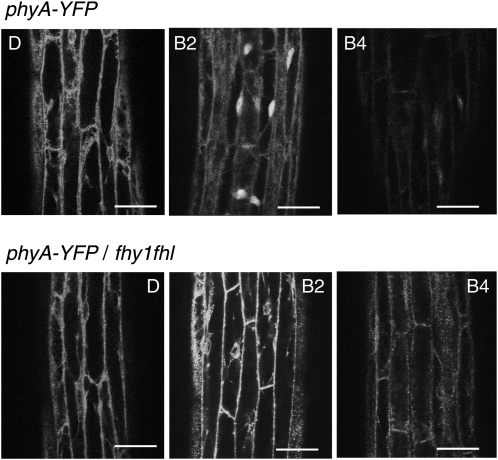
Our time-course experiment showed that nuclear phyA was more efficient than cytosolic phyA in promoting phototropism. However, our results also confirmed a previous study that demonstrated that phototropism is more effective in fhy1 fhl than in phyA (Rösler et al., 2007). A possible explanation for this result is that phyA may still enter the nucleus in fhy1 fhl when seedlings are exposed to blue light. We thus compared the subcellular localization of phyA-yellow fluorescent protein (YFP) in FHY1 FHL and fhy1 fhl mutant backgrounds both in FR light, in which FHY1 and FHL are known to be required for phyA responses and nuclear import, and in blue light (Figure 5; see Supplemental Figure 4 online) (Hiltbrunner et al., 2006; Rösler et al., 2007). Our results confirmed previous observations, because rapid nuclear import of phyA-YFP depended on FHY1 and FHL in FR light (see Supplemental Figure 4 online). Similarly, in response to blue light, nuclear accumulation of phyA was not observed in fhy1 fhl, indicating that phyA nuclear import depends on FHY1 and FHL both in FR and blue light (Figure 5; see Supplemental Figure 4 online). The reduced overall signal observed after several hours in blue light correlates with the light-regulated degradation of phyA that occurs in response to light (Figure 5; see Supplemental Figure 4 online). As expected, the decrease in signal was more rapid in blue than FR light, given that blue light leads to a greater active phytochrome/total phytochrome ratio; however, phyA-YFP was still clearly visible in the nucleus after 4 h of blue light (cf. Figure 5 and Supplemental Figure 4 online).
Figure 5.
phyA-YFP Nuclear Import Depends on FHY1 and FHL in Blue Light.
Confocal laser scanning microscopy was used to analyze 3-d-old dark-grown FHY1 FHL or fhy1 fhl seedlings transformed with PHYA-YFP. The seedlings were analyzed directly (dark) and after 2 or 4 h irradiation with blue light (0.1 μmol m−2 s−1). D, dark; B2 and B4, 2 or 4 h of blue light treatment. Bars = 50 μm.
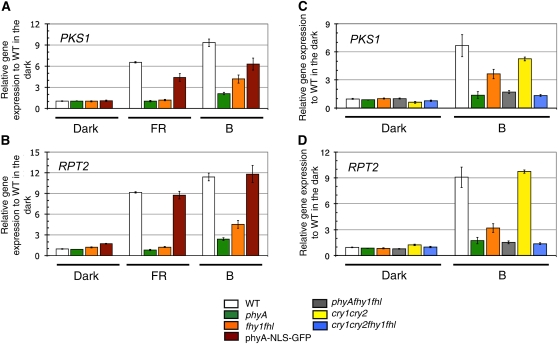
These microscopic observations cannot exclude the possibility that a small fraction of phyA still enters the nucleus in fhy1 fhl. We thus decided to test a rapid nuclear phyA response in seedlings exposed either to FR or blue light to determine whether phyA-dependent nuclear responses still operate in fhy1 fhl. Etiolated wild-type, phyA, fhy1 fhl, and phyA-NLS-GFP seedlings were thus either kept in darkness or exposed to 1 h FR or low blue light, and gene expression was analyzed by quantitative RT-PCR (qRT-PCR) in all experimental conditions. We chose these two light conditions because FR light-regulated gene expression exclusively depends on phyA, and in blue light, phyA plays an important role, but fhy1 fhl has a clearly distinct phenotype from phyA (Lariguet et al., 2006; Rösler et al., 2007; Peschke and Kretsch, 2011) (Figures 2 and 3). We analyzed the expression of RPT2 and PKS1, which are known components of phototropism signaling and are early light–induced genes (Lariguet et al., 2006; Tsuchida-Mayama et al., 2010). Both genes did not respond to FR light in phyA, whereas the response was very similar to the wild type in phyA-NLS-GFP and very similar to phyA in fhy1 fhl (Figures 6A and 6B). In response to low blue light, the expression of those two genes very strongly depended on phyA (Figures 6A and 6B). Interestingly and in contrast with the situation in FR light, fhy1 fhl showed a very distinct gene expression phenotype from phyA in blue light. Indeed, both PKS1 and RPT2 showed a significantly more robust induction in fhy1 fhl than in phyA (Figures 6A and 6B). Importantly, the blue light–regulated gene expression in fhy1 fhl was dependent on phyA, because gene expression in phyA fhy1 fhl and phyA were not significantly different (Figures 6C and 6D). Consistent with this gene expression data, phototropic bending in phyA fhy1 fhl was not more impaired than in phyA (Figure 2). These data indicate that in blue light, FHY1 and FHL largely influence phototropic bending and gene expression through phyA and that early nuclear signaling events still take place in fhy1 fhl grown in blue light.
Figure 6.
A Blue Light Treatment Leads to Significant Induction of PKS1 and RPT2 Expression in fhy1 fhl.
(A) Expression of PKS1 was measured by qRT-PCR in the wild type (WT), phyA, fhy1 fhl, and phyA-NLS-GFP. The 3-d-old etiolated seedlings were either kept in the dark, exposed for 1 h to FR light (5 μmol m−2 s−1), or exposed for 1 h to blue light (B; 0.5 μmol m−2 s−1). Data are average expression of PKS1 normalized to two control genes and expressed relative to the wild type in the dark ± 2 × se. Averages from three biological replicates with three technical replicates for each are shown.
(B) Expression of RPT2 was performed as in (A).
(C) Expression of PKS1 was performed as in (A), but in the wild type, phyA, fhy1 fhl, phyA fhy1 fhl, cry1 cry2, and cry1 cry2 fhy1 fhl.
(D) Expression of RPT2 was performed as in (C).
Cryptochromes 1 and 2 are the major photoreceptors mediating deetiolation in response to blue light (Kami et al., 2010). We tested their involvement in low blue light–induced gene expression and phototropism by analyzing cry1 cry2. The expression of PKS1 and RPT2 were only marginally affected in the cryptochrome mutant (Figures 6C and 6D). Moreover, a kinetic analysis of phototropism showed that, under these conditions, fhy1 fhl was more impaired than cry1 cry2 (see Supplemental Figure 5 online). To test whether FHY1 and FHL act in a different pathway than the cryptochromes, we analyzed phototropism and gene expression in cry1 cry2 fhy1 fhl. Interestingly, these experiments showed that the phenotype of cry1 cry2 was strongly enhanced in fhy1 fhl cry1 cry2, thus strongly suggesting that FHY1 and FHL do not act in cryptochrome signaling in blue light (Figure 6; see Supplemental Figure 5 online). Moreover, gene expression in fhy1 fhl cry1 cry2 was more strongly impaired than in fhy1 fhl, indicating that the blue light–regulated gene expression in fhy1 fhl may also partially depend on the cryptochromes (Figures 6C and 6D).
DISCUSSION
Nuclear phyA Accelerates Phototropism
By comparing phototropism in the wild type, fhy1 fhl, phyA, and phyA-NLS-GFP seedlings, we could show that nuclear phyA accelerates phototropism, whereas in fhy1 fhl, reorientation toward blue light occurred more slowly than in the wild type (Figures 2 and 3). The fact that the transcription factors HY5 and HFR1 are needed for a normal phototropic response is also consistent with the importance of nuclear signaling events in the promotion of phototropism (Figure 4). Although the phototropic phenotype of hy5 and hfr1 could result from altered gene expression in the etiolated mutants affecting cytosolic events during phototropism, collectively, our data show the importance of nuclear phyA in the promotion of phototropism (Figures 1 to 4). Under the light conditions we used to trigger phototropism, phyA plays the primary role in inducing the expression of PKS1 and RPT2, and light regulation of those genes was similar to the wild type in phyA-NLS-GFP but not in fhy1 fhl (Figure 6). Interestingly, this correlates with the prevalent function of phyA rather than the cryptochromes in the promotion of low blue light–mediated phototropism (Lariguet and Fankhauser, 2004; Tsuchida-Mayama et al., 2010) (see Supplemental Figure 5 online). Considering that overexpression of RPT2 can complement the phototropic phenotype of phyA cry1 cry2, our results strongly support the notion that nuclear phyA promotes phototropism by regulating gene expression (Figures 2, 3, and 6) (Tsuchida-Mayama et al., 2010). This is also consistent with the previously shown function of phyA in the promotion of PKS1 expression (Lariguet et al., 2006).
Phytochrome-mediated promotion of phototropism is traditionally demonstrated by pretreating etiolated seedlings with a red light pulse and returning seedlings into darkness for 1 to 2 h prior to applying unilateral blue light (Han et al., 2008). Such a treatment will lead to nuclear import of phyA, which is consistent with our findings that nuclear phyA efficiently promotes phototropism (Kircher et al., 1999; Fankhauser and Chen, 2008) (Figures 1 to 3). Moreover, our phyA-NLS and phyA-NLS-GFP lines showed a faster phototropic response than the wild type, and a red light treatment accelerated phototropism significantly in the wild type, whereas the red light promotion effect was not as strong in phyA-NLS-GFP (Figures 2B and 3; see Supplemental Figure 3 online). However, even in phyA-NLS-GFP plants, a red light pretreatment further accelerated phototropism, which is consistent with the finding that translocating phyA into the nucleus is not sufficient to induce phyA responses, because in such lines, light activation of the photoreceptor is still required (Genoud et al., 2008; Toledo-Ortiz et al., 2010). Collectively, our data indicate that phyA promotes phototropism by regulating nuclear gene expression.
Our data also confirm that phototropism in fhy1 fhl is more effective than in phyA (Figures 1 and 2) (Rösler et al., 2007). This suggests that phyA may also promote phototropism in the cytosol. Indeed, cytosolic functions of phyA have previously been reported (Rösler et al., 2010). In particular, it has been shown that phyA has an effect on the light-regulated localization of phot1. This effect might be caused by cytosolic phyA; however, the fact that a dark period is required between the red light treatment and an efficient effect on phototropism is also compatible with phyA leading to changes in gene expression leading to changes in phot1 localization (Han et al., 2008). In summary, although our studies demonstrate a more potent phototropism promotion effect of nuclear than cytosolic phyA, our studies do not rule out cytosolic effects of phyA on the regulation of hypocotyl phototropism.
Nuclear Signaling Is Still Present in fhy1 fhl, Particularly in Blue Light
The analysis of rapid light-regulated gene expression in response to FR light shows that fhy1 fhl and phyA have a very similar phenotype (Figure 6). Consistent with previous reports, these mutants are essentially blind to a 1-h FR treatment (Figure 6) (Tepperman et al., 2001; Peschke and Kretsch, 2011). The general tendency is that in fhy1 fhl, there is slightly more expression of light-induced genes than in phyA, but for most genes this is not statistically significant (Figure 6) (data not shown). This gene expression phenotype of fhy1 fhl correlates well with the morphological phenotype of fhy1 fhl, which is indistinguishable from phyA when grown in FR light (Zhou et al., 2005; Hiltbrunner et al., 2006; Rösler et al., 2007).
By contrast, light-regulated gene expression in response to low blue light is still partially functional in fhy1 fhl (Figure 6). Importantly, the gene expression phenotype in phyA was not further enhanced in phyA fhy1 fhl, indicating that in blue light as well, FHY1 and FHL act in the phyA pathway (Figure 6). This is also consistent with the phototropism phenotype of the phyA fhy1 fhl, which is not more severe than the phyA phenotype (Figure 2). Importantly, the analysis of phyA-YFP subcellular localization in fhy1 fhl reveals no difference in the pattern of phyA-YFP subcellular localization in FR or blue light (Figure 5; see Supplemental Figure 4 online), showing that in both light conditions, FHY1 and FHL are essential for robust nuclear import of phyA.
Under low blue light conditions, phyA plays a more prominent role in the regulation of gene expression than the cryptochromes (Figures 6). Importantly, in the cry1 cry2 fhy1 fhl quadruple mutant, blue light regulation of PKS1 and RPT2 was attenuated compared with in fhy1 fhl (Figure 6), indicating that the residual light-regulated gene expression in fhy1 fhl depends at least in part on the cryptochromes. Our data thus suggest that in low blue light, residual nuclear phyA signaling in fhy1 fhl contributes to light-regulated gene expression. The remaining blue light–regulated gene expression in fhy1 fhl may thus either be caused by a small amount of phyA still entering the nucleus in this mutant or cytosolic phyA initiating a signaling cascade in the cytosol that results in regulated gene expression (Neuhaus et al., 1993). An interesting question that our study helps address is why fhy1 fhl is more similar to phyA in FR than in blue light (Rösler et al., 2007) (Figures 2, 3, and 6). Light-regulated gene expression in etiolated seedlings transferred into FR light suggests that the residual phyA activity in fhy1 fhl is insufficient to cause significant changes in gene expression when the light response is exclusively regulated by phyA (Figure 6). By contrast, in blue light the low levels of phyA signaling still present in fhy1 fhl are revealed, because the combined action of multiple photoreceptors (i.e., phyA, cry1, and cry2) mediates changes in gene expression (Figure 6) (Sellaro et al., 2009).
Development of New Software for Hypocotyl Growth Measurements
To distinguish between the phenotypes of fhy1 fhl, phyA-NLS-GFP, and the wild type it was essential to perform time-course experiments of phototropic bending (Figures 1 to 3; see Supplemental Figure 3 online). In the course of this work, we developed the HypoPhen software to accelerate and standardize hypocotyl-bending measurements. To validate this semiautomatic measurement system, we have compared the output of HypoPhen with manual measurements of the same data set (see Supplemental Figure 3 online). Although the data are not exactly identical, there is very large agreement between the two measurement methods, and all our conclusions are supported by both measurement methods (Figures 2 and 3; see Supplemental Figure 3 online). Importantly, using a semiautomatic measurement system diminishes the user bias that occurs inevitably when different experimenters measure data sets. The setup used for time-lapsed imaging was inspired by previous publications (Miller et al., 2007; Wang et al., 2009; Cole et al., 2011). However, we have chosen to follow a larger number of seedlings (typically 21 to 24 per camera) to increase the throughput. Although this results in reduced image resolution of the seedlings analyzed, the compromise that we selected is still sufficient to perform tasks such as measurements of phototropic bending or growth rates. Indeed, this lower image resolution is compensated for by more sophisticated image-processing algorithms, which make the phenotyping more robust to poor image quality. For example, in contrast with HypoTrace (Wang et al., 2009) and HyDe (Cole et al., 2011), where each time point image is analyzed independently, the HypoPhen software compares successive images to assess the shape of the hypocotyl. Thus, it does not make any a priori assumption on the developmental stage of the hypocotyl and handles hypocotyls with closed cotyledons (assumed by HypoTrace), as well as open cotyledons (assumed by HyDe). It can also deal with perturbing elements, such as seed caps, leaves, and uneven illumination, that make image processing more difficult. Furthermore, our software provides the user the ability to adjust manually the detection and tracing of the hypocotyls and to save them as images for later data inspection. Perhaps more importantly, in contrast with the two aforementioned pieces of software, HypoPhen is not a black box, but is totally open-source and can be adapted and redistributed by anybody with the adequate skills. This will enable researchers to adapt it to their specific needs (e.g., for the selection of hypocotyls or the computation of bending angles and other measures). We believe that this software will be useful to the community as it becomes clear that real-time measurements are important to unravel the rapid effects of light on growth responses (Figures 1 to 3) (Cole et al., 2011).
METHODS
Plant Materials and Growth Conditions
The following genotypes of Arabidopsis thaliana were used: the wild type (Columbia-0), phyA-211 (Reed et al., 1994), fhy1-3 fhl-1 (Rösler et al., 2007), PHYApro:PHYA-GFP phyA-211, PHYApro:PHYA-NLS phyA-211, PHYApro:PHYA-NLS-GFP phyA-211 (Genoud et al., 2008), cry1 cry2, and phyA cry1 cry2 (Duek and Fankhauser, 2003). The fhy1-3 fhl-1 cry1-304 cry2-1 quadruple mutant was obtained by crossing the fhy1-3 fhl-1 (Rösler et al., 2007) and cry1-304 cry2-1 double mutants; hy5-215 (Oyama et al., 1997), a T-DNA insertion line disrupting the HFR1 open reading frame (SALK_037727), was used as an hfr1 allele; the hfr1 hy5 mutant was obtained by crossing. Transgenic lines expressing PHYApro:PHYA-YFP in phyA-211 and fhy1-3 fhl-1 background were generated by Agrobacterium-mediated transformation of phyA-211 and fhy1-3 fhl-1 plants. The T-DNA vector containing the PHYApro:PHYA-YFP construct (pPPO30A-phyA; contains a selection marker conferring resistance to Butafenacil) and the selection of transgenic plants using the herbicide Butafenacil/Inspire has been described (Rausenberger et al., 2011).
Surface-sterilized seeds were plated on one-half strength Murashige and Skoog plates with 0.8% agar, kept at 4°C in the dark for 3 d. Plates were transferred to 21 ± 1°C and exposed to 100 μmol m−2 s−1 cool white light for 6 h to induce germination and were incubated (vertically) in the dark at 21 ± 1°C until hypocotyls had the appropriate hypocotyl length (SANYO incubator). The blue light source was a light-emitting diode (λmax, 470 nm; CLF Plant Climatics GmbH).
Microscopy
To examine the subcellular relocalization of phyA-GFP, phyA-NLS-GFP, phyA-YFP, 3-d-old dark-grown seedlings or light-treated seedlings (as described in the figure legends) were placed on slides in a drop of one-half strength Murashige and Skoog medium with 0.01% agar. For confocal microscopy, Arabidopsis seedlings were observed with an inverted Zeiss confocal microscope (LSM 510 Meta INVERTED, Zeiss AXIO Vert 200 M; ×40 objective). Images were processed with Zeiss software (release 3.5; LSM Software).
Physiological Analysis for Phototropism
For long-term phototropism experiments, after induction of germination, seeds were incubated at 21 ± 1°C with unilateral blue light for 3 d. For short-term phototropism experiments, seedlings were grown in darkness typically for 54 to 60 h prior to irradiation with unilateral blue light (0.1, 1, or 10 μmol m−2 s−1). The angles relative to vertical of seedlings with a hypocotyl length of 4 to 5.9 mm at the time of the beginning of the light treatment were determined after 24 h of irradiation. For time-lapse monitoring of hypocotyl orientation, we also used etiolated seedlings with a hypocotyl length of 4 to 5.9 mm at the time of the beginning of the light treatment. Given that the speed of phototropic bending depends on the position of the cotyledons relative to the light source (see Supplemental Figure 2 online), we always used the same number of seedlings with each orientation for a measurement. Time-lapse images were acquired using a monochrome charge-coupled device camera (CV-M50IR; JAI) and infrared light-emitting diodes (FQ15603; peak emission at 940 nm, one-half bandwidth, 50 nm; Adlos AG) placed in an incubator (floraLEDS; CLF Plant Climatics GmbH). The MetaMorph software (Molecular Devices) was used to control the charge-coupled device camera system and to process images. Hypocotyl length and angles were measured using stacked images (using National Institutes of Health ImageJ software version 1.38 [http://rsb.info.nih.gov/ij/]) as described by Folta et al. (2003).
Automatic Measurement System
The HypoPhen software was developed for the semiautomatic measurement of hypocotyl bending. It takes as input time-lapsed images of growing hypocotyls and outputs bending angles. Using the first image, the user marks with the computer mouse the apical hook of the hypocotyls that need to be measured and sets a value for the image thresholding used for background removal. For the remaining images, optical flow computations are then used to track the apical hook, and a snake image processing algorithm tracks the hypocotyls. Bending values are then computed as the direction of the upper tip of the hypocotyl. The software is open-source and based on the OpenCV library (Bradski and Kaehler, 2008). It is documented and freely available at http://www.unil.ch/cbg/index.php?title=HypoPhen. Batches of infrared images displaying up to 21 hypocotyls were taken at 30-min intervals up to 24 h after continuous lateral blue light irradiation and were analyzed with HypoPhen.
To reduce the measurement noise, outliers were then removed, and the data of each hypocotyl were smoothed while ensuring a monotonous bending function (Ramsay and Silverman, 2005). This resulted in a final data set containing, for each genotype, at least 60 hypocotyls and the same number of left- and right-positioned cotyledons. Differences in kinetics between two genotypes were assessed using a Student’s t test at each time point. The significance level was set to 5%, corrected for multiple testing by the method described in Gao et al. (2008).
RNA Extraction and qRT-PCR
RNA isolation and RT-PCR were performed as previously described (Lorrain et al., 2009). After inducing germination, seedlings were grown for 3 d in the dark at 22°C and either kept in darkness for 1 h or transferred to FR light (5 μmol m−2 s−1) or blue light (0.5 μmol m−2 s−1) for 1 h. First-strand cDNA synthesis was performed with 1 μg of RNA. For qRT-PCR, 1 μL of 20-fold diluted cDNA was used.
UBC (At5g25760) and HMG1 (AT1g76490) were used as housekeeping genes for normalization of the experiments. The primers that were not previously described in Lorrain et al. (2009) are described below.
Nucleotide sequence of the new primers used in this study are as follows: HMG1_F: AAC TTT GAT ACT TTG GCA GTA GTC TTC A; HMG1_R: CGC GAT TGT GCA TTT AAC ACT T; RPT2_F: TGC AAG AAC CGG TCA ATG; and RPT2_R: TCT TGT CAC GTC GCT ATC.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PHYA (AT1G09570), PHOT1 (AT3G45780), FHY1 (AT2G37678), FHL (AT5G02200), PKS1 (AT2G02950), RPT2 (AT2G30520).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Subcellular Localization of a Constitutively Localized phyA.
Supplemental Figure 2. Phototropism Kinetics Depends on the Length of the Hypocotyl and the Orientation of the Cotyledons.
Supplemental Figure 3. Comparison of Phototropism Kinetics Using Manual Measurement and Semiautomatic Measurement with Same Time-Lapse Images.
Supplemental Figure 4. phyA Localization (phyA-YFP) in the Wild Type or fhy1 fhl under FR Light.
Supplemental Figure 5. Comparison of Phototropism Kinetics in the Wild Type, phyA, fhy1 fhl, cry1 cry2, cry1 cry2 phyA, and cry1 cry2 fhy1 fhl under Low Blue Light.
Supplementary Material
Acknowledgments
We thank the University of Lausanne Cellular Imaging Facility for help with confocal microscopy and the Lausanne Genomic Technologies Facility for help with quantitative PCR analysis. This work was supported by the University of Lausanne, a grant from the Swiss National Science Foundation (3100A0-112638) to C.F., and the SystemsX.ch grant “Plant Growth in a Changing Environment” to C.F. and S.B. C.K. was supported by a Toyobo Biotechnology Foundation postdoctoral fellowship.
AUTHOR CONTRIBUTIONS
C.K. designed the research, performed research, analyzed data, and contributed to writing the article. M.H. contributed new analytic/computational tools, designed the research, performed research, and analyzed data. M.T. performed research and analyzed data. T.G. designed the research, performed research, and analyzed data. A.H. contributed new tools. S.B. analyzed data. C.F. designed the research, analyzed data, and wrote the article.
References
- Bradski G., Kaehler A. (2008). Learning OpenCV: Computer Vision with the OpenCV library. (Sebastopol, CA: O’Reilly Media; ). [Google Scholar]
- Casal J.J. (2000). Phytochromes, cryptochromes, phototropin: Photoreceptor interactions in plants. Photochem. Photobiol. 71: 1–11 [DOI] [PubMed] [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15: 637–642 [DOI] [PubMed] [Google Scholar]
- Christie J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Christie J.M., et al. (2011). phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 9: e1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole B., Kay S.A., Chory J. (2011). Automated analysis of hypocotyl growth dynamics during shade avoidance in Arabidopsis. Plant J. 65: 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carbonnel M., Davis P., Roelfsema M.R., Inoue S., Schepens I., Lariguet P., Geisler M., Shimazaki K., Hangarter R., Fankhauser C. (2010). The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 152: 1391–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBlasio S.L., Mullen J.L., Luesse D.R., Hangarter R.P. (2003). Phytochrome modulation of blue light-induced chloroplast movements in Arabidopsis. Plant Physiol. 133: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E., Fankhauser C. (2009). Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12: 69–74 [DOI] [PubMed] [Google Scholar]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski L., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Esmon C.A., Tinsley A.G., Ljung K., Sandberg G., Hearne L.B., Liscum E. (2006). A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl. Acad. Sci. USA 103: 236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chen M. (2008). Transposing phytochrome into the nucleus. Trends Plant Sci. 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Folta K.M., Lieg E.J., Durham T., Spalding E.P. (2003). Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. Plant Physiol. 133: 1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Larner V.S., Whitelam G.C. (2005). The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 49: 653–664 [DOI] [PubMed] [Google Scholar]
- Gao X., Starmer J., Martin E.R. (2008). A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 32: 361–369 [DOI] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Takano M., Neumann R., Iino M. (2005). The Rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell 17: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I.S., Tseng T.S., Eisinger W., Briggs W.R. (2008). Phytochrome A regulates the intracellular distribution of phototropin 1-green fluorescent protein in Arabidopsis thaliana. Plant Cell 20: 2835–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Iino M. (2006). Toward understanding the ecological functions of tropisms: Interactions among and effects of light on tropisms. Curr. Opin. Plant Biol. 9: 89–93 [DOI] [PubMed] [Google Scholar]
- Inoue S., Kinoshita T., Matsumoto M., Nakayama K.I., Doi M., Shimazaki K. (2008). Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. USA 105: 5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Matsushita T., Tomokiyo Y., Matsumoto M., Nakayama K.I., Kinoshita T., Shimazaki K. (2011). Functional analyses of the activation loop of phototropin2 in Arabidopsis. Plant Physiol. 156: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoudi A.K., Gordon W.R., Wagner D., Quail P., Poff K.L. (1997). Multiple phytochromes are involved in red-light-induced enhancement of first-positive phototropism in Arabidopsis thaliana. Plant Physiol. 113: 975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T., Wada M. (2000). Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol. 41: 84–93 [DOI] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kanegae T., Hayashida E., Kuramoto C., Wada M. (2006). A single chromoprotein with triple chromophores acts as both a phytochrome and a phototropin. Proc. Natl. Acad. Sci. USA 103: 17997–18001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H., Kanegae T., Christensen S., Kiyosue T., Sato Y., Imaizumi T., Kadota A., Wada M. (2003). Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 421: 287–290 [DOI] [PubMed] [Google Scholar]
- Khurana J.P., Best T.R., Poff K.L. (1989). Influence of hook position on phototropic and gravitropic curvature by etiolated hypocotyls of Arabidopsis thaliana. Plant Physiol. 90: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L., Kircher S., Toth R., Adam E., Schäfer E., Nagy F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22: 125–133 [DOI] [PubMed] [Google Scholar]
- Kim Y.M., Woo J.C., Song P.S., Soh M.S. (2002). HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J. 30: 711–719 [DOI] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S.G., Suzuki T., Tamura K., Mochizuki N., Hara-Nishimura I., Nagatani A. (2006). Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 45: 994–1005 [DOI] [PubMed] [Google Scholar]
- Lariguet P., Fankhauser C. (2004). Hypocotyl growth orientation in blue light is determined by phytochrome A inhibition of gravitropism and phototropin promotion of phototropism. Plant J. 40: 826–834 [DOI] [PubMed] [Google Scholar]
- Lariguet P., Schepens I., Hodgson D., Pedmale U.V., Trevisan M., Kami C., de Carbonnel M., Alonso J.M., Ecker J.R., Liscum E., Fankhauser C. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. USA 103: 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. (1999). Detecting protein function and protein-protein interactions from genome sequences. Science 285: 751–753 [DOI] [PubMed] [Google Scholar]
- Miller N.D., Parks B.M., Spalding E.P. (2007). Computer-vision analysis of seedling responses to light and gravity. Plant J. 52: 374–381 [DOI] [PubMed] [Google Scholar]
- Möller B., Schenck D., Lüthen H. (2010). Exploring the link between auxin receptors, rapid cell elongation and organ tropisms. Plant Signal. Behav. 5: 601–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A., Liscum E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286: 961–964 [DOI] [PubMed] [Google Scholar]
- Nagashima A., et al. (2008). Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. Plant J. 53: 516–529 [DOI] [PubMed] [Google Scholar]
- Nagatani A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7: 708–711 [DOI] [PubMed] [Google Scholar]
- Neuhaus G., Bowler C., Kern R., Chua N.H. (1993). Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell 73: 937–952 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks B.M., Quail P.H., Hangarter R.P. (1996). Phytochrome A regulates red-light induction of phototropic enhancement in Arabidopsis. Plant Physiol. 110: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke F., Kretsch T. (2011). Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation. Plant Physiol. 155: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay J.O., Silverman B.W. (2005). Functional Data Analysis. (New York: Springer; ). [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Rösler J., Jaedicke K., Zeidler M. (2010). Cytoplasmic phytochrome action. Plant Cell Physiol. 51: 1248–1254 [DOI] [PubMed] [Google Scholar]
- Rösler J., Klein I., Zeidler M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. USA 104: 10737–10742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Kagawa T., Kasahara M., Swartz T.E., Christie J.M., Briggs W.R., Wada M., Okada K. (2001). Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Wada T., Ishiguro S., Okada K. (2000). RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Briggs W.R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R., Hoecker U., Yanovsky M., Chory J., Casal J.J. (2009). Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr. Biol. 19: 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone B.B., Stowe-Evans E.L., Harper R.M., Celaya R.B., Ljung K., Sandberg G., Liscum E. (2008). Disruptions in AUX1-dependent auxin influx alter hypocotyl phototropism in Arabidopsis. Mol. Plant 1: 129–144 [DOI] [PubMed] [Google Scholar]
- Stowe-Evans E.L., Luesse D.R., Liscum E. (2001). The enhancement of phototropin-induced phototropic curvature in Arabidopsis occurs via a photoreversible phytochrome A-dependent modulation of auxin responsiveness. Plant Physiol. 126: 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Mittmann F., Wagner G., Hughes J., Wada M. (2005). A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc. Natl. Acad. Sci. USA 102: 13705–13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Kumagai S., Muto H., Sato A., Watahiki M.K., Harper R.M., Liscum E., Yamamoto K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.S., Wang X., Quail P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Kiryu Y., Kobayashi J., Oka Y., Kim Y., Nam H.G., Mochizuki N., Nagatani A. (2010). Subcellular sites of the signal transduction and degradation of phytochrome A. Plant Cell Physiol. 51: 1648–1660 [DOI] [PubMed] [Google Scholar]
- Tsuchida-Mayama T., Sakai T., Hanada A., Uehara Y., Asami T., Yamaguchi S. (2010). Role of the phytochrome and cryptochrome signaling pathways in hypocotyl phototropism. Plant J. 62: 653–662 [DOI] [PubMed] [Google Scholar]
- Wan Y.L., Eisinger W., Ehrhardt D., Kubitscheck U., Baluska F., Briggs W. (2008). The subcellular localization and blue-light-induced movement of phototropin 1-GFP in etiolated seedlings of Arabidopsis thaliana. Mol. Plant 1: 103–117 [DOI] [PubMed] [Google Scholar]
- Wang F.F., Lian H.L., Kang C.Y., Yang H.Q. (2010). Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol. Plant 3: 246–259 [DOI] [PubMed] [Google Scholar]
- Wang L., Uilecan I.V., Assadi A.H., Kozmik C.A., Spalding E.P. (2009). HYPOTrace: Image analysis software for measuring hypocotyl growth and shape demonstrated on Arabidopsis seedlings undergoing photomorphogenesis. Plant Physiol. 149: 1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo C.W., Hangarter R.P. (2003). Second positive phototropism results from coordinated co-action of the phototropins and cryptochromes. Plant Physiol. 132: 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo C.W., Hangarter R.P. (2004). Phytochrome modulation of blue-light-induced phototropism. Plant Cell Environ. 27: 1223–1228 [Google Scholar]
- Zhou Q., Hare P.D., Yang S.W., Zeidler M., Huang L.F., Chua N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43: 356–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.