This work shows that suppressing the expression of the vesicle-inducing protein in plastids (VIPP1) in Chlamydomonas leads to aberrant structures at the origin of thylakoids and to structural defects particularly in photosystem II that render mutants sensitive to high light. The data indicate that VIPPs act in the biogenesis of thylakoid membrane core complexes, in particular the photosystems.
Abstract
The vesicle-inducing protein in plastids (VIPP1) was suggested to play a role in thylakoid membrane formation via membrane vesicles. As this functional assignment is under debate, we investigated the function of VIPP1 in Chlamydomonas reinhardtii. Using immunofluorescence, we localized VIPP1 to distinct spots within the chloroplast. In VIPP1-RNA interference/artificial microRNA cells, we consistently observed aberrant, prolamellar body-like structures at the origin of multiple thylakoid membrane layers, which appear to coincide with the immunofluorescent VIPP1 spots and suggest a defect in thylakoid membrane biogenesis. Accordingly, using quantitative shotgun proteomics, we found that unstressed vipp1 mutant cells accumulate 14 to 20% less photosystems, cytochrome b6f complex, and ATP synthase but 30% more light-harvesting complex II than control cells, while complex assembly, thylakoid membrane ultrastructure, and bulk lipid composition appeared unaltered. Photosystems in vipp1 mutants are sensitive to high light, which coincides with a lowered midpoint potential of the QA/QA− redox couple and increased thermosensitivity of photosystem II (PSII), suggesting structural defects in PSII. Moreover, swollen thylakoids, despite reduced membrane energization, in vipp1 mutants grown on ammonium suggest defects in the supermolecular organization of thylakoid membrane complexes. Overall, our data suggest a role of VIPP1 in the biogenesis/assembly of thylakoid membrane core complexes, most likely by supplying structural lipids.
INTRODUCTION
The thylakoids of chloroplasts represent an internal membrane system that is detached from the inner envelope membrane. As thylakoid lipids are not synthesized at the thylakoid membranes but rather at the chloroplast inner and outer envelope membranes and the endoplasmic reticulum, a transport system must exist that enables a flow of lipid components from these biogenic membranes to the thylakoids (Benning, 2008, 2009). Ultrastructural studies suggested lipid transport via vesicles that bud off from the inner envelope and fuse with the thylakoids (Carde et al., 1982). The M30 protein was identified in pea (Pisum sativum) as a plastid protein that is associated with both envelope and thylakoid membranes and therefore was suggested as a candidate protein for lipid transfer from envelope to thylakoids (Li et al., 1994). M30 was renamed vesicle-inducing protein in plastids1 (VIPP1) based on the phenotype observed in the Arabidopsis thaliana hcf155 mutant, which expresses M30/VIPP1 to ~20% of wild-type levels: hcf155 plants have much less and distorted thylakoids and reduced amounts of photosystem I (PSI), photosystem II (PSII), light-harvesting complex B, cytochrome b6f complex, and ATP synthase compared with wild-type plants. Moreover, hcf155 plants lack vesicles originating from the inner chloroplast envelope (Kroll et al., 2001; Aseeva et al., 2007). These observations led to the proposal that VIPP1 is essential for the formation of thylakoid membranes via vesicle traffic, a conclusion that was supported by the almost complete lack of thylakoids in a cyanobacterial vipp1 mutant strain (Westphal et al., 2001). However, in a similarly constructed cyanobacterial vipp1 mutant strain, Fuhrmann et al. (2009a) only found reduced, less well arranged thylakoid layers and reduced amounts of (trimeric) PSI. Because VIPP1 is an essential protein, the disruption of the VIPP1 gene generated merodiploid cells that still accumulated >25% of wild-type VIPP1 levels. Hence, Gao and Xu (2009) generated a cyanobacterial strain expressing VIPP1 under control of the copper-responsive petE promoter and under copper-depleted conditions observed that depletion of VIPP1 correlated first with a loss of photosynthetic activity (in particular of PSII) before thylakoid membranes were depleted. Therefore, Gao and Xu questioned the role of VIPP1 in thylakoid formation.
The picture becomes even more confusing when looking at the proposed function for the closest homolog of VIPP1 in prokaryotes, the phage shock protein A (PspA) (Joly et al., 2010). The phage shock response is induced by agents that potentially affect the integrity of the plasma membrane and normally lead to a loss of the proton motive force. Examples for inducing agents are filamentous phage infection, severe heat shock, depletion of the protein membrane insertase YidC, or blockage of the twin-Arg (TAT) or Sec translocons (Brissette et al., 1990; Kleerebezem and Tommassen, 1993; van der Laan et al., 2003; DeLisa et al., 2004). PspA in its oligomeric form was shown to suppress proton leakage from damaged membranes by directly interacting with membrane lipids phosphatidylserine and phosphatidylglycerol (PG; Kleerebezem et al., 1996; Kobayashi et al., 2007). Functional conservation between PspA and VIPP1 is suggested by the findings that both proteins improved protein export via the twin-Arg translocon pathway (DeLisa et al., 2004), and both proteins assemble into rotationally symmetric rings of >1 MD (Aseeva et al., 2004; Hankamer et al., 2004; Liu et al., 2007; Standar et al., 2008; Fuhrmann et al., 2009b). Still, some specificity for PspA and VIPP1 function must exist because both are present in cyanobacteria, but cyanobacterial PspA cannot substitute for the function of cyanobacterial VIPP1 (Westphal et al., 2001).
Several opinions exist not only regarding the function of VIPP1 but also concerning its localization. In chloroplasts of higher plants and algae, VIPP1 was localized to thylakoids and the inner envelope (Li et al., 1994; Kroll et al., 2001; Liu et al., 2005), but a localization only to inner envelopes was proposed by Aseeva et al. (2004). Moreover, VIPP1 was also found in stromal fractions (Li et al., 1994; Liu et al., 2005). In cyanobacteria, VIPP1 was initially reported to be localized exclusively to the plasma membrane (Westphal et al., 2001). However, a dual localization of VIPP1 to plasma membrane and thylakoids was reported (Srivastava et al., 2005), and, more recently, VIPP1 was also detected in the cytoplasm (Srivastava et al., 2006; Fuhrmann et al., 2009b). Eventually, these data suggest that VIPP1 is in an equilibrium between membrane-bound and soluble forms, as appears to be the case for bacterial PspA (Brissette et al., 1990; Kleerebezem and Tommassen, 1993).
VIPP1 activity appears to be tightly linked to plastidic chaperones, especially the HSP70 and HSP90 systems (Nordhues et al., 2010). VIPP1 interacts with a specialized J-domain protein termed CDJ2, which delivers VIPP1 to chloroplast HSP70B (Liu et al., 2005). HSP70B and CDJ2 catalyze the assembly and disassembly of VIPP1 oligomers (Liu et al., 2007). The interaction of VIPP1 with chloroplast HSP70 was confirmed in higher plants (Aseeva et al., 2007), and VIPP1 was found to interact also with chloroplast HSP90, which might support HSP70 during VIPP1 oligomer assembly/disassembly (Heide et al., 2009). To elucidate why the chloroplast HSP90/HSP70 systems catalyze the assembly/disassembly of VIPP1 oligomers, it is essential to understand the function of VIPP1. However, as outlined above, the functional analyses done so far in higher plants and cyanobacteria did not yet provide definitive conclusions on VIPP1 function. We therefore decided to perform a functional analysis of VIPP1 in the unicellular green alga Chlamydomonas reinhardtii, in which the interaction between VIPP1 and the chaperones was discovered. While doing so, we found that, unlike higher plants and cyanobacteria, volvocean algae contain two VIPP paralogs. In C. reinhardtii, these two paralogs share 50% identical and 65% similar residues and were termed VIPP1 and VIPP2 (S. Schmollinger and M. Schroda, unpublished results). Although the presence of two VIPP paralogs complicated the analysis, our results strongly suggest that these VIPPs are redundantly involved in the biogenesis/assembly of thylakoid membrane protein complexes at distinct sites within the chloroplast and not in vesicle-mediated thylakoid membrane formation as proposed earlier.
RESULTS
C. reinhardtii VIPP1 Is Efficiently Downregulated by RNA Interference
To investigate the function of VIPP1 in C. reinhardtii, we first used an RNA interference (RNAi) strategy to reduce VIPP1 function. To do this, we made a construct harboring a piece of genomic, intron-containing VIPP1 DNA in sense orientation followed by a complementary piece of cDNA in antisense orientation (see Supplemental Figure 1 online). Expression of the RNAi construct is driven by the strong HSP70A-RBCS2 fusion promoter (Schroda et al., 2000). The ARG7 gene present on the same vector was used as selectable marker for the transformation of the Arg-auxotrophic cw15-325 recipient strain. This construct silenced the VIPP1 gene efficiently, as shown by the reduction of VIPP1 protein to <5% of wild-type levels in ~10% of the Arg-prototrophic transformants (Figure 1A; see Supplemental Figure 4 online). However, as wild-type VIPP1 levels recovered within 1 to 6 months in VIPP1-RNAi strains the RNAi construct itself appears to be silenced, as observed earlier with other inverted repeat constructs (Yamasaki et al., 2008). Therefore, during the course of this work, we regularly had to generate fresh VIPP1-RNAi strains. Eventually, >30 independent lines were characterized. Control strains were generated by transforming the cw15-325 strain with a plasmid containing the ARG7 gene alone.
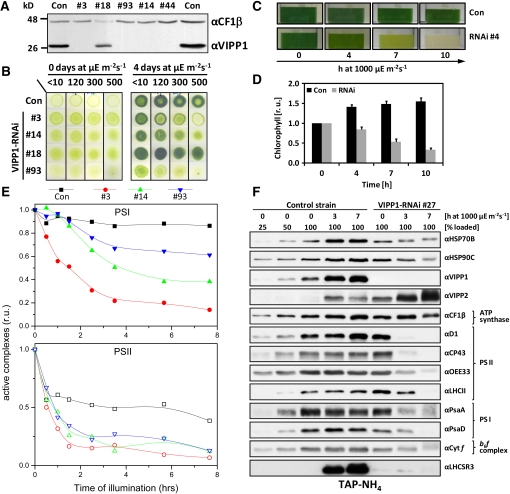
Figure 1.
Photobleaching of High Light–Exposed VIPP1-RNAi Strains.
(A) VIPP1 protein levels in VIPP1-RNAi strains are strongly reduced. Whole-cell proteins from transformants generated with VIPP1-RNAi construct pMS439 corresponding to 2 μg chlorophyll were separated on a 14% SDS-polyacrylamide gel and analyzed by immunoblotting. CF1β served as loading control.
(B) High light sensitivity of VIPP1-RNAi strains. Ten microliters containing 107 cells were spotted onto TAP-NH4 agar plates and photographed directly or after a 4-d incubation at the indicated light intensities.
(C) Bleaching of VIPP1-RNAi strains. A control strain and VIPP1-RNAi strain #4 grown in TAP-NH4 medium were exposed to a light intensity of ~1000 μE m−2 s−1 for 10 h.
(D) The chlorophyll content rapidly declines in high light–exposed VIPP1-RNAi strains. A control strain (n = 3) and five VIPP1-RNAi strains (#3, #26, #44, #49, and #93) were treated as in (A), and the chlorophyll content was determined. Chlorophyll contents are given relative to the concentrations determined prior to the shift to high light, which were set to 1. Error bars represent se. r.u., relative units.
(E) PSII and PSI in VIPP1-RNAi strains are very sensitive to high light. Equal cell densities of a control strain and VIPP1-RNAi strains #3, #14, and #93 were subjected to high light as in (A), and PSII and PSI activities were measured by determining the DCMU-sensitive and -insensitive fractions of the electrochromic shift signal.
(F) Subunits of PSII and PSI are rapidly degraded in VIPP1-RNAi strains exposed to high light intensities. Whole-cell proteins from high light–exposed control and VIPP1-RNAi strain #27 grown in TAP-NH4 medium were separated on 14% SDS-polyacrylamide gels and analyzed by immunoblotting.
VIPP1-RNAi/amiRNA Strains Are Sensitive to High Light
Despite the strong reduction of VIPP1 levels in VIPP1-RNAi strains (Figure 1A), we observed no obvious phenotypes in cells grown at low light intensities in Tris-acetate-phosphate (TAP)-NH4 medium: Cells grew normally under mixotrophic conditions (Figure 1B), had fully developed thylakoid membranes (see Figures 3C and 10A), and seemed to assemble thylakoid membrane protein complexes like control cells (see Supplemental Figure 2 online). The composition of bulk thylakoid membrane lipids in VIPP1-RNAi strains was indistinguishable from control cells (see Supplemental Figure 3 online), and we could not observe consistent changes in the number of thylakoids per granum stack (as judged from the analysis of electron micrographs from 50 cells each of two control and five VIPP1-RNAi lines). This observation was unexpected, as Arabidopsis vipp1 mutants expressing VIPP1 at ~20% of wild-type levels have an albinotic phenotype and cannot grow photoautotrophically due to a degraded thylakoid membrane system (Kroll et al., 2001; Aseeva et al., 2007).
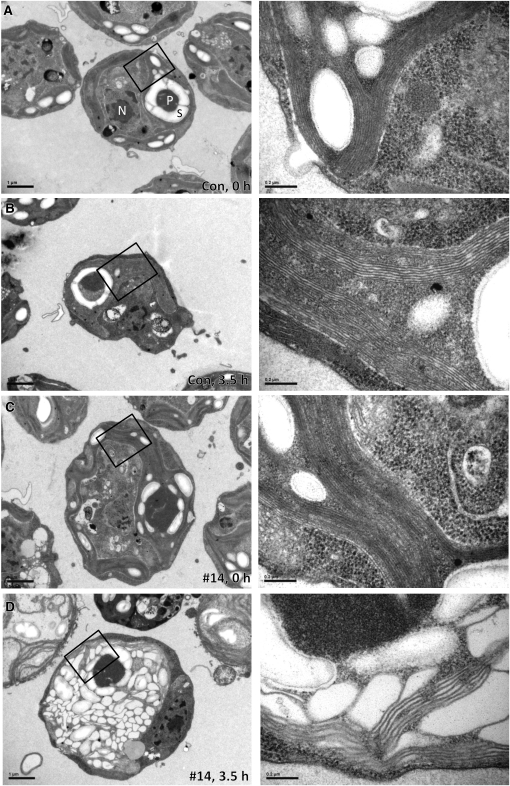
Figure 3.
Thylakoids in VIPP1-amiRNA Strains Exposed to High Light Intensities Are Extremely Swollen.
(A) Electron microscopy image of a cell from the control strain grown at low light intensities. Cells were grown at ~30 μE m−2 s−1 in TAP-NH4 medium. An overview image is shown on the left, and a zoom-in of the region demarcated by the black box is shown on the right. N, nucleus; P, pyrenoid; S, starch. Bars in overview images correspond to 1 μm and those in zoom-ins to 0.2 μm.
(B) Electron microscopy image of a cell from the control strain exposed to high light. Cells were grown at ~30 μE m−2 s−1 in TAP-NH4 medium and exposed to ~1000 μE m−2 s−1 for 3.5 h. Images were taken as in (A).
(C) Electron microscopy image of a cell from a VIPP1-amiRNA strain grown at low light intensities. VIPP1-amiRNA strain #14 was grown at ~30 μE m−2 s−1 in TAP-NH4 medium. Images were taken as in (A).
(D) Electron microscopy image of a cell from a VIPP1-amiRNA strain exposed to high light. VIPP1-amiRNA strain #14 was grown at ~30 μE m−2 s−1 in TAP-NH4 medium and exposed to ~1000 μE m−2 s−1 for 3.5 h. Images were taken as in (A).
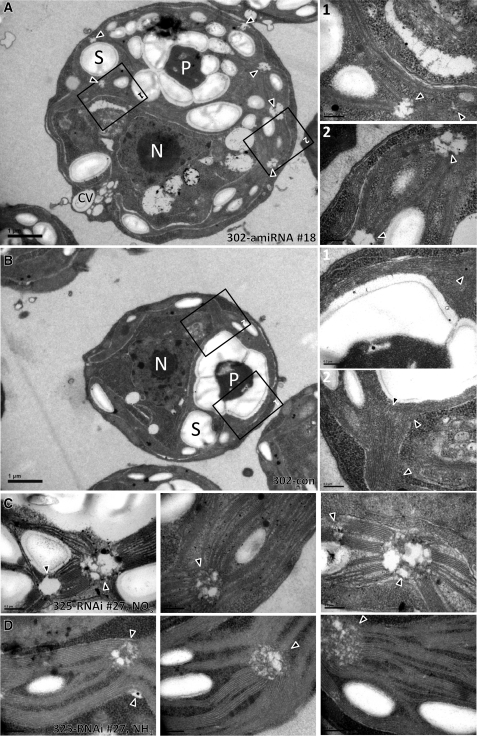
Figure 10.
VIPP1-RNAi/amiRNA Strains Harbor Aberrant Structures at the Origin of Thylakoid Membranes.
(A) Electron microscopy image of a cell from VIPP1-amiRNA strain #18. Cells were grown at ~30 μE m−2 s−1 in TAP-NH4 medium. An overview image is shown on the left, and zoom-ins of the regions demarcated by black boxes are shown on the right. Triangles indicate regions at the origin of multiple thylakoid membrane ramifications. CV, contractile vacuole; N, nucleus; P, pyrenoid; S, starch. Bars in overview images = 1 μm, those in zoom-ins = 0.2 μm.
(B) Electron microscopy image of a cell from the control strain. Cells were grown at ~30 μE m−2 s−1 in TAP-NH4 medium. Symbols are as in (A).
(C) Electron microscopy image of a cell from VIPP1-RNAi strain #27. Cells were grown at ~30 μE m−2 s−1 in TAP-NO3 medium. Abbreviations are as in (A).
(D) Electron microscopy image of a cell from VIPP1-RNAi strain #27. Cells were grown at ~30 μE m−2 s−1 in TAP-NH4 medium. Abbreviations are as in (A).
In previous work, we observed that the C. reinhardtii VIPP1 gene was strongly induced when dark-grown cells were shifted into the light (Liu et al., 2005). In C. reinhardtii, the thylakoid membranes are fully developed also in the dark; this suggested to us that VIPP1 may be of particular importance in the light. We therefore exposed control and VIPP1-RNAi strains to increasing light intensities (up to 500 μE m−2 s−1) and observed that VIPP1-RNAi strains containing very low levels of VIPP1 failed to grow and bleached at higher light intensities. By contrast, no effect of high light was observed with the control strain and with a VIPP1-RNAi strain expressing VIPP1 to ~50% of wild-type levels (#18; Figure 1B).
To substantiate this finding, we exposed mixotrophically grown control and VIPP1-RNAi strains to high light intensities (~1000 μE m−2 s−1) (Figure 1C). Within 10 h, we observed severe bleaching of VIPP1-RNAi strains. On average, the chlorophyll content declined to ~40% of starting levels, while in the control strain, it increased by ~60% during high light treatment as a consequence of growth (Figure 1D).
Assessment of PSII and PSI activities by measurements of the electrochromic shift revealed a fast decline of PSII activity in high light–treated VIPP1-RNAi strains, with activities dropping to ~15 to 35% of the initial values already after 90 min and to ~10% after 7.5 h at high light (Figure 1E). In the control, PSII activity dropped only to ~50% of the initial value. Also, PSI activities declined in high light–treated VIPP1-RNAi strains but generally not as fast as PSII activities. The 7.5-h high light treatment resulted only in an ~10% drop of PSI activity in the control, but the loss of PSI activity in the VIPP1-RNAi strains ranged between ~40 and ~90% and correlated with the residual VIPP1 levels present in these strains (Figure 1E; see Supplemental Figure 4 online).
Protein gel blot analyses revealed that the D1, CP43, and OEE33 subunits of PSII were reduced by >75% after 3 h at high light (Figure 1F). Interestingly, light-harvesting complex II (LHCII) levels in VIPP1-RNAi cells grown under low light conditions appeared to be higher than in control cells, and it took 7 h at high light until LHCII levels were reduced by >50%. A reduction of PSI core subunits PsaA and PsaD in VIPP1-RNAi cells by more than half was observed only after 7 h at high light and therefore was not as fast as the loss of the PSII core subunits, which is in line with the spectroscopic data. Levels of CF1β and cytochrome f, subunits of the ATP synthase and cytochrome b6f complex, respectively, were barely reduced in high light–exposed VIPP1-RNAi strains. No decline in any of the investigated subunits was observed in high light–treated control cells.
VIPP1 and VIPP2 were expressed constitutively in control cells grown at low light intensities, and levels of both proteins increased strongly during exposure to high light (Figure 1F). While high light treatment led to an increase in levels of plastidic chaperones HSP70B and HSP90C and of the LHCSR3 protein in control cells, none of these proteins increased in high light–treated VIPP1-RNAi strains. As the genes coding for these three proteins are nuclear encoded, this finding suggests that VIPP1-RNAi strains might be defective in a retrograde high light signaling pathway. The existence of more than one high light signaling pathway is implied by the observation that VIPP1-RNAi strains are still capable of inducing VIPP2 expression in response to high light exposure.
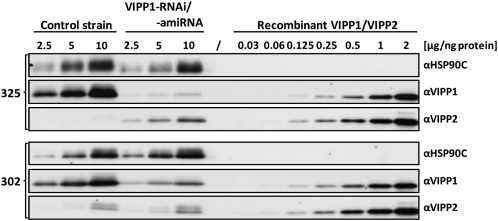
Interestingly, already under low light conditions, VIPP1-RNAi cells expressed VIPP2 at much higher levels than control cells, and they increased VIPP2 expression further during exposure to high light intensities (Figure 1F). Apparently, VIPP1-RNAi cells try to compensate for the lack of VIPP1 by expressing additional VIPP2, but VIPP2 cannot completely substitute VIPP1. This may have two reasons, which are not mutually exclusive: First, VIPP2 may be functionally redundant with VIPP1, but its expression levels may be too low to completely compensate for the VIPP loss in VIPP1-RNAi strains. Second, VIPP2 overexpression in VIPP1-RNAi strains may replenish the total VIPP pool, but VIPP2 function may be distinct from that of VIPP1 and, therefore, VIPP2 cannot substitute VIPP1. To distinguish between these possibilities, we needed to get an estimate of the relative cellular expression levels of VIPP1 and VIPP2. For this, we separated total cell proteins next to dilution series of purified, recombinant VIPP1 and VIPP2 and immunodetected VIPP1 and VIPP2 with specific antisera. Exposure times were chosen such that signals from recombinant VIPP1 and VIPP2 were about equal, thus allowing for a direct comparison of VIPP1 and VIPP2 signals from total cell proteins (Figure 2). This procedure allowed us to estimate that the total VIPP pool size in VIPP1-RNAi cells of the cw15-325 background was ~25% of that in control cells, with a VIPP1:VIPP2 ratio of ~1:4.
Figure 2.
Examination of VIPP1/2 Levels in VIPP1-RNAi and -amiRNA Strains by Immunoblotting.
Transformants generated with empty vectors (control strains) or VIPP1-RNAi and VIPP1-amiRNA constructs in the cw15-325 (#12) and cw15-302 (#18) strain backgrounds, respectively, were grown in TAP-NH4 medium. Total protein from the transformants and purified recombinant VIPP1 and VIPP2 at the indicated protein concentrations were separated on 7.5 to 15% SDS-polyacrylamide gels and analyzed by immunoblotting using antisera against VIPP1 and VIPP2. HSP90C served as loading control.
Although these data support the first scenario, we could not exclude the second one as the two VIPPs are expressed at strongly different ratios in VIPP1-RNAi cells compared with control cells. We therefore decided to repress VIPP1 in a strain background (cw15-302) that we knew to be unable to increase VIPP2 expression levels in response to VIPP1 repression. For this, we used an artificial microRNA (amiRNA) approach to also rule out off-target effects potentially caused by the inverted repeat construct (Molnar et al., 2009; see Supplemental Figure 5A online). As shown in Figure 2, VIPP2 in the cw15-302 strain background is expressed to higher levels than in the cw15-325 background and VIPP2 expression is not elevated in response to VIPP1 repression. However, VIPP1 in the cw15-302 background in several independent transformants could be repressed to only ~25% of wild-type VIPP1 levels such that the ratio of VIPP1:VIPP2 was ~2:1. Exposure of VIPP1-amiRNA strains to high light intensities again resulted in photobleaching (see Supplemental Figure 5B online). These results suggested that VIPP1 and VIPP2 are largely functionally redundant and that high light sensitivity is caused by a reduction in plastidic VIPP levels independent of its composition. Moreover, high light sensitivity apparently is not caused by off-target effects of the RNAi construct.
Because of the proposed link between VIPPs and chloroplast biogenesis, we investigated the effect of high light treatment on thylakoid membrane ultrastructure in VIPP1-depleted strains. To this end, we used transmission electron microscopy on control and VIPP1-amiRNA strains prior to and after a 3.5-h exposure to high light intensities. While the high light treatment hardly affected thylakoid structure in control cells, thylakoids in most cells of the VIPP1-amiRNA strains were extremely swollen (Figure 3).
In summary, VIPP1 is the major VIPP in C. reinhardtii. Apparently, the total VIPP pool cannot be constitutively repressed to below ~25% of wild-type levels. Whereas VIPP1-RNAi/amiRNA strains showed no obvious phenotypes under low light conditions, they turned out to be highly sensitive to high light, as manifested by chlorophyll bleaching, loss of photosystem activities, and thylakoid swelling. PSII was most sensitive in VIPP1-RNAi strains, closely followed by PSI, while ATP synthase and cytochrome b6f complex were only mildly affected, if at all. Finally, high light–induced accumulation of the HSP90C and HSP70B chaperones and of LHCSR3 normally observed in control cells was abolished in VIPP1-RNAi cells.
VIPP1-RNAi Cells Are More Severely Photoinhibited and Repair Photodamaged PSII More Slowly Than Control Cells
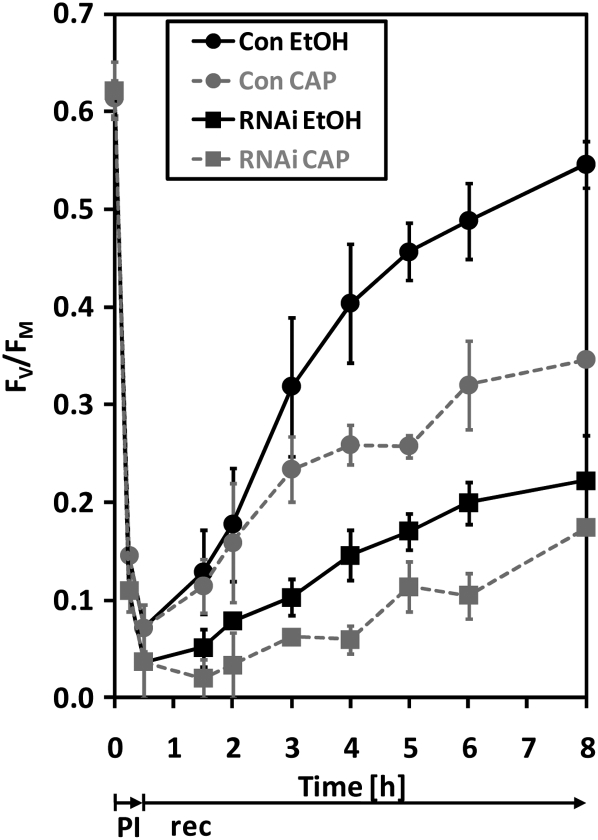
The observed sensitivity of VIPP1-RNAi/amiRNA strains to high light raises two questions: First, is it a secondary effect that occurs only after prolonged exposure to high light (e.g., as a consequence of thylakoid swelling)? Second, is the fast loss of PSII activity and core subunits during high light treatment caused by the inability of VIPP1-RNAi strains to repair photodamaged D1 protein? To answer these questions, we exposed control and VIPP1-RNAi strains to ~1800 μE m−2 s−1 for 30 min in the presence and absence of chloramphenicol, an inhibitor of organellar protein biosynthesis, and monitored PSII recovery from photoinhibition at low light intensities. As shown in Figure 4, loss of PSII maximum quantum efficiency was more severe in VIPP1-RNAi cells than in control cells and its recovery, both in the presence and absence of chloramphenicol, was faster in control than in VIPP1-RNAi strains. PSII recovery in the presence of chloramphenicol is attributable to reversibly photodamaged reaction centers, whereas the additional PSII recovery in the absence of chloramphenicol is derived from PSII centers from which irreversibly damaged D1 was exchanged by de novo synthesized D1 (Ohad et al., 1990; Schroda et al., 1999). In VIPP1-RNAi strains, we observed that additional PSII was recovered in cells carrying out organellar protein biosynthesis compared with cells lacking organellar protein biosynthesis. We conclude that, compared with the control, VIPP1-RNAi strains are also more sensitive to a short application of very high intensity light. VIPP1-RNAi strains are generally capable of repairing photodamaged D1, but the protein biosynthesis-dependent repair rate of PSII is slower in VIPP1-RNAi strains than in controls.
Figure 4.
VIPP1-RNAi Cells Are More Severely Photoinhibited at Very High Light and Replace Photodamaged D1 by de Novo–Synthesized D1 Slower Than Control Cells.
Cultures of control and VIPP1-RNAi strain #80 grown in TAP-NH4 medium were split into two parts. One part was supplemented with ethanol-dissolved chloramphenicol (CAP) to a final concentration of 100 μg/mL, while the other received the same volume of pure ethanol. The four cultures were exposed to ~1800 μE m−2 s−1 for 30 min (photoinhibition [PI]) and shifted back to ~30 μE m−2 s−1 (recovery [rec]). Maximum quantum efficiency of PSII during photoinhibition and recovery was measured with a PAM fluorometer as variable fluorescence (FV = FM − F0) normalized to FM. Shown is the average of two independent experiments; error bars represent se.
PSII repair requires the movement of photodamaged PSII from grana stacks to stroma-exposed thylakoid regions (Goral et al., 2010). Another process requiring extensive movement of protein complexes within the thylakoid membrane is state transitions. This process is triggered by an imbalance in excitation energy between the photosystems and leads, upon overexcitation of PSII, to the phosphorylation of LHCII by the STT7 kinase and the movement of LHCII to PSI (Depège et al., 2003; Bellafiore et al., 2005). The remigration of LHCII in case of PSI overexcitation requires the dephosphorylation of LHCII. State transitions are accompanied by thylakoid membrane rearrangements, and VIPP1 was suggested as a candidate protein that might mediate these rearrangements (Chuartzman et al., 2008). To test whether VIPP1-RNAi strains are impaired in state transitions, we followed state transitions in control and VIPP1-RNAi strains by pulse amplitude–modulated (PAM) fluorometry and 77K fluorescence emission spectroscopy. Both measurements indicated that VIPP1-RNAi strains were not impaired in state transitions (see Supplemental Figure 6 online), hence ruling out the possibility that the high light sensitivity of VIPP1-RNAi strains originated from a generally reduced capacity for lateral movement of thylakoid membrane complexes. However, we cannot rule out the possibility that membrane complex mobility is impaired in swollen thylakoids.
Thylakoid Swelling Enhances but Does Not Cause the Sensitivity of VIPP1-RNAi Strains to High Light
Swelling of thylakoids was observed in C. reinhardtii wild-type cells exposed to very high light intensities of 5000 μE m−2 s−1 (Topf et al., 1992) and in ATP synthase mutants exposed to 70 μE m−2 s−1 (Majeran et al., 2001). Swelling was shown to depend on the acidification of the thylakoid lumen (i.e., on the formation of a ΔμH+) (Majeran et al., 2001) and was suggested to depend on the presence of ammonium in the growth medium (Topf et al., 1992). The mechanism is that ammonium ions in the stroma become deprotonated to a certain extent and cross the thylakoid membrane as uncharged ammonia. In the acidified lumen, ammonia becomes reprotonated and accumulates as ammonium ions and may thereby generate an increased osmotic pressure within the lumen.
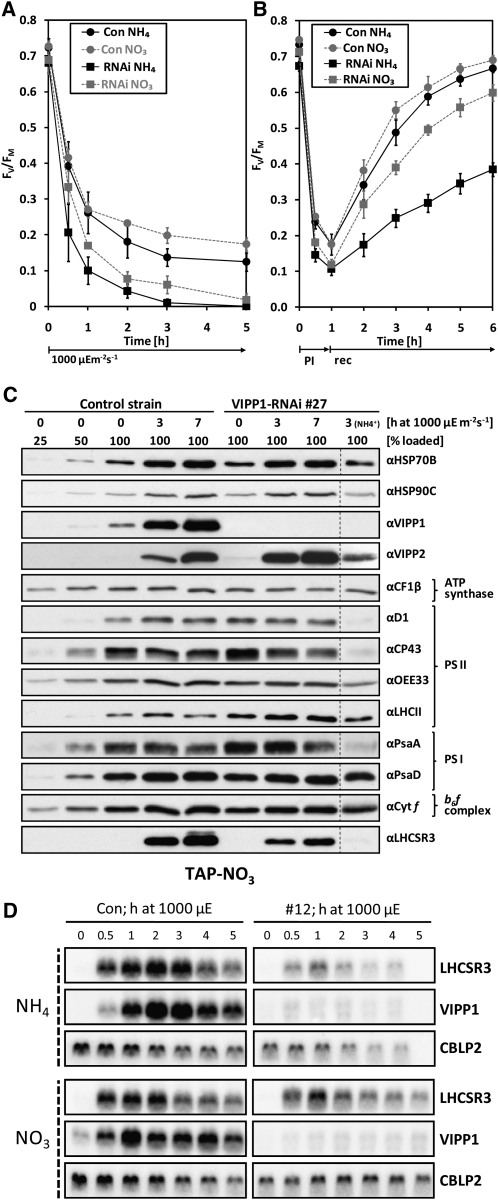
If the high light sensitivity of VIPP1-RNAi/amiRNA strains was caused by thylakoid swelling, which only occurs in the presence of ammonium, this phenotype should be abolished when mutant strains are grown on nitrate as nitrogen source. To test this prediction, we took advantage of the fact that the cw15-325 strain used as recipient for the VIPP1-RNAi constructs, unlike most C. reinhardtii laboratory strains, is able to use nitrate as nitrogen source. We exposed control and VIPP1-RNAi strains grown on nitrate to high light intensities (~1000 μE m−2 s−1) and observed that bleaching was retarded when compared with VIPP1-RNAi strains grown on ammonium (see Supplemental Figure 7 online). Also, the loss of PSII maximum quantum efficiency was slower when VIPP1-RNAi cells were grown on nitrate compared with ammonium but still faster and more severe than in control cells grown on either nitrogen source (Figure 5A). Similar effects were observed when cells were exposed to very high light intensities (~1800 μE m−2 s−1) for 60 min and allowed to recover at low light: The extent of PSII photodamage was highest and recovery slowest in VIPP1-RNAi cells grown on ammonium, followed by VIPP1-RNAi cells grown on nitrate, control cells grown on ammonium, and control cells grown on nitrate (Figure 5B). Interestingly, PSII maximum quantum efficiency in control cells was also less severely affected by high light when cells were grown on nitrate as a nitrogen source compared with ammonium.
Figure 5.
High Light Sensitivity Is Alleviated but Not Abolished in VIPP1-RNAi Strains Grown on Nitrate.
(A) PSII in VIPP1-RNAi strains is less high light sensitive in cells grown on nitrate compared with ammonium. Control (Con) and VIPP1-RNAi strains #12 and #27 were grown in TAP-NO3 or TAP-NH4 medium. Cells were exposed to ~1000 μE m−2 s−1 and maximum quantum efficiency of PSII was measured with a PAM fluorometer as described in Figure 4. Shown is the average of two independent experiments. Error bars represent se.
(B) PSII in VIPP1-RNAi strains is less sensitive to photoinhibition in cells grown on nitrate compared with ammonium. Control and VIPP1-RNAi strains #5, #27, and #41 were grown in TAP-NO3 or TAP-NH4 medium. Cells were exposed to ~1800 μE m−2 s−1 for 1 h (photoinhibition [PI]) and shifted back to ~30 μE m−2 s−1 (recovery [rec]). Maximum quantum efficiency of PSII was measured with a PAM fluorometer as described in Figure 4. Shown is the average of four independent experiments. Error bars represent se.
(C) Subunits of PSII and PSI are less prone to degradation in high light–exposed VIPP1-RNAi strains grown on nitrate. Whole-cell proteins from nitrate-grown control and VIPP1-RNAi strain #27 exposed to ~1000 μE m−2 s−1 for 7 h were separated on 14% SDS-polyacrylamide gels and analyzed by immunoblotting. For comparison, whole-cell proteins from VIPP1-RNAi strain #27 grown on ammonium and exposed to high light for 3 h was loaded next to the other samples.
(D) RNA gel blot analysis of high light–exposed control and VIPP1-RNAi strains. Control and VIPP1-RNAi strain #12 were grown in TAP-NO3 or TAP-NH4 medium. Cells were exposed to ~1000 μE m−2 s−1 for 5 h, and RNA was extracted from samples taken at the indicated time points and subjected to RNA gel blot analysis. CBLP2 served as loading control.
To test whether the reduced high light sensitivity of VIPP1-RNAi strains grown on nitrate correlated with reduced thylakoid swelling, we took 1015 electron microscopy images from control and VIPP1-RNAi cells, grown on nitrate or ammonium, prior to and after a 3- to 7-h exposure to high light intensities (~1000 μE m−2 s−1). The observed thylakoid phenotypes were sorted into the categories “ordered,” “disordered,” and “swollen” (examples for these categories are given in Supplemental Figure 8 online). As summarized in Table 1, cells with swollen thylakoids were only observed in VIPP1-RNAi strains grown on ammonium. Here, 8% of the cells displayed swollen thylakoids already at low light intensities (30 μE m−2 s−1) and the fraction of cells with swollen thylakoids increased to 68% after the 3 h of high light exposure. Even after 7 h of high light exposure, we observed no thylakoid swelling in VIPP1-RNAi cells grown on nitrate. However, at that time, 36% of the mutant cells were in the process of lysis, whereas this was the case for only 1 to 2% of control cells (see Supplemental Figure 9 online). These results indicate that thylakoid swelling is not causing lysis of high light–exposed VIPP1-RNAi cells grown on nitrate.
Table 1.
Categorization of Thylakoid Structure
| Condition/Strain | TAP-NH4 | TAP-NO3 | ||||
| Ordered | Disordered | Swollen | Ordered | Disordered | Swollen | |
| High light | ||||||
| Con (LL) | 100 | 0 | 0 | 99 | 1 | 0 |
| Con (180′ HL) | 100 | 0 | 0 | 98 | 2 | 0 |
| RNAi #27 (LL) | 92 | 0 | 8 | 92 | 8 | 0 |
| RNAi #27 (180′ HL) | 27 | 5 | 68 | 100 | 0 | 0 |
| Photoinhibition | ||||||
| Con (60′ PI) | 87 | 0 | 11 | 98 | 2 | 0 |
| Con (60′ rec) | 99 | 0 | 1 | 100 | 0 | 0 |
| RNAi #27 (60′ PI) | 46 | 16 | 38 | 92 | 8 | 0 |
| RNAi #27 (60′ rec) | 74 | 3 | 23 | 100 | 0 | 0 |
| Heat shock | ||||||
| Con (25°C) | 96 | 0 | 4 | n.d. | n.d. | n.d. |
| Con (40°C) | 100 | 0 | 0 | n.d. | n.d. | n.d. |
| RNAi #32 (25°C) | 90 | 1 | 9 | n.d. | n.d. | n.d. |
| RNAi #32 (40°C) | 70 | 9 | 21 | n.d. | n.d. | n.d. |
Transmission electron microscopy images were taken on cells from control (Con) and VIPP1-RNAi strains #27 and #32 grown in TAP-NH4 or TAP-NO3 medium under the following conditions: LL, low light intensities of ~30 μE m−2 s−1; 180′ HL, 180 min at high light intensities of ~1000 μE m−2 s−1; 60′ PI, 60 min at photoinhibitory light of ~1800 μE m−2 s−1; 60′ rec, 60 min at ~30 μE m−2 s−1 for recovery from photoinhibition; 25°C, 25°C and ~30 μE m−2 s−1; 40°C, 40°C for 1 h at ~5 μE m−2 s−1; n.d., not determined. On average, 101 images per strain and condition were analyzed and sorted into three categories of thylakoid structure for which examples are shown in Supplemental Figure 8 online. Values are in percentages.
A similar picture emerged when we took another 800 electron microscopy images to monitor thylakoid swelling in control and VIPP1-RNAi cells that were exposed to very high light intensities (~1800 μE m−2 s−1) for 60 min and allowed to recover at low light. Here, 38% of ammonium-grown VIPP1-RNAi cells contained swollen thylakoids after 60 min at very high light, and this fraction declined to 23% after 60 min of recovery at low light intensities (Table 1). Interestingly, swollen thylakoids were also observed in 11% of ammonium-grown control cells exposed to very high light (thus corroborating the observations from Topf et al., 1992) and this fraction declined to 1% after 60 min of recovery. No thylakoid swelling was observed in nitrate-grown control and VIPP1-RNAi cells exposed to very high light intensities (Table 1).
Also, at the protein level, we found that exposure to high light (~1000 μE m−2 s−1) had less severe effects on VIPP1-RNAi cells when they were grown on nitrate as the nitrogen source compared with cells grown on ammonium (cf. Figures 1F and 5C). After 7 h of high light exposure, we found a clear decline only in levels of CP43 and perhaps PsaA. While chaperones HSP70B and HSP90C appeared to be normally induced by high light, induction of the LHCSR3 protein still was less pronounced in VIPP1-RNAi cells compared with control cells. Likewise, the strong decline of PSII subunits and, to a lesser extent, of PSI subunits observed after exposure of ammonium-grown VIPP1-RNAi cells to very high light intensities (~1800 μE m−2 s−1) was virtually absent upon exposure of nitrate-grown VIPP1-RNAi cells to very high light (cf. Supplemental Figures 10A and 10B online). However, similar to VIPP1-RNAi cells continuously exposed to ~1000 μE m−2 s−1, VIPP1-RNAi cells exposed for 60 min to ~1800 μE m−2 s−1 were still deficient in the full induction of LHCSR3 protein expression, despite growth on nitrate. And in contrast with nitrate-grown VIPP1-RNAi cells exposed to ~1000 μE m−2 s−1, cells exposed to ~1800 μE m−2 s−1 still appeared to be impaired in the full induction of the HSP70B and HSP90C chaperones and still experienced reduction of initially elevated LHCII levels (see Supplemental Figure 10B online).
Taken together, our observations show that thylakoid swelling is indeed only observed in high light–treated cells grown on ammonium as nitrogen source. Moreover, thylakoid swelling enhances, but does not cause, the sensitivity of VIPP1-RNAi strains to high light.
VIPP1-RNAi Strains Are Impaired in the Perception/Transmission of the High Light State
We wondered whether the reduced accumulation of the LHCSR3 protein in high light–treated VIPP1-RNAi strains was due to a defect in the perception/transmission of the high light conditions or due to a defect in the conversion of the LHCSR3 transcript into a stable protein, for example, by its impaired insertion into the thylakoid membrane. In the former case, we expected a reduced induction of the LHCSR3 transcript in response to high light, and in the latter case, we expected no effect. To distinguish between these possibilities, we exposed cells from control and VIPP1-RNAi strains to high light (~1000 μE m−2 s−1) and analyzed LHCSR3 transcripts by RNA gel blot analysis. As shown in Figure 5D, induction of the LHCSR3 gene was strongly reduced in VIPP1-RNAi strains grown on ammonium as nitrogen source, while differences between control and VIPP1-RNAi strains in the high light–induced expression of LHCSR3 were less pronounced in cells grown on nitrate. Similarly, the reduction in LHCSR3 transcript accumulation observed in VIPP1-RNAi strains compared with a control strain after exposure to very high light (~1800 μE m−2 s−1) was less pronounced in cells grown on nitrate instead of ammonium (see Supplemental Figure 10C online). In control cells grown on either nitrogen source, VIPP1 transcripts strongly accumulated in response to high light exposure, whereas they were almost completely repressed in VIPP1-RNAi cells.
These data suggest that the reduced accumulation of LHCSR3 protein observed in high light–exposed VIPP1-RNAi strains compared with the control strain is mainly based on the reduced expression of the LHCSR3 gene. This in turn suggests that VIPP1-RNAi strains are defective in the retrograde signaling of the high light state, which is particularly true for cells grown on ammonium but to a lesser extent also for cells grown on nitrate.
Specific Photosynthesis Parameters Are Affected in VIPP1-RNAi Strains
Although we have shown that thylakoid swelling occurs only in high light–exposed cells grown on ammonium and not in cells grown on nitrate as nitrogen source, it is not clear why swelling is so much more severe in VIPP1-RNAi/amiRNA cells compared with control cells. Thylakoid swelling in high light–treated mutant cells may result from their ability to generate a higher proton-motive force (pmf) than control cells, caused, for example, by a reduced ATP synthase activity (Majeran et al., 2001). Alternatively, thylakoid swelling may have resulted from the reduced ability of VIPP1-RNAi/amiRNA strains to counteract the osmotic pressure generated in the thylakoids upon high light exposure in the presence of ammonium.
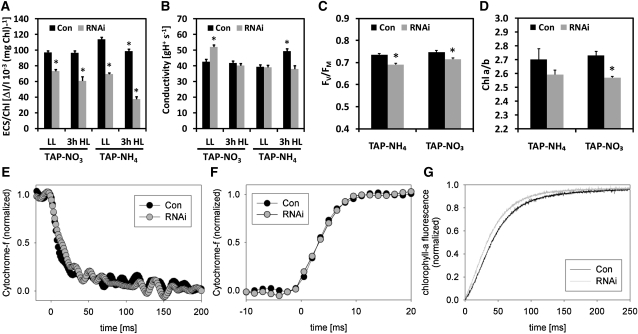
To test the first hypothesis, we used electrochromic shift measurements to compare control and VIPP1-RNAi strains in respect of the pmf generated at saturating light intensities and with regard to thylakoid membrane conductivity (corresponding to the ATP synthase activity). These measurements were performed with cells grown under mixotrophic conditions using ammonium and nitrate as nitrogen sources prior to and after a 3-h exposure to high light intensities of ~1000 μE m−2 s−1. As shown in Figures 6A and 6B, maximum thylakoid membrane energization in saturating light was significantly lower in VIPP1-RNAi cells than in control cells under all conditions tested, while ATP synthase activity was never lower than that of the untreated control. Hence, thylakoid swelling in high light–exposed VIPP1-RNAi strains is unlikely to be caused by an increased pmf.
Figure 6.
VIPP1-RNAi Strains Are Affected in Some Photosynthesis Parameters.
(A) Thylakoid membrane energization. Control (Con) and VIPP1-RNAi strains #2-7, #2-19, #3-30, #4-19, and #4-49 were grown in TAP-NO3 or TAP-NH4 medium and kept in low light (LL) of ~30 μE m−2 s−1 or exposed to high light (HL) of ~1000 μE m−2 s−1 for 3 h. Maximum thylakoid membrane energization was then determined by measuring the ECS in saturating light. Shown is the average of eight and six measurements on the control strain, and of 12 and 10 measurements on VIPP1-RNAi strains grown on NO3 and NH4, respectively. Error bars represent se. Chl, chlorophyll.
(B) ATP synthase activity. The activity of the ATP synthase was inferred from the decay kinetic of the ECS signal during a short dark interval using the same cells as described in (A).
(C) Maximum quantum efficiency of PSII. FV/FM was measured after 5 min of far-red illumination, followed by 10 min of dark adaptation of the cell suspension using control and VIPP1-RNAi strains #4, #9, #40, #53, #72, #111, and #129 grown in low light. Shown is the average of four and two measurements on the control strain, and of 13 and 10 measurements on VIPP1-RNAi strains. Error bars represent se.
(D) Chlorophyll a/b ratios. Chlorophyll was extracted with 80% acetone from the same cells as described in (C), and the chlorophyll concentration was determined spectrophotometrically. Asterisks indicate the significance of the difference to untreated controls grown on nitrate and ammonium as nitrogen source, respectively (t test, P value ≤ 0.05).
(E) Cytochrome f reduction kinetics. Control and VIPP1-RNAi strains #4, #9, and #72 were grown in TAP-NH4 medium. Cytochrome f reduction was initiated by switching off saturating red light at time point zero. The fully reduced state of cytochrome f in the dark was normalized to zero and the fully oxidized state to one. Kinetics were recorded four times for each strain. Averages for the control and all three VIPP1-RNAi strains are shown.
(F) Cytochrome f oxidation kinetics. Control and VIPP1-RNAi strains #27, #29, and #30 were grown in TAP-NH4 medium. Cytochrome f oxidation was initiated by switching on a saturating red light pulse at time point zero. Six replicates of the control strain and two each for the VIPP1-RNAi strains were recorded and averages plotted. Again, the fully reduced state of cytochrome f in the dark was normalized to zero and the fully oxidized state to one.
(G) Fluorescence induction kinetics. Control and VIPP1-RNAi strains #27, #29, und #30 were grown in TAP-NH4 medium. Dark-adapted cells trapped in state 1 were supplemented with DCMU, and fluorescence was induced by switching on the light. Six replicates of the control strain and two each for the VIPP1-RNAi strains were recorded and averages plotted. The F0 values were normalized to zero and the FM values to one.
To identify defects in VIPP1-RNAi strains that might cause their increased light sensitivity and the apparently reduced ability of their thylakoids to withstand an increased osmotic pressure, we performed more biophysical measurements. We observed that PSII maximum quantum efficiency was mildly but significantly lower in VIPP1-RNAi compared with control cells (Figure 6C), while cytochrome f reduction and oxidation kinetics were unaltered, indicating similar electron transport capacities in control and VIPP1-RNAi strains (Figures 6E and 6F). Moreover, the chlorophyll a/b ratio was lower in VIPP1-RNAi strains than in controls (Figure 6D), which may result from the higher levels of chlorophyll b–rich LHCII in VIPP1-RNAi strains (Figures 1F and 5C; see Supplemental Figures 10A and 10B online). A larger antenna cross section in VIPP1-RNAi strains was also supported by the faster fluorescence induction kinetics in mutant versus control cells (Figure 6G). The reduced sigmoidicity of the induction curve in VIPP1-RNAi strains also indicated that cooperativity between PSII centers was lower in VIPP1-RNAi cells compared with control cells. Thermoluminescence measurements revealed a reduced maximum temperature of the Q-band in VIPP1-RNAi cells compared with control cells, a phenotype observed in cells grown on either ammonium or nitrate as nitrogen source (Table 2). Following a saturating single turnover flash at low temperature, a thermoluminescence band, named Q-band, is detected upon heating in the region of 8 to 12°C. This luminescence arises from the recombination of the charge pairs S2QA−with S2 being an oxidation state of the Mn cluster and QA− the semireduced primary quinone acceptor in PSII (Rutherford et al., 1982). The downshift of the maximum temperature of the Q-band in VIPP1-RNAi cells reflects a lowering of the midpoint potential of the redox couple QA/QA− (Krieger-Liszkay and Rutherford, 1998). A lower midpoint potential of QA was shown to increase the yield of 1O2 generation in the light (Fufezan et al., 2007) and therefore might represent the mechanism underlying high light sensitivity of VIPP1-RNAi cells.
Table 2.
Temperature of the Q-Band
| Strain | Tmax (°C) | |
| TAP-NO3− | TAP-NH4+ | |
| Wild type | 11.5 ± 0.5 | 11.1 ± 0.5 |
| VIPP1-RNAi | 7.4 ± 1.2 | 9.0 ± 1.0 |
Cells were grown in TAP medium containing either ammonium or nitrate as nitrogen source at low light intensities of ~30 μE m−2 s−1. Values represent averages ± sd from thermoluminescence measurements on two independent control and four independent VIPP1-RNAi lines (#1-29, #2-29, #4-49, and #3-30).
We conclude that thylakoid swelling in ammonium-grown, high light–exposed VIPP1-RNAi cells is not caused by a higher pmf but rather by a disturbance of thylakoid membrane organization. The defect causing this disturbance also might be responsible for reduced PSII maximum quantum efficiency, lowered midpoint potential of the QA/QA− redox couple, reduced PSII cooperativity, and increased antenna cross section in VIPP1-RNAi strains.
PSII in VIPP1-RNAi Strains Is Sensitive to Heat Shock
If VIPP1-RNAi strains indeed have a defect in thylakoid membrane organization, we can expect phenotypes to also be induced by treatments that do not involve high light intensities but similarly represent a threat to thylakoid membrane integrity. Heat shock at 40 to 42°C, combined with low light intensities that are harmless for PSII at 25°C, was demonstrated previously to result in loss of PSII activity in C. reinhardtii (Schuster et al., 1988). This finding was explained by cross-linking of the D1 protein with other proteins, presumably induced by the overproduction of radicals by heat-stressed, illuminated PSII.
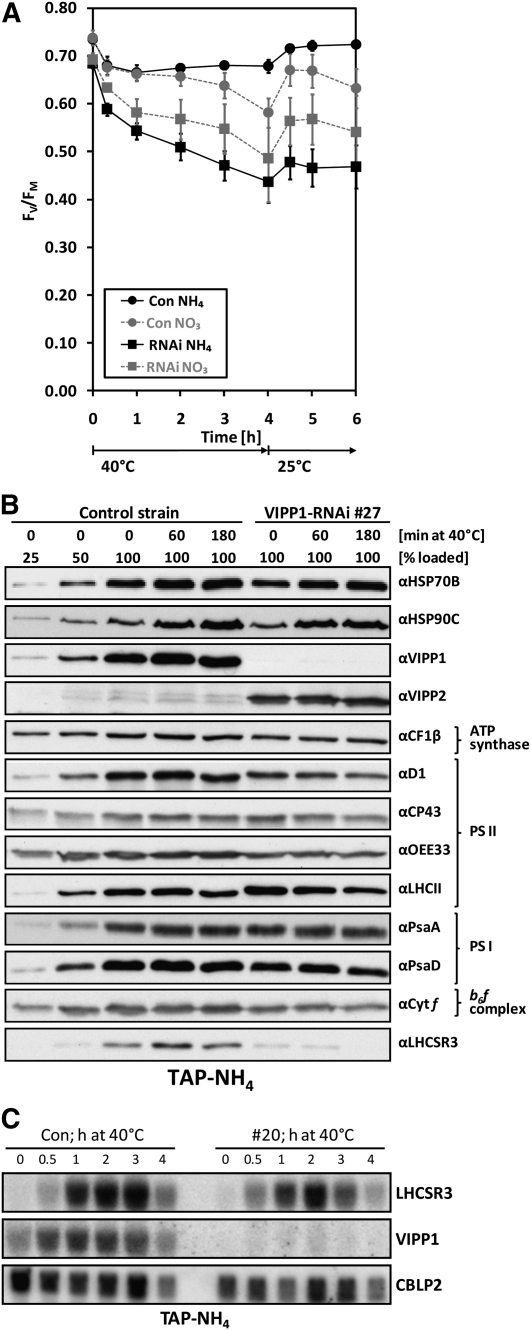
To test whether PSII was more sensitive to heat stress in VIPP1-RNAi cells compared with control cells, we shifted mutant and control cells from 25 to 40°C at low light intensities (~5 μE m−2 s−1) for 4 h and back to 25°C for 2 h for recovery and monitored PSII maximum quantum efficiency. This experiment was done with cells grown mixotrophically on ammonium or nitrate as nitrogen sources (Figure 7A). In control cells grown on ammonium, variable fluorescence/maximum fluorescence (FV/FM) declined already within the first 20 min at 40°C from 0.73 to 0.68 but recovered fast and almost completely after shifting cells back to 25°C. The loss of PSII maximum quantum efficiency in nitrate-grown control cells followed kinetics similar to those observed for ammonium-grown control cells within the first hour at 40°C, but, surprisingly, the FV/FM value in nitrate-grown control cells declined further to 0.58 after 4 h at 40°C and did not fully recover at 25°C. In ammonium-grown VIPP1-RNAi cells heat-shocked for 4 h, the FV/FM value dropped dramatically from 0.68 to 0.44 and at 25°C recovered only to 0.47. This effect was less drastic in nitrate-grown VIPP1-RNAi cells, in which the FV/FM value declined from 0.69 to 0.49 after 4 h at 40°C and recovered maximally to 0.57 at 25°C.
Figure 7.
PSII of VIPP1-RNAi Strains Is Highly Susceptible to Heat Stress.
(A) PSII maximum quantum efficiency of control and VIPP1-RNAi strains exposed to heat stress. Control and VIPP1-RNAi strains #5, #20, #27, and #41 were grown in TAP-NO3 or TAP-NH4 medium, and cells were exposed to 40°C at ~5 μE m−2 s−1. FV/FM was measured over time with a PAM fluorometer as described in Figure 4. Shown is the average of five and seven independent experiments for control and VIPP1-RNAi strains, respectively. Error bars represent se.
(B) PSII subunits are only mildly affected by heat stress. Whole-cell proteins were extracted from control and VIPP1-RNAi strain #27 grown and heat-stressed as described in (A). Whole-cell proteins were separated on 14% SDS-polyacrylamide gels and analyzed by immunoblotting.
(C) RNA gel blot analysis of heat-stressed control and VIPP1-RNAi strains. Control and VIPP1-RNAi strain #20 were grown and heat stressed as described in (A). RNA was extracted from samples taken at the indicated time points and subjected to RNA gel blot analysis. CBLP2 served as loading control.
To pick up changes induced by heat shock at the ultrastructural level, we took 400 electron microscopy images of ammonium-grown control and VIPP1-RNAi strains prior to and after a 60-min heat shock at 40°C. Interestingly, the fraction of VIPP1-RNAi cells containing swollen thylakoids increased from 9% under nonstress conditions to 21% after heat shock (Table 1). Heat stress hardly affected levels of thylakoid membrane complex subunits, no matter on which nitrogen source cells were grown (Figure 7B; see Supplemental Figure 11A online). While in control and VIPP1-RNAi strains levels of the VIPP2 protein were strongly induced by high light, they did not further increase during heat stress (cf. Figures 1F and 7B). Also, in contrast with the high light treatments, the HSP70B and HSP90C chaperones appeared to be normally induced by heat stress in VIPP1-RNAi strains grown on either nitrogen source (Figure 7B; see Supplemental Figure 11A online). In control cells grown on ammonium or nitrate, heat stress led to a slight and transient increase in levels of the LHCSR3 protein. By contrast, we observed no increase in LHCSR3 levels in VIPP1-RNAi strains after 60 min at 40°C but a strong decline of LHCSR3 after 180 min of heat stress. Also, the induction of the LHCSR3 gene by heat shock was reduced in VIPP1-RNAi strains (Figure 7C; see Supplemental Figure 11B online).
In summary, the dramatic effect of heat stress combined with very low light intensities on PSII maximum quantum efficiency in VIPP1-RNAi strains further supports the notion that VIPP1 depletion affects the structural organization of thylakoid membrane protein complexes. Again, this defect appears to have a secondary effect on retrograde signaling triggering the induction of the LHCSR3 gene by heat stress.
Quantitative Proteomics Reveals Reduced Levels of Thylakoid Membrane Core Complexes but Increased LHCII Levels in VIPP1-RNAi Cells
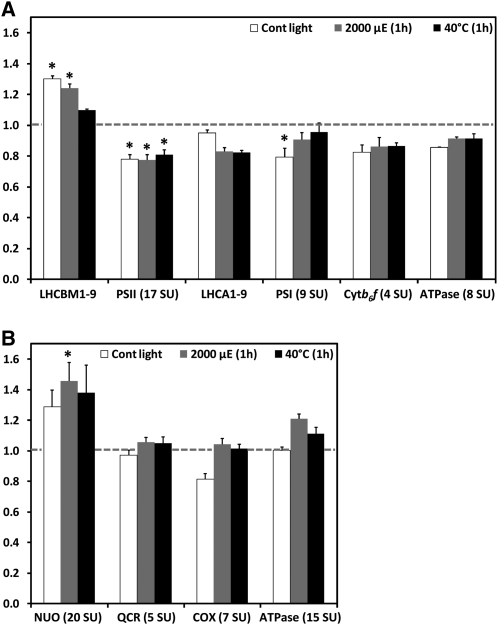
The conclusion that thylakoid membranes in VIPP1-RNAi/amiRNA strains must harbor structural defects prompted us to get a more quantitative picture of alterations in the abundance of thylakoid membrane protein complexes. The method of choice for this is shotgun proteomics using 15N metabolic labeling, by which a much higher quantification accuracy can be achieved than with quantitative immunoblotting (Mühlhaus et al., 2011). Using this technique, we compared the accumulation of the main protein complexes of the thylakoid membranes in control and VIPP1-RNAi strains that were grown on 15NO3 and 14NO3 as nitrogen sources, respectively (cells were grown on nitrate to avoid the pleiotropic effects caused by thylakoid swelling in cells grown on ammonium). Cells were kept under nonstress conditions or were exposed to 1-h high light (2000 μE m−1 s−1) and heat stress (40°C) treatments. For comparison, we also quantified the relative abundances of protein complexes involved in the mitochondrial respiratory chain. For each of the measured complexes, average values determined for multiple subunits are shown (see Supplemental Data Set 1 online for details). As shown in Figure 8A, thylakoid membranes from unstressed VIPP1-RNAi cells contained 30% more LHCII, but ~20% less PSII, PSI, and cytochrome b6f complex, and 14% less ATP synthase than control cells, while levels of LHCI were unchanged. High light and heat stress led to a 5 to 15% decline in levels of LHCs and to less reduced levels of PSI (5 to 9%) and ATP synthase (8%) in VIPP1-RNAi cells compared with stressed control cells, while levels of PSII and cytochrome b6f complex remained unchanged. Interestingly, levels of the mitochondrial NADH:ubiquinone oxidoreductase complex were 30% higher and those of the cytochrome oxidase 19% lower in VIPP1-RNAi cells compared with control cells, while levels of the ubiquinone-cytochrome c reductase complex and the ATP synthase were unchanged (Figure 8B). High light and heat stress treatments led to increases in levels of all respiratory chain complexes in VIPP1-RNAi cells relative to control cells. Hence, it appears possible that VIPP1-RNAi cells attempt to compensate for the slightly reduced capacity of their thylakoid membrane core complexes by higher levels of the NADH:ubiquinone oxidoreductase complex.
Figure 8.
Levels of Thylakoid Membrane Core Complexes Are Slightly Lower in VIPP1-RNAi Cells Than in Control Cells.
(A) Ratios of thylakoid membrane core complexes in VIPP1-RNAi cells relative to control cells. A control strain and VIPP1-RNAi strain #111 were metabolically labeled using 15NO3 and 14NO3, respectively, as nitrogen source and maintained at ~30 μE m−2 s−1 and 25°C for 1 h (Cont light), exposed to photoinhibitory light of ~2000 μE m−2 s−1 for 1 h at 25°C, or heat-shocked at 40°C for 1 h at ~5 μE m−2 s−1. After mixing control and VIPP1-RNAi cells from each treatment at a 1:1 ratio, proteins in membrane-enriched fractions were separated by SDS-PAGE and digested tryptically in gel. Peptides were eluted, desalted, and analyzed by liquid chromatography-MS/MS. Peptide identification and quantification was performed as described previously (Mühlhaus et al., 2011). Quantification values for single core complex subunits (SU) were computed from quantified peptides, and the average ratio of light (VIPP1-RNAi) to heavy (control) subunits was calculated for the different core complexes. Error bars represent se, and asterisks indicate the significance of the difference of the ratio from one (assuming equal variance; t test, P value ≤ 0.05).
(B) Ratios of respiratory chain core complexes in VIPP1-RNAi relative to control cells. Ratios of respiratory chain core complexes were determined as described in (A).
VIPP1-RNAi/amiRNA Strains Harbor Aberrant Structures at the Origin of Thylakoid Membranes
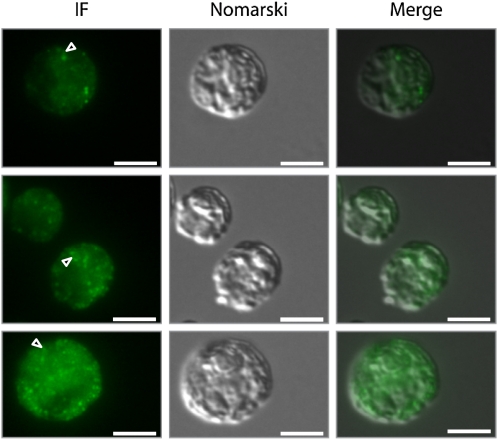
Finally, we set out to gather information on the localization of VIPP1 in C. reinhardtii by immunofluorescence microscopy using an affinity-purified antibody against VIPP1. As shown in Figure 9, VIPP1 was detected as diffuse material throughout the chloroplast but also in distinct dot-like structures that sometimes also appeared to extend into rods. This localization pattern is in agreement with our previous finding that C. reinhardtii VIPP1 may form rod-like structures, which by the help of molecular chaperones are kept in equilibrium with ring-like structures and smaller assembly states (Liu et al., 2007). We reasoned that rod-like structures potentially formed by VIPP1 might be visible in electron micrographs and therefore revisited the electron micrographs taken from control and VIPP1-RNAi/amiRNA strains. Unfortunately, we were unable to distinguish potential VIPP1 rods or rings from thylakoid membranes in control cells. However, we observed aberrant structures in VIPP1-RNAi/amiRNA cells that occurred particularly in regions from which multiple thylakoid membrane layers emerge and which may well correspond to the dot-like structures observed by immunofluorescence microscopy (Figure 10A). The aberrant structures were often found around the pyrenoid but were not restricted to that area. They were very rarely observed in control cells and occurred abundantly and consistently in VIPP1-RNAi and amiRNA cells (cw15-325 and cw15-302 backgrounds, respectively) grown under low light or high light on ammonium or nitrate (Table 3, Figure 10).
Figure 9.
Immunofluorescence Microscopy Detects VIPP1 in Distinct Punctae and as Diffuse Material in the Chloroplast.
Control cells from the cw15-325 background were grown at ~30 μE m−2 s−1 in TAP-NH4 medium and fixed and processed for immunofluorescence (IF) microscopy as described in Methods. The signal recognized by the affinity-purified anti-VIPP1 antibody is shown in green. Triangles indicate potential rod-like extensions. Bars = 5 μm.
Table 3.
Quantification of Aberrant PLB-Like Structures in VIPP1-RNAi/amiRNA Cells
| Strain | Growth Conditions | ||
| cw15-325 | LL, NH4 | 3.5 h HL, NH4 | LL, NO3 |
| Control | 0 | n.d. | 1 |
| VIPP1-RNAi #27 | 44 | n.d. | 64 |
| VIPP1-RNAi #41 | 63 | n.d. | 89 |
| cw15-302 | |||
| Control | 2 | 4 | n.d. |
| VIPP1-amiRNA #14 | 48 | 69 | n.d. |
| VIPP1-amiRNA #18 | 63 | 50 | n.d. |
| VIPP1-amiRNA #20 | 51 | 48 | n.d. |
Cells were grown in TAP medium containing either ammonium or nitrate as nitrogen source at low light (LL) intensities of ~30 μE m−2 s−1 or at high light intensities (HL) of ~1000 μE m−2 s−1. Fifty electron micrographs of each line were analyzed. n.d., not determined.
In summary, we could localize VIPP1 to diffuse material and to distinct spots within the chloroplast. As judged from the distribution of these spots, they may correspond to regions from which multiple thylakoid layers emerge and which are aberrant in VIPP1-RNAi/amiRNA cells.
DISCUSSION
Here, we present a thorough characterization of phenotypes arising in C. reinhardtii as a consequence of VIPP1 depletion mediated by inverted repeat and amiRNA constructs. VIPP1 expression was repressed in two different strain backgrounds: In strain cw15-325, VIPP1 levels were reduced to below 5% of wild-type levels, which, however, resulted in the upregulation of VIPP2 to ~20% of total VIPP levels (Figure 2). In strain cw15-302, VIPP2 was not upregulated in response to VIPP1 repression, but VIPP1 only could be reduced to ~25% of wild-type levels. Despite the different contributions of VIPP1 and VIPP2 to the residual VIPP pool in the two strain backgrounds, phenotypes were the same (Figures 1, 3, and 10; see Supplemental Figure 5B online). This allows us to conclude that VIPP1 and VIPP2 largely are functionally redundant and that the total VIPP pool in C. reinhardtii cannot be constitutively repressed to below ~25% of wild-type levels, similar to the situations in Arabidopsis and cyanobacteria (Kroll et al., 2001; Westphal et al., 2001; Fuhrmann et al., 2009a). VIPP1 obviously is the major VIPP species in C. reinhardtii, a conclusion that is supported by the finding that repression of VIPP2 in strain cw15-325 did not result in high light sensitivity or other obvious phenotypes (M. Rütgers and M. Schroda, unpublished results). Very similar phenotypes obtained with VIPP1-RNAi and -amiRNA constructs also widely rule out off-target effects in the RNAi lines (Ossowski et al., 2008).
Aberrant Structures at the Origin of Multiple Thylakoid Membrane Layers in VIPP1-RNAi/amiRNA Cells Suggest Deficits in Thylakoid Biogenesis
The most striking observation we consistently made in C. reinhardtii VIPP1-RNAi/amiRNA cells under all growth conditions tested is the occurrence of aberrant structures in regions from which bundles of thylakoids emerge and often project into different directions of the chloroplast (Figure 10, Table 3). Some of these aberrant structures were reminiscent of prolamellar body (PLB)–like structures observed in dark-grown C. reinhardtii y-1 (yellow-in-the-dark) mutants (Hoober and Blobel, 1969; Friedberg et al., 1971). y-1 mutants are defective in nuclear genes that are involved in the posttranscriptional expression or accumulation of the CHLL subunit of the light-independent protochlorophyllide reductase, leading to a block in chlorophyll synthesis in the dark (Cahoon and Timko, 2000). Hence, PLB-like structures are likely made of thylakoid membrane precursors in which the biogenesis of photosynthetic protein complexes is arrested or at least retarded. The aberrant structures in VIPP1-RNAi/amiRNA cells were frequently (but not exclusively) located close to the pyrenoid in regions previously suggested to house translation zones (T-zones; Figure 10) (Uniacke and Zerges, 2007). T-zones were postulated to represent areas where de novo PSII biogenesis and the regulated transport of newly assembled PSII complexes to thylakoid membranes occur. If indeed the aberrant, PLB-like structures in VIPP1-RNAi/amiRNA cells correspond to T-zones, they might be caused by a problem during photosystem biogenesis/assembly.
In cyanobacteria, groups of thylakoids converge at peripheral cytoplasmic points without any apparent connection to the plasma membrane (Kunkel, 1982). Thylakoids at these points are attached to so-called thylakoid centers, which are 30 to 50 nm wide tubular structures of up to 1 μm length that are composed of subunits generating a 14-fold rotational symmetry (Kunkel, 1982; van de Meene et al., 2006). Thylakoid centers have been postulated to be linked to a membrane fraction with a density intermediate to that of thylakoid and plasma membranes (Hinterstoisser et al., 1993). It has also been postulated that these are the sites at which protein/pigment complexes are assembled and incorporated into photosynthetic membranes and that potentially are related to T-zones in C. reinhardtii (Nickelsen et al., 2011). A coincidence too striking to be overlooked is that recombinant VIPP1 from Arabidopsis, C. reinhardtii, and Synechocystis forms ring-like structures with an outer diameter of 25 to 37 nm, a 12- to 17-fold rotational symmetry, and the capacity to assemble into rod-like structures of up to 1.4 μm (Aseeva et al., 2004; Liu et al., 2007; Fuhrmann et al., 2009b). Hence, it is tempting to speculate that thylakoid centers in fact are VIPP1 rods, which again may correspond to microtubule-like structures reported in plastids of diverse algal and plant species in various types of tissues (Liu et al., 2007, and references therein). Thylakoid centers may have moved from a peripheral position in cyanobacteria to more central ones in chloroplasts and still serve as sites of thylakoid biogenesis. This speculation is supported by the localization of VIPP1 by immunofluorescence to distinct spots within the chloroplast that sometimes appear to extend into rod-like structures (Figure 9). Support also comes from the biochemical localization of VIPP1 to low-density membranes (Liu et al., 2005) that are membranes intermediate between envelope and thylakoids at which thylakoid protein biogenesis might occur (Zerges and Rochaix, 1998). Depletion of VIPP1 may impair the formation of proper rods, thereby leading to disordered thylakoid centers and eventually resulting in the aberrant structures observed. Alternatively, rods may be formed normally but soluble VIPP1 is depleted and not available for chaperone-mediated cycling between soluble and complexed forms (Liu et al., 2007). If this process was required for the transport of building blocks to T-zones for thylakoid biogenesis (see below), the resulting tailback may be the cause for the aberrant structures observed, comparable to the generation of PLB-like structures by the absence of chlorophyll in the y-1 mutant.
Is VIPP1 Involved in the Biogenesis of Thylakoid Membranes or of Thylakoid Membrane Core Complexes?
The aberrant, PLB-like structures at the origin of thylakoids point to a deficit in thylakoid biogenesis. However, is the formation of the lipid bilayer itself affected, as proposed previously (Kroll et al., 2001; Westphal et al., 2001; Aseeva et al., 2007), or is the biogenesis/assembly of the photosynthesis complexes affected, as suggested by Gao and Xu (2009)?
Similar to what has been reported for a VIPP1 depleted Arabidopsis mutant (Kroll et al., 2001; Aseeva et al., 2007), we find no change in lipid composition, but we do observe slightly lower levels of fully assembled photosystems, cytochrome b6f complex, and ATP synthase (by 14 to 20%) in nonstressed C. reinhardtii VIPP1-RNAi strains (Figure 8A; see Supplemental Figures 2 and 3 online). In contrast with the earlier reports, however, we find neither an obvious reduction of thylakoid membranes nor changes in the number of membranes per granum (Figures 3 and 10). We rather find levels of LHCII increased by 30%, which correlates with a lower chlorophyll a/b ratio and faster fluorescence induction kinetics (Figures 6D, 6G, and 8A). Hence, there appears to be no limitation of thylakoid membranes in VIPP1-RNAi/amiRNA strains for housing protein complexes, which points to a role of VIPP1 in the biogenesis/assembly of core complexes rather than in the formation of the membranes themselves. These data are in support of those from Gao and Xu (2009), who showed in Synechocystis that depletion of VIPP1 first affected PSII activity, then PSI activity, and once photosystem activities were lost, thylakoids degenerated. A role for VIPP1 in core complex biogenesis/assembly is also supported by the previous finding that VIPP1 interacts with Alb3.2 (Göhre et al., 2006). Alb3.2 was suggested to play a role in PSI and PSII assembly, as it was found to directly interact with photosystem subunits. Moreover, both photosystems accumulated to reduced levels when Alb3.2 was downregulated by RNAi, while cytochrome b6f and ATP synthase were barely affected. Therefore, it appears possible that VIPP1 supports Alb3.2 during photosystem assembly.
Phenotypes in Stressed C. reinhardtii VIPP1-RNAi/amiRNA Strains Point to Defects in the Structural Organization of Thylakoid Membrane Complexes
If VIPP1 is involved in the biogenesis/assembly of thylakoid membrane core complexes, which step may be affected in VIPP1 depleted cells? We believe that the pleiotropic phenotypes associated with VIPP1 depletion under stress conditions provide some hints to answer this question.
Prolonged exposure of cells to light intensities of ~1000 μE m−2 s−1 resulted in a much more drastic decline of PSII and PSI activities in VIPP1-RNAi strains compared with a control strain. PSII activities were more severely and more rapidly affected than PSI activities. Loss of photosystem activity in VIPP1-RNAi strains was accompanied by photobleaching of chlorophyll and loss of subunits of both photosystems and of LHCII, while cytochrome b6f complex and ATP synthase were hardly affected (Figure 1; see Supplemental Figure 4 online). Also, a short exposure to very high light intensities (~1800 μE m−2 s−1) resulted in more severe photoinhibition of VIPP1-RNAi strains and slower recovery compared with a control strain (Figure 4). Accordingly, VIPP1-RNAi strains under these conditions also suffered from a more drastic loss of PSII and PSI subunits than control cells (see Supplemental Figure 10A online).
The sensitivity of VIPP1-RNAi strains to both high light treatments was drastically increased in cells grown on ammonium instead of nitrate as nitrogen source and correlated with extensive thylakoid swelling (Figures 1, 3, and 5, Table 1; see Supplemental Figure 10 online). Thylakoid swelling is likely caused by osmosis where ammonium, accumulating in the acidified lumen, acts as an osmoticum and potentially also as a denaturing agent. Interestingly, induction of LHCSR3 and HSP90C/HSP70B was more strongly impaired in VIPP1-RNAi cells grown on ammonium compared with cells grown on nitrate. HSP70B was shown to be involved in photoprotection and repair of PSII during and after photoinhibition (Schroda et al., 1999), and LHCSR3 was shown to be essential for nonphotochemical quenching in C. reinhardtii (Peers et al., 2009). It is unclear how the thylakoid defects in the VIPP1-RNAi strains impair the expression of LHCSR3 and the chaperones, as the signals triggering their induction are still unknown. However, similar defects in LHCSR3 induction and increased sensitivity to high light treatments, as described here for the VIPP1-RNAi strains, were recently reported for knockdown mutants of the chloroplast calcium sensor protein CAS (Petroutsos et al., 2011). CAS is a thylakoid membrane protein and could be involved in a Ca2+-dependent step of light stress signal transduction. Therefore, one possible scenario might be that thylakoid swelling in ammonium-grown VIPP1-RNAi cells either impairs CAS binding to the thylakoids or precludes the correct perception of the signal triggering the induction of photoprotective mechanisms.
In C. reinhardtii, thylakoid swelling was observed only in severely photoinhibited wild-type cells or in ATP synthase mutants (Topf et al., 1992; Majeran et al., 2001) (Table 1) (i.e., under conditions where a large ΔpH/pmf was generated in cells grown on ammonium). It is important to stress that thylakoid swelling occurs in VIPP1-RNAi strains exposed to high light intensities despite the fact that they are capable of generating only a lower pmf than control cells (Figure 6A). Hence, VIPP1-RNAi strains appear to have a structural defect in their thylakoid membranes that impairs their ability to withstand the osmotic pressure normally generated by the diffusion of water toward the ammonium accumulating in the acidified lumen. This structural defect is most likely also the cause for the drastically increased thermosensitivity of PSII in VIPP1-RNAi strains compared with control cells (Figure 7A).
In conclusion, our compiled evidence suggests that VIPP1 depletion affects the structural organization of the thylakoid membrane complexes, in particular that of the photosystems. However, at which level could this structural defect occur? In grana stacks of higher plants, interactions between protein supercomplexes, especially PSII and LHCII, result in the formation of semicrystalline arrays (Kirchhoff et al., 2007; Daum et al., 2010). Comparable semicrystalline PSII arrays have also been observed in Synechocystis (Folea et al., 2008), but so far, their existence has not been addressed in C. reinhardtii thylakoids. A disturbance in the formation of such semicrystalline PSII-LHCII complexes should not cause obvious changes in thylakoid ultrastructure or in the assembly or basic function of thylakoid membrane protein complexes. However, such arrays are expected to contribute to the rigidity of thylakoid membranes. If a depletion of VIPP1 was causing a deficit in the formation of higher order structures like the semicrystalline PSII-LHCII arrays, this would explain the thylakoid swelling phenotype observed in high light–exposed VIPP1-RNAi/amiRNA cells grown on ammonium as nitrogen source. It would also explain the reduced cooperativity of PSII centers suggested by the less sigmoid fluorescence induction curves in VIPP1-RNAi cells compared with control cells (Figure 6G).
VIPP1 Might Support Alb3.2 in Photosystem Assembly by Delivering Small Hydrophobic Polypeptides or Lipids
From the interaction of VIPP1 with Alb3.2, the occurrence of aberrant structures at the origins of thylakoids, and the reduced accumulation of core complexes in VIPP1-RNAi/amiRNA cells, we propose a role for VIPP1 in the biogenesis/assembly of thylakoid membrane core complexes. From the thylakoid swelling phenotype in high light and the sensitivity of PSII to heat stress, we proposed that depletion of VIPP1 may lead to deficits in the structural organization of the thylakoid membrane complexes. But how can we bring both functions together, and how can we explain the increased high light sensitivity of nitrate-grown VIPP1-RNAi strains, which occurs independent of thylakoid swelling?
An explanation that may account for all observations made is that VIPP1 supports Alb3.2 during photosystem assembly by delivering small hydrophobic photosystem subunits or specific structural lipids. Small hydrophobic proteins may be loaded onto VIPP1 at the TIC complex in the chloroplast inner envelope, as suggested previously as a general pathway for thylakoid-located photosynthesis proteins (Jouhet and Gray, 2009a, 2009b). Specific lipids may be loaded onto VIPP1 directly at their site of synthesis at the inner envelope (Benning, 2008). Loading of small proteins/specific lipids onto VIPP1 and their release into an Alb3.2-containing photosystem assembly complex at thylakoid origins might be achieved by the chaperone-mediated assembly/disassembly of VIPP1 oligomers (Liu et al., 2007), which is supported by the finding that levels of VIPP1, CDJ2, and HSP70B were increased twofold in Alb3.2-RNAi strains (Göhre et al., 2006). In this scenario, the pleiotropic phenotypes associated with VIPP1 depletion would arise from photosystems containing reduced amounts of small subunits or structural lipids and therefore are incompletely assembled or impaired in the formation of thylakoid membrane protein supercomplexes with the consequences described above.
The idea of VIPP1 being a carrier for small hydrophobic PSI and PSII subunits appears unlikely, as there exists no evidence that VIPP1 or PspA interact with proteins other than the HSP90C/HSP70B/CDJ2/CGE1 (co-)chaperones (Heide et al., 2009) or regulatory subunits of the psp operon, respectively (Joly et al., 2010). Moreover, it is difficult to imagine that VIPP1 would similarly exhibit specificity for small subunits of PSI and PSII. Finally, although levels of small subunits PSBR, PSBW, PSAG, PSAH, and PSAL appear to be lower under some conditions tested (see Supplemental Data Set 1 online), this is not consistently observed under all conditions, which would be expected if their delivery to the respective photosystem was generally perturbed. By contrast, there is no doubt that VIPP1 and PspA interact with membranes and a direct interaction of PspA with PG and phosphatidylserine was demonstrated (Kobayashi et al., 2007). Furthermore, at least one of the four lipid species present in thylakoid membranes (monogalactosyl-diacylglycerol [MGDG], digalactosyl-diacylglycerol [DGDG], sulfoquinovosyl-diacylglycerol [SQDG], and PG) was found to be an integral component of thylakoid membrane complexes.
More specifically, 11 MGDGs, seven DGDGs, five SQDGs, and two PGs were identified in the crystal structure of PSII from Thermosynechococcus elongatus (Guskov et al., 2009), and PSII isolated from spinach (Spinacia oleracea) and rice (Oryza sativa) was found to contain approximately three MGDGs, approximately two DGDGs, approximately one SQDG, and approximately two PGs (Sakurai et al., 2006). The two PGs were localized to the cytoplasmic side close to the QB site (Guskov et al., 2009), and reduced electron transfer rates from QA to QB upon PG depletion suggested a role of PG in maintaining the structural integrity of the QB site (Gombos et al., 2002). Moreover, PG was shown to be associated with the D1 protein and to be important for PSII dimerization (Kruse and Schmid, 1995; Kruse et al., 2000). Three PGs and one MGDG were identified in the crystal structure of PSI from Synechococcus elongatus (Jordan et al., 2001). One MGDG was found in the crystal structure of the cytochrome b6f complex (Stroebel et al., 2003). Finally, one PG and one DGDG were found in LHCII, where PG is intimately involved in trimerization (Standfuss et al., 2005). Hence, VIPP1 would need to be specific for only a single lipid species to serve in the assembly of both photosystems.
If VIPP1 is indeed acting as a lipid chaperone during photosystem assembly, which lipid may it be supplying? There are three reasons to favor PG: (1) PspA interacts specifically with phospholipids and PG is the only phospholipid in the thylakoids. (2) PG is present in both photosystems. (3) The other three lipid classes are uniformly distributed in the membranes and PSII complexes, but PG is 2 to 5 times more abundant in PSII dimers (Sakurai et al., 2006; Domonkos et al., 2008), thus suggesting the need for an active enrichment process. In several studies, the synthesis pathway for PG was disrupted in Synechocystis, C. reinhardtii, or Arabidopsis (Domonkos et al., 2008). Strikingly, the resulting phenotypes to a large part resemble those observed in VIPP1-depleted lines presented in this and earlier studies: To start with Synechocystis, the pgsA and cdsA mutants, disrupted in genes encoding phosphatidylglycerolphosphate synthase and cytidine 5′-diphosphate diacylglycerol synthase, respectively, died unless PG was added exogenously. Depletion of exogenously supplied PG led to the loss of PSII and PSI activities (Hagio et al., 2000; Sato et al., 2000, 2004) and thus is similar to what Gao and Xu (2009) observed as a consequence of VIPP1 depletion and what we observed in high light–treated VIPP1-RNAi cells. PG-depleted Synechocystis mutants were highly susceptible to photobleaching, which was explained by an increased formation of singlet oxygen as a consequence of reduced electron transfer from QA to QB that enhances the occurrence of the dangerous triplet state of chlorophyll (Sakurai et al., 2003; Domonkos et al., 2008). Moreover, loss of PSI activity upon PG depletion was found to come along with an accumulation of PSI monomers at the expense of PSI trimers (Domonkos et al., 2004), similar to the observation of Fuhrmann et al. (2009a) in a vipp1 merodiploid strain.
The Arabidopsis pgp1 mutant, defective in the gene encoding phosphatidylglycerolphosphate synthase and accumulating only 12% of wild-type PG levels, is unable to grow photoautotrophically, has pale, yellow-green leaves, and its chloroplasts contain only few, swollen thylakoid membranes without granum structure (Hagio et al., 2002). This phenotype indeed resembles that of the Arabidopsis Δvipp1 mutant (Kroll et al., 2001; Aseeva et al., 2007).
Only the phenotype of the C. reinhardtii mf1 and mf2 mutants, in which the deficiency of PG leads to the nearly complete absence of PSII cores due to a severely reduced translation of D1 and CP47 (Maroc et al., 1987; Pineau et al., 2004), was much more severe than that observed in the VIPP1-RNAi strains reported here. Also not consistent with a simple lack of PG in PSII of VIPP1-RNAi cells is our finding that the midpoint potential of the QA/QA− redox couple in PSII is lowered (Table 2). This points to a problem with the integrity of the QA site (Fufezan et al., 2007) instead of the QB site, which was affected by PG depletion in cyanobacteria (Gombos et al., 2002). Certainly, a more negative midpoint potential of the QA/QA− redox couple has been shown to increase the yield of 1O2 (Krieger-Liszkay and Rutherford, 1998; Fufezan et al., 2007) and most likely is causing the high light sensitivity of the VIPP1-RNAi cells. Phenotypes that very closely resemble those of the VIPP1-RNAi strains were observed in the C. reinhardtii hf-2 mutant, which is defective in the biosynthesis of SQDG. The lack of SQDG had no obvious effect on the assembly of PSI, PSII, and LHCII. While PSI activity was unaffected, PSII activity was reduced by ~40% (Sato et al., 1995). Moreover, PSII activity was more sensitive to photoinhibition and highly sensitive to heat stress (Minoda et al., 2002; Sato et al., 2003). These compiled observations indicate that at least in C. reinhardtii, the phenotypes in VIPP1-RNAi/amiRNA cells cannot be explained by a simple lack of PG in the photosystems. However, they do point to a structural problem potentially caused by an incomplete or inaccurate incorporation of PG into the photosystems, thus producing PSII centers that are similarly labile as those completely devoid of SQDG.
A New Hypothesis for a Role of VIPP1 Different from That of a Vesicle-Inducing Protein
In summary, we propose here a new hypothesis for the function of VIPP1 in the chloroplast. This hypothesis claims that VIPP1 rods are in fact the thylakoid centers found in cyanobacteria and organize sites at which thylakoid biogenesis occurs. Chaperone-mediated dynamic cycling between soluble, smaller VIPP1 assembly states and rods may serve to provide structural lipids for core complex assembly, which occurs in collaboration with Alb3.2. Depletion of VIPP1 slows down this assembly process and leads to the aberrant PLB-like structures observed at the origin of thylakoid layers. Moreover, it leads to an incomplete/inaccurate incorporation of structural lipids into thylakoid membrane core complexes, in particular the photosystems. This directly impairs their structure and function (i.e., the integrity of the QA site in PSII) and affects the formation of supercomplexes, like the semicrystalline PSII-LHCII arrays, thus leading to a higher susceptibility to photoinhibition and reduced ability to withstand an increased lumenal turgor. The pronounced high light inducibility of VIPP1 and the reduced capacity of VIPP1-RNAi cells to repair photodamaged PSII by de novo D1 synthesis also indicate a role of VIPP1 during the PSII repair cycle, where a supply of structural lipids will also be needed.
There is no doubt that this hypothesis is still speculative. However, in contrast with the earlier proposal that VIPP1 acts as a vesicle inducing protein, it accounts for most phenotypes observed in VIPP1-depleted cyanobacteria, algae, and higher plants and for most of the localization studies and biochemical and structural observations made so far. For certain, this hypothesis provides a basis for new experimental approaches to eventually understand the enigmatic process of thylakoid biogenesis.
METHODS
Algal Strains and Culture Conditions
Chlamydomonas reinhardtii strains cw15-325 (cwd, mt+, and arg7−) and cw15-302 (cwd, mt+, nit1−and arg7−), both kindly provided by R. Matagne (University of Liège, Belgium), were used as recipient strains for transformation. Both strains were transformed with pCB412 (containing only the wild-type ARG7 gene) to generate control strains. Strain cw15-325 was transformed with pMS439 (containing ARG7 and the VIPP1-RNAi construct) and strain cw15-302 with pMS552 (containing ARG7 and the VIPP1-amiRNA construct). pCB412 was linearized with XhoI, pMS439 with SpeI, and pMS552 with HindIII. One microgram of DNA was used for transformation with glass beads (Kindle, 1990). Strains were grown mixotrophically in TAP medium (Harris, 2008) supplemented with 7.5 mM NH4Cl or 4 mM KNO3 on a rotary shaker at 25°C at ~30 μE m−2 s−1. Fifteen hours before each experiment, cultures were diluted with TAP medium containing the respective nitrogen source to obtain equal cell densities of ~4 × 106 cells/mL when the experiment started. Cell numbers were determined using a Z2 Coulter Counter (Beckman Coulter).
Vector Construction and Screening for VIPP1-Underexpressing Transformants
Construction of the VIPP1-RNAi vector (pMS439) was described previously (Heide et al., 2009). The miRNA targeting C. reinhardtii VIPP1 was designed according to the detailed instructions given by Molnar et al. (2009) using the WMD2 tool at http://wmd2.weigelworld.org (Ossowski et al., 2008). Resulting oligonucleotides 5′-ctagtCGCGACCTGATTAAGAAGCGAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTCGCATCTTAATCAGGTCGCGg-3′ and 5′-ctagcCGCGACCTGATTAAGATGCGAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTCGCTTCTTAATCAGGTCGCGa-3′ (uppercase letters indicate miRNA*/miRNA sequences) were annealed by boiling and slowly cooling down in a thermocycler and ligated into SpeI-digested pChlamiRNA2, yielding pMS552. Screening for correct constructs was done as described by Molnar et al. (2009). VIPP1-underexpressing transformants were initially identified by analyzing ~40 transformants by immunoblotting. After discovering their sensitivity to high light, candidates for VIPP1-underexpressing strains were selected among ~60 transformants that were streaked on TAP-NH4 agar plates and showed bleaching after exposure to ~400 μE m−2 s−1 for 4 d. Strong reduction of VIPP1 levels in high light–sensitive transformants was then verified by immunoblotting.
Protein Gel and Gel Blot Analyses
Protein extraction and quantification, SDS-PAGE, semidry blotting, and detection by enhanced chemiluminescence were performed as described previously (Schroda et al., 1999). Antisera were against HSP90C (Willmund and Schroda, 2005), HSP70B (Schroda et al., 1999), VIPP1 (Liu et al., 2005), VIPP2 (S. Schmollinger and M. Schroda, unpublished results), LHCSR3 (Naumann et al., 2007), PsaD (Naumann et al., 2005), LHCII (Vallon et al., 1986), CP43 (Vallon et al., 1985), cytochrome f (Pierre and Popot, 1993), PsbA (Agrisera, AS05 084), PsaA (Agrisera, AS06 172), OEE33 (Agrisera, AS06 142-33), and CF1β (Lemaire and Wollman, 1989). Depending on the abundance of the antigen in the cell and the quality of the antiserum, different amounts of whole-cell proteins were loaded per SDS-polyacrylamide minigel: 15.3 μg for the detection of HSP90C, VIPP2, CP43, OEE33, PsaA, and cytochrome f; 3.8 μg for HSP70B, PsaD, and LHCSR3; and 0.8 μg for VIPP1, CF1β, D1, and LHCII.
RNA Gel Blot Analyses
RNA extraction, RNA gels, blotting, and hybridization were performed as described previously (Liu et al., 2005). Probes used for hybridization were the ~1-kb full-length cDNA of CBLP2, a 2-kb NheI-AatII cDNA fragment containing the HSP70B coding region, a 1.4-kb BamHI fragment containing part of the VIPP1 coding region, and a 0.34-kb RT-PCR product containing part of the LHCSR3 coding region (Naumann et al., 2007).
High Light, Photoinhibition, and Heat Shock Experiments
For the exposure to high light (~1000 μE m−2 s−1), 150 mL of cell cultures were transferred to 400-mL beakers and placed onto spots on a rotary shaker that were illuminated by Osram HLX 250W 64663 Xenophot bulbs. A glass plate nonpermissive to infrared and UV irradiation was placed between light sources and beakers and cooled by ventilation. Beaker positions of all simultaneously treated cultures were swapped at regular intervals during the high light treatment to ensure equal illumination. The beakers were placed on a water-cooled metal plate to maintain a temperature of 25°C in the cell cultures. The same setup was used for photoinhibition, but cultures were exposed to ~1800 μE m−2 s−1. For heat shock treatments, cell cultures were incubated and agitated in a water bath at 40°C and ~5 μE m−2 s−1. Light intensities were determined using a Luxmeter (Heinz Walz Mess- und Regeltechnik).
Quantitative Proteomics
The control strain was grown in TAP supplemented with K15NO3 (≥98%; NLM-765; Cambridge Isotope Laboratories), and VIPP1-RNAi strain #111 was grown in TAP supplemented with K14NO3. Cells were grown to a density of ~4 × 106 cells/mL. The cultures were split into three parts: One part was incubated at ~30 μE m−2 s−1 and 25°C for 1 h (unstressed), the second part was treated at ~2000 μE m−2 s−1 and 25°C for 1 h (photoinhibition), and the third part was exposed to 40°C at ~5 μE m−2 s−1 for 1 h (heat shock). After mixing 15N-labeled with 14N-labeled cells from each treatment at a ratio of 1:1 (based on same cell numbers), membrane-enriched fractions were prepared by freeze-thawing and centrifugation (30 min at 16,000g and 4°C). Pellets were resuspended in lysis buffer (50 mM NH4HCO3, 1 mM DTT, and 1 mM NaCl), sonicated, and centrifuged again as above. The pellet was resuspended with lysis buffer, and proteins corresponding to 20 μg chlorophyll were mixed with 2× SDS sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol, and 0.005% bromophenol blue), boiled for 60 s, and separated on a 12% SDS-polyacrylamide gel. The three lanes were cut into 14 gel bands each, and proteins therein were digested with 12.5 ng/μL sequencing grade modified trypsin (Promega) for 16 h at 37°C. The eluted peptides from the gel slices were desalted, separated, and measured by liquid chromatography–mass spectrometry essentially as described by Mühlhaus et al. (2011) with the following alterations: For HPLC separation a gradient ramping from 0 to 35% B (A, 2% acetonitrile and 0.5% acetic acid; B, 80% acetonitrile and 0.5% acetic acid) after 45 min and then to 100% B after 50 min was employed. Data were acquired in the positive polarity mode in data-dependent fashion: one survey scan from 300 to 2000 mass-to-charge ratio in the orbitrap analyzer (resolution of 60,000 at 400 mass-to-charge ratio; target filling of 106 ions) was followed by tandem mass spectrometry (MS/MS) scans in the linear ion trap (target filling of 104 ions) of the seven most intense precursor ions detected in the survey scan. Singly charged ions were rejected for MS/MS analysis, and we allowed two MS/MS events for each ion before putting it on the exclusion list for 180 s. Identification of peptide sequences and quantification were performed by the Integration of Mass Spectrometry Identification and Quantification Software (Mühlhaus et al., 2011). The light-to-heavy ratios calculated for all unique peptides were used to determine the relative abundance of the corresponding proteins. Unequal mixing of labeled and unlabeled proteins in each sample was corrected for by normalizing the light-to-heavy ratio determined for each peptide in a sample by the median of light-to-heavy ratios of all peptides in that sample. The median of the normalized light-to-heavy ratios of all peptides from one protein was taken as the relative abundance of that protein. Quantification values of core complex subunits are listed in Supplemental Data Set 1 online. Sequences and quantification values of all identified peptides are presented in Supplemental Data Set 2 online.
Chlorophyll a Fluorescence Measurements
To monitor PSII maximum quantum efficiency during high light exposure and heat stress, the FV/FM ratio was determined from intact cells at 25°C using the PAM-100 fluorometer (Heinz Walz). Basal fluorescence (F0) was detected after dark adaptation for 2 min and far-red adaptation for an additional 2 min (the measurements of Figure 6 were all done with a Dual-PAM-100 after 5 min of far-red illumination, followed by 10 min of dark adaptation). FM was determined with a single 4000 μE m−2 s−1 light pulse. For chlorophyll a fluorescence induction curves, cells equivalent to 10 μg/mL chlorophyll were used. Cells were first illuminated with far-red light to induce state 1, followed by the addition of 100 μM DCMU and incubation in darkness for 15 min. To keep cells in state 1, the sample was strongly stirred (aerobic conditions). Then, chlorophyll a fluorescence induction was measured at 25 μE light intensity with a temporal resolution of 100 μs.
Spectroscopy
The time course of changes in PSI and PSII activities during light exposure was followed using a JTS spectrophotometer (Bio-Logic). PSI and PSII activities were estimated from changes in the amplitude of the fast phase (100 μs) of the electrochromic absorption shift (ECS) signal (at 520 to 545 nm) upon excitation with a saturating laser flash (Bailleul et al., 2010). The ECS spectral change linearly follows the number of light-induced charge separations within the reaction centers (Witt, 1979). Thus, PSII contribution can be calculated from the decrease in the signal amplitude upon addition of DCMU (20 μM) and hydroxylamine (1 mM) that irreversibly block PSII charge separation once the sample has been preilluminated (Joliot and Delosme, 1974). Conversely, PSI activity was estimated as the fraction of the signal that was insensitive to these inhibitors. For the measurement of pmf and ATP synthase activity, the ECS was used as an in vivo probe of the pmf across the thylakoid membrane (Kramer et al., 2003; Takizawa et al., 2007; Bailleul et al., 2010). The difference transmission signal was measured with a KLAS-100 LED-array spectrophotometer (Heinz Walz), which allows the simultaneous measurement of light-induced difference absorption signals at eight wavelength pairs between 505 and 570 nm. The first two wavelength pairs at 505 to 520 nm and 520 to 535 nm are dominated by the electrochromic shift transmittance changes arising from the xanthophyll cycle. The other wavelength pairs (540 to 545 nm, 545 to 550 nm, 550 to 554 nm, 554 to 559 nm, 559 to 563 nm, and 563 to 570 nm) are required to completely deconvolute the ECS signal from scattering effects, the C550 pheophytin signal, and from redox changes of the cytochromes f and b6. The deconvoluted transmission signals (ΔI/I) were normalized to the chlorophyll content of the algal suspension. The maximum amplitude of the ECS (ECST) was used as a measure for the light-induced pmf across the thylakoid membrane. Cells were resuspended in 20% (w/v) Ficoll and illuminated for 5 min prior to each measurement to allow photosynthesis to reach steady state. ECST was determined after illuminating the suspension with saturating light (1500 μE m−2 s−1), which then was interrupted by a short interval of darkness (12 s), and the dark-interval relaxation kinetics of the ECS were measured. To determine ATP synthase activity, the fast phase (between 0 and 500 ms after the end of actinic illumination) of the dark interval relaxation kinetics was fitted with a single exponential decay function. The reciprocal value of the half-time of the ECS decay, the thylakoid conductivity (gH+), is a measure of ATP synthase activity, as fast ECS decay is exclusively attributable to proton efflux through the ATP synthase (Cruz et al., 2001; Baker et al., 2007). Cytochrome f reduction was measured in parallel to the ECS decay kinetics. For cytochrome f oxidation measurements cell numbers equivalent to 15 to 20 μg/mL chlorophyll were used. Cells were resuspended in 20% (w/v) Ficoll. Cells were preilluminated for 5 min to fully activated the Calvin cycle, followed by the addition of 10 μM DCMU, to inhibit PSII. Cytochrome f was oxidized by a saturating red light pulse (1660 μE m−2 s−1).
Thermoluminescence Measurements
Thermoluminescence was measured with a home-built apparatus. To measure thermoluminescence arising from the S2QA− charge recombination (Q-band), C. reinhardtii cells were incubated in the presence of 10 μM DCMU for 5 min in the dark at 20°C and then flashed with one saturating xenon flash at −5°C. For luminescence detection, the samples were warmed at a constant rate (0.4°C s−1) from 1 to 70°C.
Indirect Immunofluorescence Microscopy
Control transformants in the cw15-325 background were grown in TAP-NH4 media and fixed and stained as described previously (Cole et al., 1998; Pérez-Pérez et al., 2010). The primary antibody was affinity purified from polyclonal anti-VIPP1 antiserum (Liu et al., 2005; Willmund and Schroda, 2005) and used in a 1:250 dilution. A fluorescein isothiocyanate–labeled goat anti-rabbit antibody (1:500; Sigma-Aldrich) was used as secondary antibody. Preparations were photographed on a DM6000B microscope (Leica Microsystems) with an ORCA-ER camera (Hamamatsu) and the Leica Application Suite Advanced Fluorescence software package (Leica).
Transmission Electron Microscopy
Cells were fixed for 2 h (at room temperature) to 14 h (at 4°C) in 100 mM sodium cacodylate containing 2.5% glutaraldehyde at pH 7.2. After two washes with 100 mM Na-cacodylate, pH 7.2, and two washes with distilled water, the cells were osmicated in 1% OsO4 in distilled water for 1 h at 4°C. After two washes with distilled water, cells were resuspended in 20% BSA (w/v) and incubated for 20 min at room temperature. Cells were centrifuged for 10 min at 16,000g, and the cell pellet was incubated for 30 min at room temperature with 2.5% glutaraldehyde in distilled water at pH 7.4. After two washes with distilled water, the cell pellet was cut into pieces (1 to 2 mm3), dehydrated using increasing concentrations of ethanol and embedded in glycid ether 100 (formerly Epon 812; Serva) with propylene oxide as intermediate solvent following standard procedures. Polymerization was performed for 48 h at 65°C. Ultrathin sections (~60 to 70 nm) were cut with a diamond knife (type ultra 35°; Diatome) on an EM UC6 ultramicrotome (Leica) and mounted on single-slot Pioloform-coated copper grids (Plano). The sections were stained with uranyl acetate and lead citrate (Reynolds, 1963) and viewed with a JEM-2100 (Jeol) or EM 902A (Carl Zeiss) transmission electron microscope (both operated at 80 kV). Micrographs were taken using a 4080 × 4080-pixel or 1350 × 1040-pixel charge-coupled device camera (UltraScan 4000 or Erlangshen ES500W, respectively; Gatan) and Gatan Digital Micrograph software (version 1.70.16). Image brightness and contrast were adjusted and figures assembled using Adobe Photoshop 8.0.1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: VIPP1, AAU06582 (GenBank), Cre13.g583550 (Phytozome); and VIPP2, Cre11.g468050 (Phytozome).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Construct Used for the Suppression of VIPP1 by RNAi.
Supplemental Figure 2. Major Thylakoid Membrane Protein Complexes Are Fully Assembled in VIPP1-RNAi Strains.
Supplemental Figure 3. Bulk Membrane Lipid Composition in VIPP1-RNAi Strains Is Indistinguishable from That of Control Strains.
Supplemental Figure 4. Subunits of PSII and PSI Are Rapidly Degraded in VIPP1-RNAi Strains Exposed to High Light Intensities.
Supplemental Figure 5. High Light Sensitivity Is Also Observed in VIPP1-amiRNA Strains Unable to Induce VIPP2 Expression.
Supplemental Figure 6. VIPP1-RNAi Strains Are Not Impaired in State Transitions.
Supplemental Figure 7. Delayed Bleaching of VIPP1-RNAi Strains Grown on Nitrate.
Supplemental Figure 8. Categories of Thylakoid Structure.
Supplemental Figure 9. Lysis of High Light–Exposed VIPP1-RNAi Cells Grown on Nitrate Appears Not to Be Caused by Thylakoid Swelling.
Supplemental Figure 10. VIPP1-RNAi Strains Are Less Sensitive to Photoinhibition When Grown on Nitrate Compared with Ammonium.
Supplemental Figure 11. VIPP1-RNAi Strains Are Less Sensitive to Heat Shock When Grown on Nitrate Compared with Ammonium.
Supplemental Data Set 1. Quantification Results for Subunits of Thylakoid Membrane and Respiratory Chain Core Complexes.
Supplemental Data Set 2. Sequences and Quantification Results for All Peptides Identified by Quantitative Shotgun Proteomics.
Supplementary Material
Acknowledgments
We thank Olivier Vallon (Institut de Biologie Physico-Chimique, Paris, France) for the antisera against CF1β, cytochrome f, and LHCII, Michael Hippler (University of Münster, Germany) for antisera against PsaD and LHCSR3, Jochen Golecki (University of Freiburg, Germany) for first survey electron microscopy images, and Rita Grotjahn and Franziska Richter (University of Bayreuth, Germany) for help with sample preparation for transmission electron microscopy and the evaluation of electron microscopy images. This work was supported by the Max Planck Society, by grants from the Deutsche Forschungsgemeinschaft (Schr 617/2-4 and 617/5-1), and by the Bundesministerium für Bildung und Forschung (Systems Biology Initiative FORSYS; Project GoFORSYS).
AUTHOR CONTRIBUTIONS
A.N. performed all growth, protein gel, and RNA gel analyses with the cw15-325 strains. M.A.S., G.F., T.R., A.K.-L., A.N., S. Schönfelder, and H.L. performed the biophysical measurements. A.-K.U., B.S., and S.G. took all electron micrographs and did the statistical evaluations. S. Schönfelder and M.R. performed all growth and protein gel analyses with the cw15-302 strains. F.S. performed the lipid analyses and the mass spectrometry measurements, and T.M. evaluated the mass spectrometry data. S. Schmollinger cloned the VIPP1-amiRNA construct, generated recombinant VIPP2, and took immunofluorescence micrographs with J.L.C. M.S. conceived and coordinated the project, prepared most of the figures, and wrote the article.
References
- Aseeva E., Ossenbühl F., Eichacker L.A., Wanner G., Soll J., Vothknecht U.C. (2004). Complex formation of Vipp1 depends on its alpha-helical PspA-like domain. J. Biol. Chem. 279: 35535–35541 [DOI] [PubMed] [Google Scholar]
- Aseeva E., Ossenbühl F., Sippel C., Cho W.K., Stein B., Eichacker L.A., Meurer J., Wanner G., Westhoff P., Soll J., Vothknecht U.C. (2007). Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 45: 119–128 [DOI] [PubMed] [Google Scholar]
- Bailleul B., Cardol P., Breyton C., Finazzi G. (2010). Electrochromism: A useful probe to study algal photosynthesis. Photosynth. Res. 106: 179–189 [DOI] [PubMed] [Google Scholar]
- Baker N.R., Harbinson J., Kramer D.M. (2007). Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ. 30: 1107–1125 [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Benning C. (2008). A role for lipid trafficking in chloroplast biogenesis. Prog. Lipid Res. 47: 381–389 [DOI] [PubMed] [Google Scholar]
- Benning C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25: 71–91 [DOI] [PubMed] [Google Scholar]
- Brissette J.L., Russel M., Weiner L., Model P. (1990). Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87: 862–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon A.B., Timko M.P. (2000). yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. Plant Cell 12: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carde J.-P., Joyard J., Douce R. (1982). Electron microscopic studies of envelope membranes from spinach plastids. Biol. Cell 44: 315–324 [Google Scholar]
- Chuartzman S.G., Nevo R., Shimoni E., Charuvi D., Kiss V., Ohad I., Brumfeld V., Reich Z. (2008). Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell 20: 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J.A., Sacksteder C.A., Kanazawa A., Kramer D.M. (2001). Contribution of electric field (Delta psi) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. control of pmf parsing into Delta psi and Delta pH by ionic strength. Biochemistry 40: 1226–1237 [DOI] [PubMed] [Google Scholar]
- Daum B., Nicastro D., Austin J., II, McIntosh J.R., Kühlbrandt W. (2010). Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell 22: 1299–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisa M.P., Lee P., Palmer T., Georgiou G. (2004). Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186: 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- Domonkos I., Laczkó-Dobos H., Gombos Z. (2008). Lipid-assisted protein-protein interactions that support photosynthetic and other cellular activities. Prog. Lipid Res. 47: 422–435 [DOI] [PubMed] [Google Scholar]
- Domonkos I., et al. (2004). Phosphatidylglycerol is essential for oligomerization of photosystem I reaction center. Plant Physiol. 134: 1471–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folea I.M., Zhang P., Aro E.M., Boekema E.J. (2008). Domain organization of photosystem II in membranes of the cyanobacterium Synechocystis PCC6803 investigated by electron microscopy. FEBS Lett. 582: 1749–1754 [DOI] [PubMed] [Google Scholar]
- Friedberg I., Goldberg I., Ohad I. (1971). A prolamellar body-like structure in Chlamydomonas reinhardi. J. Cell Biol. 50: 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fufezan C., Gross C.M., Sjödin M., Rutherford A.W., Krieger-Liszkay A., Kirilovsky D. (2007). Influence of the redox potential of the primary quinone electron acceptor on photoinhibition in photosystem II. J. Biol. Chem. 282: 12492–12502 [DOI] [PubMed] [Google Scholar]
- Fuhrmann E., Bultema J.B., Kahmann U., Rupprecht E., Boekema E.J., Schneider D. (2009b). The vesicle-inducing protein 1 from Synechocystis sp. PCC 6803 organizes into diverse higher-ordered ring structures. Mol. Biol. Cell 20: 4620–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann E., Gathmann S., Rupprecht E., Golecki J., Schneider D. (2009a). Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Xu X. (2009). Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity before the loss of thylakoid membranes. FEMS Microbiol. Lett. 292: 63–70 [DOI] [PubMed] [Google Scholar]
- Göhre V., Ossenbühl F., Crèvecoeur M., Eichacker L.A., Rochaix J.D. (2006). One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos Z., Várkonyi Z., Hagio M., Iwaki M., Kovács L., Masamoto K., Itoh S., Wada H. (2002). Phosphatidylglycerol requirement for the function of electron acceptor plastoquinone Q(B) in the photosystem II reaction center. Biochemistry 41: 3796–3802 [DOI] [PubMed] [Google Scholar]
- Goral T.K., Johnson M.P., Brain A.P., Kirchhoff H., Ruban A.V., Mullineaux C.W. (2010). Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: Effects of photoinhibition and protein phosphorylation. Plant J. 62: 948–959 [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Hagio M., Gombos Z., Várkonyi Z., Masamoto K., Sato N., Tsuzuki M., Wada H. (2000). Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol. 124: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagio M., Sakurai I., Sato S., Kato T., Tabata S., Wada H. (2002). Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol. 43: 1456–1464 [DOI] [PubMed] [Google Scholar]
- Hankamer B.D., Elderkin S.L., Buck M., Nield J. (2004). Organization of the AAA(+) adaptor protein PspA is an oligomeric ring. J. Biol. Chem. 279: 8862–8866 [DOI] [PubMed] [Google Scholar]
- Harris E.H. (2008). The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use. (San Diego, CA: Elsevier/Academic Press; ). [Google Scholar]
- Heide H., Nordhues A., Drepper F., Nick S., Schulz-Raffelt M., Haehnel W., Schroda M. (2009). Application of quantitative immunoprecipitation combined with knockdown and cross-linking to Chlamydomonas reveals the presence of vesicle-inducing protein in plastids 1 in a common complex with chloroplast HSP90C. Proteomics 9: 3079–3089 [DOI] [PubMed] [Google Scholar]
- Hinterstoisser B., Cichna M., Kuntner O., Peschek G.A. (1993). Cooperation of plasma and thylakoid membranes for the biosynthesis of chlorophyll in cyanobacteria: the role of thylakoid centers. J. Plant Physiol. 142: 407–413 [Google Scholar]
- Hoober J.K., Blobel G. (1969). Characterization of the chloroplastic and cytoplasmic ribosomes of Chlamydomonas reinhardi. J. Mol. Biol. 41: 121–138 [DOI] [PubMed] [Google Scholar]
- Joliot P., Delosme R. (1974). Flash-induced 519 nm absorption change in green algae. Biochim. Biophys. Acta 357: 267–284 [DOI] [PubMed] [Google Scholar]
- Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., Stumpf M.P., Buck M. (2010). Managing membrane stress: The phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34: 797–827 [DOI] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Jouhet J., Gray J.C. (2009a). Interaction of actin and the chloroplast protein import apparatus. J. Biol. Chem. 284: 19132–19141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J., Gray J.C. (2009b). Is chloroplast import of photosynthesis proteins facilitated by an actin-TOC-TIC-VIPP1 complex? Plant Signal. Behav. 4: 986–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H., Haase W., Wegner S., Danielsson R., Ackermann R., Albertsson P.A. (2007). Low-light-induced formation of semicrystalline photosystem II arrays in higher plant chloroplasts. Biochemistry 46: 11169–11176 [DOI] [PubMed] [Google Scholar]
- Kleerebezem M., Crielaard W., Tommassen J. (1996). Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15: 162–171 [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Tommassen J. (1993). Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol. Microbiol. 7: 947–956 [DOI] [PubMed] [Google Scholar]
- Kobayashi R., Suzuki T., Yoshida M. (2007). Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66: 100–109 [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Cruz J.A., Kanazawa A. (2003). Balancing the central roles of the thylakoid proton gradient. Trends Plant Sci. 8: 27–32 [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A., Rutherford A.W. (1998). Influence of herbicide binding on the redox potential of the quinone acceptor in photosystem II: Relevance to photodamage and phytotoxicity. Biochemistry 37: 17339–17344 [DOI] [PubMed] [Google Scholar]
- Kroll D., Meierhoff K., Bechtold N., Kinoshita M., Westphal S., Vothknecht U.C., Soll J., Westhoff P. (2001). VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse O., Hankamer B., Konczak C., Gerle C., Morris E., Radunz A., Schmid G.H., Barber J. (2000). Phosphatidylglycerol is involved in the dimerization of photosystem II. J. Biol. Chem. 275: 6509–6514 [DOI] [PubMed] [Google Scholar]
- Kruse O., Schmid G.H. (1995). The role of phosphatidylglycerol as a functional effector and membrane anchor of the D1-core peptide from photosystem II-particles of the cyanobacterium Oscillatoria chalybea. Z. Naturforsch. C 50: 380–390 [PubMed] [Google Scholar]
- Kunkel D.D. (1982). Thylakoid centers: structures associated with the cyanobacterial photosynthetic membrane system. Arch. Microbiol. 133: 97–99 [Google Scholar]
- Lemaire C., Wollman F.A. (1989). The chloroplast ATP synthase in Chlamydomonas reinhardtii. I. Characterization of its nine constitutive subunits. J. Biol. Chem. 264: 10228–10234 [PubMed] [Google Scholar]
- Li H.M., Kaneko Y., Keegstra K. (1994). Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol. Biol. 25: 619–6328061316 [Google Scholar]
- Liu C., Willmund F., Golecki J.R., Cacace S., Hess B., Markert C., Schroda M. (2007). The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 50: 265–277 [DOI] [PubMed] [Google Scholar]
- Liu C., Willmund F., Whitelegge J.P., Hawat S., Knapp B., Lodha M., Schroda M. (2005). J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 16: 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Olive J., Drapier D., Vallon O., Wollman F.A. (2001). The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol. 126: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroc J., Tremolieres A., Garnier J., Guyon D. (1987). Oligomeric form of the light-harvesting chlorophyll a+b protein complex CPII, phosphatidylglycerol, Δ3-trans-hexadecenoic acid and energy transfer in Chlamydomonas reinhardtii, wild-type and mutants. Biochim. Biophys. Acta 893: 91–99 [Google Scholar]
- Minoda A., Sato N., Nozaki H., Okada K., Takahashi H., Sonoike K., Tsuzuki M. (2002). Role of sulfoquinovosyl diacylglycerol for the maintenance of photosystem II in Chlamydomonas reinhardtii. Eur. J. Biochem. 269: 2353–2358 [DOI] [PubMed] [Google Scholar]
- Molnar A., Bassett A., Thuenemann E., Schwach F., Karkare S., Ossowski S., Weigel D., Baulcombe D. (2009). Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Mühlhaus T., Weiss J., Hemme D., Sommer F., Schroda M. (2011). Quantitative shotgun proteomics using a uniform 15N-labeled standard to monitor proteome dynamics in time course experiments reveals new insights into the heat stress response of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 10: M110.004739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007). Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B., Stauber E.J., Busch A., Sommer F., Hippler M. (2005). N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B., Stengel A., Schottkowski M., Soll J., Ankele E. (2011). Biogenesis of the cyanobacterial thylakoid membrane system—An update. FEMS Microbiol. Lett. 315: 1–5 [DOI] [PubMed] [Google Scholar]
- Nordhues A., Miller S.M., Muhlhaus T., Schroda M. (2010). New insights into the roles of molecular chaperones in Chlamydomonas and Volvox. Int. Rev. Cell Mol. Biol. 285: 75–113 [DOI] [PubMed] [Google Scholar]
- Ohad I., Adir N., Koike H., Kyle D.J., Inoue Y. (1990). Mechanism of photoinhibition in vivo. A reversible light-induced conformational change of reaction center II is related to an irreversible modification of the D1 protein. J. Biol. Chem. 265: 1972–1979 [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Peers G., Truong T.B., Ostendorf E., Busch A., Elrad D., Grossman A.R., Hippler M., Niyogi K.K. (2009). An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Florencio F.J., Crespo J.L. (2010). Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152: 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D., Busch A., Janssen I., Trompelt K., Bergner S.V., Weinl S., Holtkamp M., Karst U., Kudla J., Hippler M. (2011). The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23: 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre Y., Popot J.L. (1993). Identification of two 4-kDa miniproteins in the cytochrome b6f complex from Chlamydomonas reinhardtii. C. R. Acad. Sci. III 316: 1404–1409 [PubMed] [Google Scholar]
- Pineau B., Girard-Bascou J., Eberhard S., Choquet Y., Trémolières A., Gérard-Hirne C., Bennardo-Connan A., Decottignies P., Gillet S., Wollman F.A. (2004). A single mutation that causes phosphatidylglycerol deficiency impairs synthesis of photosystem II cores in Chlamydomonas reinhardtii. Eur. J. Biochem. 271: 329–338 [DOI] [PubMed] [Google Scholar]
- Reynolds E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford A.W., Crofts A.R., Inoue Y. (1982). Thermoluminescence as a probe of photosystem II photochemistry. The origin of the flash-induced glow peaks. Biochim. Biophys. Acta Bioenergetics 682: 457–465 [Google Scholar]
- Sakurai I., Hagio M., Gombos Z., Tyystjarvi T., Paakkarinen V., Aro E.M., Wada H. (2003). Requirement of phosphatidylglycerol for maintenance of photosynthetic machinery. Plant Physiol. 133: 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai I., Shen J.R., Leng J., Ohashi S., Kobayashi M., Wada H. (2006). Lipids in oxygen-evolving photosystem II complexes of cyanobacteria and higher plants. J. Biochem. 140: 201–209 [DOI] [PubMed] [Google Scholar]
- Sato N., Aoki M., Maru Y., Sonoike K., Minoda A., Tsuzuki M. (2003). Involvement of sulfoquinovosyl diacylglycerol in the structural integrity and heat-tolerance of photosystem II. Planta 217: 245–251 [DOI] [PubMed] [Google Scholar]
- Sato N., Hagio M., Wada H., Tsuzuki M. (2000). Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc. Natl. Acad. Sci. USA 97: 10655–10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Sonoike K., Tsuzuki M., Kawaguchi A. (1995). Impaired photosystem II in a mutant of Chlamydomonas reinhardtii defective in sulfoquinovosyl diacylglycerol. Eur. J. Biochem. 234: 16–23 [DOI] [PubMed] [Google Scholar]
- Sato N., Suda K., Tsuzuki M. (2004). Responsibility of phosphatidylglycerol for biogenesis of the PSI complex. Biochim. Biophys. Acta 1658: 235–243 [DOI] [PubMed] [Google Scholar]
- Schroda M., Blöcker D., Beck C.F. (2000). The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21: 121–131 [DOI] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Wollman F.A., Beck C.F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Even D., Kloppstech K., Ohad I. (1988). Evidence for protection by heat-shock proteins against photoinhibition during heat-shock. EMBO J. 7: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R., Battchikova N., Norling B., Aro E.M. (2006). Plasma membrane of Synechocystis PCC 6803: A heterogeneous distribution of membrane proteins. Arch. Microbiol. 185: 238–243 [DOI] [PubMed] [Google Scholar]
- Srivastava R., Pisareva T., Norling B. (2005). Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5: 4905–4916 [DOI] [PubMed] [Google Scholar]
- Standar K., Mehner D., Osadnik H., Berthelmann F., Hause G., Lünsdorf H., Brüser T. (2008). PspA can form large scaffolds in Escherichia coli. FEBS Lett. 582: 3585–3589 [DOI] [PubMed] [Google Scholar]
- Standfuss J., Terwisscha van Scheltinga A.C., Lamborghini M., Kühlbrandt W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 24: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D., Choquet Y., Popot J.L., Picot D. (2003). An atypical haem in the cytochrome b(6)f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Takizawa K., Cruz J.A., Kanazawa A., Kramer D.M. (2007). The thylakoid proton motive force in vivo. Quantitative, non-invasive probes, energetics, and regulatory consequences of light-induced pmf. Biochim. Biophys. Acta 1767: 1233–1244 [DOI] [PubMed] [Google Scholar]
- Topf J., Gong H., Timberg R., Mets L., Ohad I. (1992). Thylakoid membrane energization and swelling in photoinhibited Chlamydomonas cells is prevented in mutants unable to perform cyclic electron flow. Photosynth. Res. 32: 59–69 [DOI] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2007). Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19: 3640–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O., Wollman F.A., Olive J. (1985). Distribution of intrinsic and extrinsic subunits of the PSII protein complex between appressed and non-appressed regions of the thylakoid membrane: An immunocytochemical study. FEBS Lett. 183: 245–250 [Google Scholar]
- Vallon O., Wollman F.A., Olive J. (1986). Lateral distribution of the main protein complexes of the photosynthetic appartus in Chlamydomonas reinhardttii and in spinach: An immunocytochemical study using inteact thylakoid membranes and PS II enriched membrane preparation. Photobiochem. Photobiophys. 12: 203–220 [Google Scholar]
- van de Meene A.M., Hohmann-Marriott M.F., Vermaas W.F., Roberson R.W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184: 259–270 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Urbanus M.L., Ten Hagen-Jongman C.M., Nouwen N., Oudega B., Harms N., Driessen A.J., Luirink J. (2003). A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. USA 100: 5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S., Heins L., Soll J., Vothknecht U.C. (2001). Vipp1 deletion mutant of Synechocystis: A connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl. Acad. Sci. USA 98: 4243–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F., Schroda M. (2005). HEAT SHOCK PROTEIN 90C is a bona fide Hsp90 that interacts with plastidic HSP70B in Chlamydomonas reinhardtii. Plant Physiol. 138: 2310–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt H.T. (1979). Energy conversion in the functional membrane of photosynthesis. Analysis by light pulse and electric pulse methods. The central role of the electric field. Biochim. Biophys. Acta 505: 355–427 [DOI] [PubMed] [Google Scholar]
- Yamasaki T., Miyasaka H., Ohama T. (2008). Unstable RNAi effects through epigenetic silencing of an inverted repeat transgene in Chlamydomonas reinhardtii. Genetics 180: 1927–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W., Rochaix J.D. (1998). Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 140: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.