Abstract
Recent advances in sequencing technology have enabled the rapid generation of billions of bases at relatively low cost. A crucial first step in many sequencing applications is to map those reads to a reference genome. However, when the reference genome is large, finding accurate mappings poses a significant computational challenge due to the sheer amount of reads, and because many reads map to the reference sequence approximately but not exactly. We introduce Hobbes, a new gram-based program for aligning short reads, supporting Hamming and edit distance. Hobbes implements two novel techniques, which yield substantial performance improvements: an optimized gram-selection procedure for reads, and a cache-efficient filter for pruning candidate mappings. We systematically tested the performance of Hobbes on both real and simulated data with read lengths varying from 35 to 100 bp, and compared its performance with several state-of-the-art read-mapping programs, including Bowtie, BWA, mrsFast and RazerS. Hobbes is faster than all other read mapping programs we have tested while maintaining high mapping quality. Hobbes is about five times faster than Bowtie and about 2–10 times faster than BWA, depending on read length and error rate, when asked to find all mapping locations of a read in the human genome within a given Hamming or edit distance, respectively. Hobbes supports the SAM output format and is publicly available at http://hobbes.ics.uci.edu.
INTRODUCTION
DNA sequencing is an indispensable tool in many areas of biology and medicine. Recent technological breakthroughs in high-throughput sequencing have made it possible to sequence billions of bases quickly and cheaply. For instance, the HiSeq platform from Illumina can produce 6 billion 100 bp reads within only 11 days. The SOLiD system from Life Technologies can generate over 20 GB of DNA sequences per day. These technological advances have opened the door for personal genome sequencing, and the creation of a number of new tools for studying diseases, genomes and epigenomes.
Mapping the reads from high-throughput sequencers to a reference sequence often represents the first step in the computational analysis of sequencing data in many applications. The enormous amount of reads produced from the sequencers poses a great challenge on the speed and the accuracy of read alignment programs for two major reasons. First, the reference sequence can be very large. For instance, the human genome is about 3 billion base pairs long. Mapping a billion reads to the human genome amounts to check 3 × 1018 candidate locations. Second, due to sequencing errors and/or genetic variations, many reads map to the reference sequence approximately but not exactly, and therefore, to map a read to the reference sequence, read mapping programs should allow a certain number of mismatches between the read and a candidate location. Although a number of high-performance read alignment programs have been developed, it still took days or even weeks (depending on different mapping criteria) to align billions of reads to a large reference genome on a single desktop. As the sequencing technology is progressing toward generating longer reads, and thus requiring read-mapping programs to be able to handle more mismatches, the need for faster and more accurate read alignment programs is greater than ever.
Related work
Existing approaches to the read-mapping problem can be broadly classified into two categories: trie-based methods and gram-based methods. In the first group, most of the popular packages use the Borrows-Wheeler Transform (BWT) (1) and usually FM index (2) to encode their trie, e.g. Bowtie (3), BWA (4), SOAP2 (5). These packages have a very small memory footprint (∼2thinsp;GB), and are very efficient for finding a few mappings for short reads with not too many mismatches. They use backtracking to allow mismatches during the tree traversal, and therefore, their performance deteriorates as the read length and the number of mismatches increase. BWT-based packages are typically not designed for finding a large number of mappings per read.
The gram-based methods follow a filter-and-verify paradigm. Using grams, they first identify a set of candidate mappings, and then verify the true distance for those candidates to remove false positives. The candidate-generation step is often supported by an inverted index on grams (from the reads and/or the reference sequence), leading to a relatively large memory footprint. Early packages like SSAHA (6) and BLAST (7) had long mapping times, infeasible for large data sets. Newer packages like Maq (8), RMAP (9), ZOOM, SHRiMP (11), RazerS (12), mrsFAST (13), and mrFAST-CO (14) offer significant improvements, but they do not consistently outperform BWT-based methods. We will show that Hobbes outperforms both existing gram-based and BWT-based methods.
Hobbes
Applications may differ in their requirements on a read-mapping package. Sometimes finding a couple of mappings per read is sufficient, but other times the application may need all mapping positions. For instance, in RNA-seq applications, due to the occurrence of homologous genes and multiple RNA isoforms originated from the same gene, finding all mapping positions will be necessary for quantifying the expression level of a particular gene isoform (15). Similarly, in ChIP-seq applications, finding all mapping positions is a necessary step for characterizing protein binding patterns in repeat regions of a genome (16,17).
Hobbes is a gram-based read mapper, supports Hamming and edit distance and is efficient in both of those situations. Hobbes is about 2–10 times faster than state-of-the-art packages when finding all mappings per read, and performs comparably when looking for a few mappings. Hobbes is also at least as accurate as other packages.
In the following sections, we identify two performance bottlenecks of existing gram-based approaches, and make two major contributions to overcome them: first, we present a novel technique for judiciously choosing a small set of grams of each read to generate candidate mappings. Second, we develop a cache- and CPU-efficient filter for removing false positive mappings during the traversal of inverted lists.
MATERIALS AND METHODS
We first discuss how to map reads to the reference sequence with a given Hamming-distance threshold, and later extend our solution to support edit distance. Our approach is based on generating overlapping q-grams of the reference sequence, and constructing an inverted index of those q-gram positions. To map a read, we generate its q-grams, and access the inverted index to compute a superset of all mapping positions. We then remove false-positive positions by computing the real distance of the read to the subsequences starting at those positions in the reference sequence. Next, we summarize the basic q-gram method focusing on Hamming distance, although some techniques directly apply to edit distance as well.
Basic Q-gram method
For a positive integer q, the q-grams of a sequence are all its overlapping substrings of length q. For example, the 3-grams of a sequence s = TGCCCTA are G(s) = {(1,TGC), (2,GCC), (3,CCC), (4,CCT), (5,CTA)}.
Approximate subsequence matching using q-grams is based on the following intuition: if two sequences are similar, then they share a certain number of q-grams. For the Hamming distance this idea has been formalized as ‘count filtering’ (18).
Count filtering
If two sequences r and s are within Hamming distance d, then their q-gram sets G(s) and G(r) share at least the following number of q-grams:
| (1) |
The lower bound T on common grams in the above equation is based on the observation that a character substitution can affect at most q grams, and hence d substitutions can affect at most d * q grams.
Gram filtering variants
Other variants of filtering use multiple patterns based on the pigeonhole principle (19), non-overlapping grams (6), gapped grams (20,21) or variable-length grams (22).
Q-gram inverted index
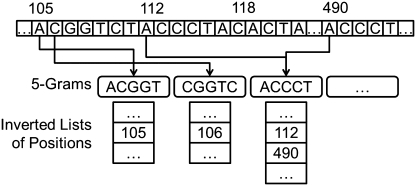
Finding substrings in a reference sequence that share at least T q-grams with a given read can be facilitated with an inverted index on q-gram positions, explained as follows for Hamming distance. Figure 1 shows an example of a 5-gram inverted index. To map a read ACGGTCTTCCCTACGGT within Hamming distance d = 2 and T = 17 − 5 + 1 − 2*5 = 3, we first look up the read's 5-grams in the inverted index. Notice that only the grams ACGGT and CGGTC (underlined in the read) are present in the index. We traverse their inverted lists, and normalize each element relative to the position of the corresponding gram in the read. For example, the 5-gram CGGTC appears at position 2 in the read, so the relative position of the element on CGGTC's inverted list is 106 − 2 + 1 = 105. In this way, we can count how many of the read's grams are contained in substrings of the reference sequence starting at a fixed position (position 105, in this example). The gram ACGGT appears twice in the read, and we treat each occurrence as a separate list. Its appearance at position 1 yields a normalized list of {105 − 1 + 1 = 105, 118 − 1 + 1 = 118}, and a list {105 − 14 + 1 = 92, 118 − 14 + 1 = 105} for position 14. Next, we count the occurrences of each element on the normalized lists. The positions 92 and 118 are pruned according to the count filter, because their occurrences do not meet the lower bound of T = 3. Position 105 has a count of 3, and therefore it is a candidate answer whose Hamming distance to the read still needs to be computed.
Figure 1.
Excerpt of a reference sequence and a portion of its 5-gram inverted index. The inverted lists of the 5-grams ACGGT, CGGTC and ACCCT are shown, each containing a sorted list of positions in the reference sequence at which the respective 5-gram appears.
Performance issues of q-gram counting
For a long reference sequence, the above approach for mapping reads could suffer from the following performance problems:
CPU intensive gram counting: using all of a read's relevant inverted lists for gram counting can be expensive if there are many of them, or if some of the lists are very long. The cost of gram counting is directly related to the total number of elements on those inverted lists.
Cache misses during candidate verification: CPU caches are very small but fast memories that act as intermediaries to main memory. Transferring new data into the cache (a ‘cache miss’) can last hundreds of CPU cycles. Accessing random portions of a very long sequence has low locality, and hence it can become a performance bottleneck due to cache misses.
In the ‘Materials and Methods’ section, we present a new technique to judiciously select a few grams from a read to overcome the first performance issue. To tackle the second issue, we develop a novel filter based on augmenting the inverted lists with additional information.
Judiciously selecting Q-grams from reads
In this section, we aim to reduce the cost of processing inverted lists to generate candidate mappings. We present a new technique to judiciously select a few grams from a read to minimize the number of corresponding inverted lists, and the number of inverted-list elements that need to be considered for mapping the read.
Existing methods for q-gram selection
First, we briefly summarize the main ideas already developed in the literature to reduce the number of grams.
Prefix filter
Consider a read s with a gram set G(s) and the lower bound T in Equation (1). Let the ‘prefix gram set’ of read s be the |G(s)| − T + 1 least frequent grams in G(s), i.e. with the shortest inverted lists. A candidate mapping must share at least one gram with the prefix gram set of s, because otherwise it could only reach a maximum count of T − 1 (23).
Shortened prefix
Xiao et al. (24) use the positions of q-grams to reduce the size of the prefix gram set in the context of edit distance-based joins. Their solution imposes a global ordering on the grams based on their frequency to achieve a consistent notion of prefix gram set across all strings, and constructs a q-gram inverted index on prefix grams on-the-fly. To improve the index-construction time (the dominating factor), they reduce the prefix gram set of each string to the first i ≤ |G(s)| − T + 1 grams in the global ordering which contain d + 1 non-overlapping grams, where d is a given edit distance threshold. Inspired by their work, we develop a new method to judiciously select a few q-grams for probing our index.
Optimal q-gram prefix selection
Recall that a substitution can affect at most q grams [Equation (1)]. The insight that these q affected grams must be overlapping lead us to develop the following lower bound based on the positions of q-grams.
Lemma 1. (Position-Based Prefix) —
Given a sequence s and its q-gram set G(s), let P be a subset of d + 1 non-overlapping q-grams from G(s). Then each sequence within Hamming distance d of s must have a gram in P.
The intuition of the lemma is as follows. A substitution at position p can affect at most q overlapping grams, namely those starting from a position in [p − q − 1, p]. Since non-overlapping grams are at least q positions apart from each other, d substitutions can affect at most d non-overlapping grams. Among d + 1 non-overlapping grams, at least one gram remains unaffected by d substitutions. Since this analysis is true for every subset P of d + 1 non-overlapping grams of G(s), we can generalize the lower bound as follows.
Lemma 2. (Generalized Position-Based Prefix) —
A sequence r within Hamming distance d of sequence s must share at least k grams with every subset of d + k non-overlapping q-grams of s.
Optimal prefix selection
We want to select a set of prefix grams that is optimal in the sense that (i) it refers to a minimum number of inverted lists and (ii) those inverted lists have the minimum total number of elements. The position-based prefix described above satisfies (i), but a read could have many possible sets of d + 1 non-overlapping q-grams. To satisfy (ii) we develop the following dynamic programming algorithm to select that set of d + 1 non-overlapping q-grams from the read (Supplementary Data), which minimizes the total number of corresponding inverted-list elements.
Subproblem
Let 1 ≤ i ≤ d + 1 and 1 ≤ j ≤ |G(s)| − d*q be two integers. Let M(i, j) be a lower bound on the sum of the lengths of the inverted lists of i non-overlapping grams starting from a position no greater than j + (i − 1)*q. Our goal is to compute M(d + 1, |G(s)| − d*q).
Initialization
Let L[p] denote the inverted list corresponding to the q-gram at position p, and L[p].len its length. We initialize the row M(0, j) to zero, and the column and M(i, 0) to infinity.
Recurrence function
| (2) |
Example —
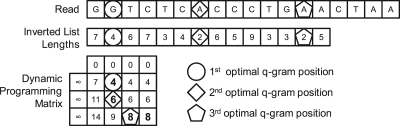
Figure 2 shows an example of finding an optimal q-gram prefix of a read s = GGTCTCACCCTGAACTAA, gram length q = 5 and Hamming distance threshold d = 2. An optimal set of positions of the d + 1 = 3 non-overlapping q-grams are highlighted, including the cell in the matrix from which the q-gram position can be inferred. For example, ‘4’ is the minimum value in the first row, and since its first appearance is in Column 2, we can infer that the first optimal q-gram position is at 2 + (1 − 1)*q = 2.
Figure 2.
Example of our dynamic programming algorithm for finding an optimal set of prefix grams for a read GGTCTCACCCTGAACTAA, gram length q = 5 and Hamming distance d = 2. Optimal gram positions are highlighted with a circle, a diamond, and a pentagon.
Complexity
The complexity of the algorithm for finding an optimal prefix for a read s with length |s| and Hamming distance d is O(|s|d). Notice that the actual cost of the algorithm decreases when we increase d and q, because there are fewer sets of non-overlapping grams to choose from. For example, in Figure 2 we need to populate 12 matrix cells for a read of length 18. Our experiments show that the algorithm performs very well for good d and q values in real data sets.
Cache-efficient filtering of candidate mappings
A straightforward implementation of the q-gram-based filter and verification procedure can lead to poor performance due to cache misses. Since the reference sequence is often much larger (e.g. 3 GB for the human genome) than the CPU cache (e.g. 12 MB L3 cache for an Intel Xeon X5670), each verification of a candidate position likely causes a cache miss. Since most candidate positions typically are false positives, paying a cache miss for fetching an irrelevant substring of the reference sequence is very wasteful.
In this section, we present a cache- and compute-efficient filter for removing false-positive candidate mappings without accessing the reference sequence. The main idea is to augment the inverted lists with additional filtering data, such that it is in the CPU cache during the traversal of an inverted list.
Mapping q-gram neighborhoods to bitvectors
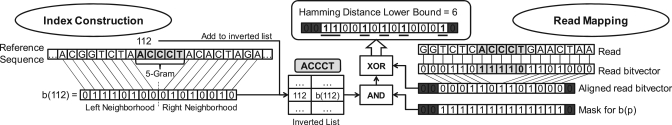
We attach to each inverted-list element an encoding of its corresponding neighboring characters in the reference sequence using 1 bit per character. The left-hand side of Figure 3 illustrates this procedure on an exemplary 5-gram at position 112 in a reference sequence. We use 16 bits to encode the eight characters to the left and right of the 5-gram ACCCT. Since we only access ACCCT's inverted list if ACCCT is also contained in the read we are processing, it is unnecessary to include ACCCT itself in the bitvector. The size of the bitvectors is a tunable parameter, and we use 16 bits for this example.
Figure 3.
Adding bitvectors to a q-gram inverted index (left), and pruning candidates mappings with them (right), using a mapping of A, T ⇒ 0, and C, G ⇒ 1. The left portion shows how to encode the left and right neighborhood of a 5-gram ACCCT at position 112 in the reference sequence as a 16-bit bitvector, mapping A, T ⇒ 0, and C, G ⇒ 1. Both the position 112 and its bitvector b(112) are inserted into ACCCT's inverted list. The right portion shows how to prune candidate mappings of a read GGTCTCACCCTGAACTAA from ACCCT's inverted list. The dark gray boxes indicate invalid bits we must ignore, based on ACCCT's position in the read. The light gray boxes highlight the matching q-gram ACCCT.
It might seem that using a single bit per character reduces the filtering capability by 50% because we map strings of a four-letter alphabet (A, C, T, G) to a two-letter alphabet (0, 1). But, it is well known that not every character substitution is equally likely on real data (25). For example, Table 1 shows the frequency of character substitutions we gathered in a simple experiment, as follows. For each of the 35 bp reads we computed the optimal gram prefix and traversed the corresponding inverted lists to obtain candidate mapping positions. Next, we recorded the frequency of character substitutions during the verification of these candidate positions. Since ‘A → G’ and ‘T → C’ are the most frequent substitutions, our results suggest the following encoding: A, T ⇒ 0 and C, G ⇒ 1, such that the characters of the most frequent substitutions get different bit values.
Table 1.
Frequency of character substitutions using 2 million 35 bp reads on hg18
| Read character | hg18 character | No. of substitutions |
|---|---|---|
| A | T | 687 276 051 |
| A | G | 1 382 950 075 |
| A | C | 559 937 841 |
| T | G | 395 839 922 |
| T | C | 1 232 657 183 |
| G | C | 393 616 199 |
The results suggest a mapping: A, T ⇒ 0 and C, G ⇒ 1.
Apart from representing more characters with a fixed number of bits, our bitvector encoding also allows CPU-efficient filtering of candidate positions, as follows:
Candidate filtering using bitvectors
Let us revisit the example in Figure 2. After computing an optimal gram prefix, we need to traverse their corresponding inverted lists to find candidate mappings. Suppose we begin with the list of gram ACCCT, since it is the shortest list of those in the optimal prefix.
The right-hand side of Figure 3 illustrates how we use the bitvectors for filtering. Before scanning ACCCT's inverted list, we map the read to a bitvector. Next, we ‘shift away’ the bits of the matching q-gram ACCCT in the read's bitvector to align the positions of its bits with those in the reference-sequence bitvectors. Recall that we omitted the q-gram itself when generating bitvectors for the reference sequence. This shifting produces invalid bits at both ends of the read bitvector shown as dark boxes. These invalid bits represent portions of the bitvectors from the reference sequence that we cannot use for pruning candidates. For example, since there are only six characters to left of ACCCT in the read, we should ignore two of the 8 bits representing the left neighborhood of ACCCT's occurrences in the reference sequence. We generate a bitmask to remove those invalid bits from each bitvector in ACCCT's inverted list.
Now that we have aligned the read's bitvector with the bitvectors in the inverted list, and generated a bitmask to remove invalid bits from those bitvectors, we start scanning ACCCT's inverted list. For each candidate position, we use the read's bitvector and the candidate's bitvector to compute a lower bound on the Hamming distance between their corresponding original sequences, as follows. First, we do a ‘bitwise-AND’ operation between the bitmask and the candidate's bitvector. Then, we do a ‘bitwise-XOR’ operation between the resulting bitvector and the read's bitvector to produce a final bitvector. In the final bitvector, a bit is set to 1 if and only if the original character at the corresponding position in the read is different from the corresponding character in the reference sequence. We prune a candidate if the number of 1-bits in the final bitvector exceeds our Hamming distance threshold. We determine the number of 1-bits in the final bitvector with a single CPU instruction, popcount, supported by most modern CPUs.
In summary, our new bitvector-filtering technique eliminates candidate mappings without accessing the reference sequence, thus avoiding expensive cache misses. In addition, our filter only requires a handful of CPU instructions per inverted-list element, namely bitwise-AND, bitwise-XOR, popcount and a final comparison with the Hamming distance threshold.
Supporting insertions and deletions
Allowing insertions and deletions (indels) is important for mapping longer reads, because both sequencing errors and genetic variations can result in the deletion/insertion of bases and the chance of this happening increases as reads become longer. However, finding mappings with indels is computationally more challenging. Hobbes implements the following two methods for mapping reads with indels.
Hamming distance tends to be sufficient for shorter reads, whereas edit distance becomes important for long reads. Ultimately, the user must decide whether the added benefit of edit distance offsets its computational cost.
Non-heuristic mapping
To find all mappings of a read while allowing indels, we can use the optimal prefix grams as described in the preceding section for generating candidate mapping positions. However, the bitvector filter mentioned above is specific to Hamming distance. A similar filter for indels is possible, and we leave this direction for future work. To verify a candidate position, we conceptually consider those substrings with all possible starting and ending positions (based on the edit-distance threshold), and compute their edit distances to the read. For each candidate position, we report the substring with the lowest edit distance. This approach tends to be very slow, and the following heuristics can significantly improve the mapping performance.
Seed extension approach
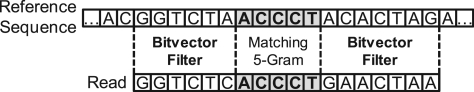
Again, we begin with the optimal prefix grams for finding candidate positions. Next, we introduce two heuristics to improve performance: first, we fix the starting position for verification, but shift it to the left once, if the initial position yielded no match. Second, we apply the bitvector filter to the neighborhood of matching grams in the reference sequence, as show in Figure 4.
Figure 4.
Seed extension approach with indels. We prune candidate positions by applying the bitvector filter on the neighborhood of matching grams.
Since most differences are due to substitutions, our intuition is that if the neighborhood already has a high Hamming distance to the corresponding substring in the read, then the candidate is probably not a match. The filter could remove valid mappings if those apparent substitutions are caused by inconveniently located indels. On the other hand, this effect is mitigated by using multiple grams. It is somewhat unlikely that the neighborhoods of all those grams have a high Hamming distance for true mappings. The bitvector-filter threshold on the neighborhood is a tuning parameter. We found that by setting it to 2/3 of the original edit distance, we capture most mappings while retaining high speed.
Letter count filter
Hobbes uses the letter-count filter described in Ref. (27) to quickly detect whether the two sequences can be within a given edit-distance threshold during the verification step. The main idea of the filter is to count the number of occurrences of each character in both sequences, and establish a lower bound on the edit distance between the two sequences using the differences of those character counts, as follows. We divide the frequencies of all base pairs into two groups. The first group contains the base pairs that are more common in the read, and the second group contains those that are more common in the candidate. For each group, we create a sum of the frequency differences of corresponding base pairs in that group, denoted by Δ1 and Δ2, respectively. The grouping ensures that an edit operation can change each Δ by at most 1, establishing max(Δ1, Δ2) as a lower bound on the edit distance between two sequences. For example, consider sequences s1 = AAACCTG and s1 = CCCCTTGG, Δ1 = (3 − 0) = 3 (A is more frequent in s1) and Δ2 = (4 − 2) + (2 − 1) + (2 − 1) = 4 (C, G and T are more frequent in s2). Based on the letter count filter, a lower bound on the edit distance between s1 and s2 is max(3, 4) = 4.
To prune candidates that survive the letter count filter, we also employ standard q-gram counting.
Supporting paired-end alignment
Hobbes supports the alignment of paired-end reads. Many next-generation sequencing technologies provide the user with paired-end reads that contain extra information about the relative position of two reads with respect to each other. To align paired-end reads, Hobbes initially considers each read of a pair separately and finds the set of candidate locations for each read. For example, given a read pair (r1, r2), Hobbes first finds the candidate location sets C1 and C2 corresponding to read r1 and r2, respectively. During the next step, for each candidate c1 ∈ C1 Hobbes performs verification only if there is a c2 ∈ C2 that satisfies the paired-end alignment constraints (appropriate orientation and distance) with respect to c1.
Hobbes provides the option of reporting the alignments of each read in a paired-end read separately if no paired alignments are found.
Implementation details
In this section, we discuss implementation details of Hobbes.
Treatment of N characters
We ignore q-grams with at least one N character. As a consequence, our inverted index does not contain those q-grams. When generating q-grams for a read, we may generate fewer than d + 1 non-overlapping q-grams (since we ignore q-grams with N characters). In our current implementation, we cannot find any mapping for such a read, although we could rely on those other q-grams to find some mappings (we leave this for future work). In all other cases, we treat N's as mismatches. Note that we can deal with reads containing N's (as long as there are enough non-overlapping q-grams), and a read can map to substrings in the reference sequence containing N's even though the inverted index does not contain q-grams with N's (we may reach such a position via a different, regular q-gram).
Hashing q-grams
We employ a collision-free hash function to map q-grams to integers, as follows. Each character {A, C, T, G} is encoded as 2 bits, and the concatenation of all such 2-bits corresponding to a q-gram forms the q-gram's hash code. With this scheme, 32-bit integers can support hashing q-grams up to length 32/2 = 16. For longer q-grams, we use 64-bit integers.
Hamming-distance verification
Before computing the actual Hamming distance between two sequences using a character-by-character comparison, we do a significantly faster chunk-by-chunk comparison, typically with a chunk size of 4 bytes. If more than d chunks differ, then the two sequences cannot be within Hamming distance d, and we avoid the character-by-character comparison.
Edit-distance verification
After a candidate passes the letter count filter, we compute the real edit distance between two sequences using SeqAn's (27) implementation of Myer's bit-parallel algorithm (28).
Cache prefetching during verification
Our bitvector filter can dramatically reduce the number of cache misses by pruning false positives without accessing the reference sequence. However, the number of surviving candidates can still be in tens of thousands. Once a candidate has passed the bitvector filter, we cannot avoid a cache miss for the distance verification. But we can mitigate the cost by prefetching a future candidate's data into the cache, thus overlapping the verification of the current candidate and the data transfer from memory to cache for the future candidate. We have found that for our CPU architecture and our set of experiments, the best performance is achieved when prefetching the data for candidate number c + 2 before verifying candidate number c.
RESULTS
Implementation and setup
We implemented Hobbes in C++, and compiled it with GCC 4.4.3. All experiments were run on a machine with 96 GB of RAM, and dual quad-core Intel Xeons X5670 (8 cores total) at 2.93 GHz, running a 64-bit Ubuntu OS. Note that Hobbes performs best on CPUs that support the popcount instruction. We used GCC's built-in functions for popcount and cache prefetching. Hobbes is freely available at http://hobbes.ics.uci.edu, and can output its results in SAM format for analysis with SAMTools.
Other read mappers and data
We compared Hobbes with the following packages:
Bowtie (3) is a BWT-based short read aligner, and is efficient for finding few mappings per read (1 by default) with a very small memory footprint. Bowtie performs a depth-first search on the index and stops when the first qualified mapping is found.
BWA (4) is also a BWT-based program, and supports gapped alignment, while Bowtie does not. BWA uses a backtracking search similar to Bowtie's to handle mismatches. By default, BWA adopts an iterative strategy to accelerates its performance, at the price potentially losing mappings. To report all feasible mappings, we disable the iterative search (−N option) in our experiments.
mrsFast (13) and mrFast-CO (14) are recent gram-based packages for gapped and ungapped alignment, respectively. They index both the reference genome and the reads. mrsFAST uses an efficient all-to-all list comparison algorithm, while mrFAST-CO follows a seed-and-extend strategy.
RazerS2 (12) builds a gram-based index on the reads, and performs gram counting while scanning over the reference sequence. RazerS2 has been reported to be very accurate in finding all mappings for typical read lengths. We set RazerS2's max − hit parameter to 300 000 000 to get all mappings.
Data
We conducted our experiments using reads with 35, 51, 76 and 100 bp. The 35 bp reads are taken from the YH database (http://yh.genomics.org.cn), the 51 and 76 bp reads come from the DDBJ DNA Data Bank of Japan (DDBJ) repository (ftp://ftp.ddbj.nig.ac.jp) with entry DRX000359 and DRX000360, respectively, and the 100 bp reads are from specimen HG00096 of the 1000 genome project (http://www.1000genomes.org/data). In all cases, we used the human genome with NCBI HG18 as our reference genome. As we do alignments read by read, the performance of Hobbes is almost linearly proportional to the number of reads. So in the following we mainly test the performance of Hobbes and other read mappers using datasets with 500K reads randomly chosen from the above-mentioned databases.
Index construction and memory footprint
We use an inverted index of overlapping q-grams on the reference genome. As described earlier, to avoid cache misses, we augment the inverted lists with bit vectors representing the neighboring characters of the corresponding q-grams. The index size is linearly dependent on the size of the reference sequence and the chosen bit-vector size. By default, Hobbes uses 16-bit vectors, resulting in a total index size of 21 GB for hg18. We used bitvector sizes of 16 and 32 bits for our experiments with edit and Hamming distance, respectively. Using a single thread, it took Hobbes 20 min to build an index on hg18, whereas Bowtie and BWA needed 114 and 56 min, respectively. In addition, Hobbes has a tight-knit multithreaded framework that parallelizes both indexing and mapping. On multicore machines, users can build an index as large as hg18 in a few minutes. Since Hobbes does alignment read by read, its memory requirement is independent on the number of input reads.
Results using hamming distance
All mappings
We configured the packages to find all mappings per read. Table 2 shows the mapping time, the fraction of reads with at least one mapping, and the total number of mappings for various read lengths and hamming distances. We observe that Hobbes is up to five times faster than other packages (even 100 times faster than RazerS2), while producing comparable mappings. For example, on 35 bp reads, Hobbes is more than four times faster than Bowtie*, which is the fastest among all other listed programs; on 51 bp and 76 bp reads, Hobbes is about three times faster than our closest competitor BWA. Moreover, Hobbes maps slightly more reads than BWA in that setting. Among the tested programs, mrsFAST and RazerS2 consistenly achieved the best mapping quality. Hobbes delivers a similar quality, while outperforming mrsFAST and RazerS2 in mapping speed by a factor of up to 10 and 200, respectively.
Table 2.
Results of mapping 500K single-end reads against HG18
| Read length (Hamming) | 35 bp (two errors) |
51 bp (three errors) |
76 bp (three errors) |
76 bp (four errors) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Algorithm | Time (h:m) | Reads mapped (%) | Total mappings (million) | Time (h:m) | Reads mapped (%) | Total mappings (million) | Time (h:m) | Reads mapped (%) | Total mappings (million) | Time (h:m) | Reads mapped (%) | Total mappings (million) |
| Bowtie* | 0:28 | 76.61 | 492.6 | 0:34 | 91.93 | 317.1 | 0:16 | 91.44 | 73.4 | NA | NA | NA |
| Bowtie | 0:54 | 76.61 | 492.6 | 0:50 | 91.93 | 317.1 | 0:18 | 91.44 | 73.4 | NA | NA | NA |
| BWA | 0:30 | 76.61 | 492.6 | 0:24 | 91.61 | 277.6 | 0:10 | 91.36 | 71.1 | 0:18 | 92.47 | 115.2 |
| mrsFAST | 0:43 | 76.61 | 492.6 | 0:59 | 91.93 | 317.1 | 0:50 | 91.44 | 73.4 | 1:10 | 92.69 | 127.2 |
| RazerS2 | 6:38 | 76.61 | 488.5 | 7:58 | 91.93 | 316.9 | 8:58 | 91.44 | 73.4 | 8:08 | 92.69 | 127.2 |
| Hobbes | 0:06 | 76.61 | 492.6 | 0:08 | 91.93 | 317.1 | 0:03 | 91.44 | 73.4 | 0:07 | 92.69 | 127.2 |
We used Bowtie in its default mode and an optimized mode (Bowtie*) where we set offrate = 0 for maximum speed. Reads mapped: the fraction of reads with at least one mapping; Total mappings: the total number of mapped locations in the reference; NA: Bowtie does not support >3 mismatches.
Few mappings
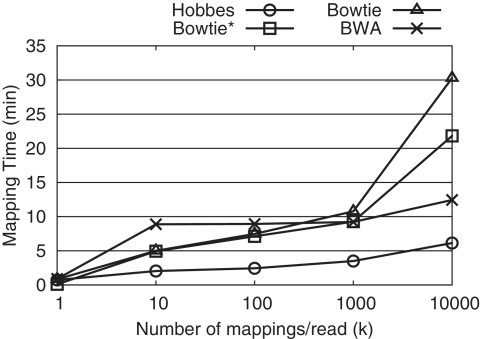
Some applications may require all mappings per read, and others only a few mappings. Most tools are optimized for only one of those cases. For example, Bowtie focuses on finding a few mappings per read, and is very good at it. Therefore, the comparison in Table 2 somewhat disfavors those packages not designed for finding all mappings. To accommodate the few-mappings use case, Hobbes efficiently supports finding any number of mappings. Figure 5 shows the mapping times of BWA, Bowtie and Hobbes for a varying number of requested mappings per read (k). We see that Hobbes performs comparably to Bowtie for very small k, but as k increases Hobbes begins to outperform other packages by a growing margin.
Figure 5.
Maximun number of mappings k per read versus mapping time on 51 bp reads with Hamming distance 3. We omitted RazerS2 due to its long mapping time, and mrsFAST because it only supports finding all mappings.
Results using edit distance
Supporting edit distance is significantly more challenging than Hamming distance, and is becoming increasingly important as reads get longer and tend to contain indels. Hobbes implements a seed extension approach to align reads within a given edit distance threshold to take advantage of the two optimization strategies we have developed (‘Materials and Methods’ section). Although unlike the Hamming distance mapping, the seed extension approach cannot guarantee to find all correct mapping locations, we will show that by using multiple seeds the mapping quality of Hobbes can be achieved to be nearly optimal. Bowtie does not support edit distance, so we compared Hobbes with BWA, mrFast-CO and RazerS2. We configured the packages to find all mappings per read.
All mappings
Table 3 lists our experimental results. We observed that Hobbes is about twice as fast as BWA and mrFast-CO, and over seven times faster than RazerS2. Notice that the performance gap increases with longer reads and higher edit distances. On 76 bp reads, RazerS2 could map slightly more reads than the other packages; however, the mapping took an order of magnitude more time than Hobbes. The speed of mrFast-CO and BWA was similar, mapped slightly fewer reads. Hobbes was about twice as fast as mrFast-CO and BWA while mapping more reads. On 100 bp reads, RazerS2 had the best quality but a comparably slow mapping speed; BWA become slower than RazerS2 and lost quality at the same time; the quality of Hobbes was very close to RazerS2. Compared with mrFast-CO, Hobbes could map more reads and was about 1.5 times as fast. In addition, the current version of mrFast-CO is limited to edit distance 6, which is problematic for mapping even longer reads, while Hobbes does not have such a limitation.
Table 3.
Results of mapping 500K single-end reads against HG18
| Read length (edit distance) | 76 bp (four errors) |
100 bp (six errors) |
||||
|---|---|---|---|---|---|---|
| Algorithm | Time (h:m) | Reads mapped (%) | Total mappings (million) | Time (h:m) | Reads mapped (%) | Total mappings (million) |
| BWA | 02:33 | 94.06 | 141.1 | 22:54 | 92.16 | 79.4 |
| mrFast-CO | 02:45 | 94.28 | 142.3 | 03:47 | 92.39 | 96.3 |
| RazerS2 | 12:26 | 94.32 | 143.6 | 17:14 | 92.50 | 96.4 |
| Hobbes | 01:46 | 94.30 | 145.8 | 02:48 | 92.47 | 100.7 |
Evaluation on simulated data
We simulated reads from the human genome using the wgsim program (http://github.com/lh3/wgsim), and then ran Hobbes to map those reads back to the same human genome. We used the default setting of wgsim, in which the mismatched bases are chosen randomly with a mismatch rate of 2% per base, and 15% of polymorphisms are indels with their sizes drawn from a geometric distribution with mean of 1.43.
Since Hobbes is only guaranteed to be exact for Hamming distance, we use the simulated data to examine the mapping quality of Hobbes using edit distance. We use two metrics to measure the accuracy of mapping: one is the fraction of reads with at least one mapping and the other is the mapping error rate. We say a read is aligned correctly if the true location of the read starts at the same location as one of its mappings. The mapping error rate is defined to be the fraction of mapped reads that are aligned incorrectly.
Table 4 shows the performance of Hobbes as compared to other programs in the case of edit distance. In terms of speed, Hobbes is the clear winner—about 3.5 times faster than BWA and 6 times faster than RaserS2 for 100 bp reads. In terms of the fraction of reads mapped, Hobbes is slightly less than the best program, RaserS2, but the margin is small with a difference of only 0.02% for both 76 bp and 100 bp reads. Hobbes achieves the best mapping error rate of 0.22%. These results demonstrate that although Hobbes sacrifices some accuracy for speed, its mapping quality is comparatively better than the other packages tested.
Table 4.
Results of mapping 500K simulated reads
| Read length (edit distance) | 76 bp (four errors) |
100 bp (six errors) |
||||
|---|---|---|---|---|---|---|
| Algorithm | Time (h:m) | Reads mapped (%) | Error rate (%) | Time (h:m) | Reads mapped (%) | Error rate (%) |
| BWA | 01:22 | 96.05 | 2.17 | 07:55 | 97.09 | 1.78 |
| mrFast-CO | 02:15 | 97.84 | 3.43 | 03:22 | 99.43 | 3.63 |
| RazerS2 | 10:08 | 97.90 | 0.98 | 12:59 | 99.50 | 1.15 |
| Hobbes | 01:08 | 97.88 | 0.22 | 02:20 | 99.48 | 0.22 |
Paired-End alignment
We compared Hobbes with other state-of-the-art packages using paired-end reads.
Hamming distance
For Hamming distance, Hobbes is guaranteed to find all correct mappings. Table 5 summarizes the results of Hobbes and several programs for mapping reads of various lengths and Hamming distance thresholds to the human reference genome. We focus on the speed of mapping since the quality of mapping is similar among different programs. Hobbes is close to Bowtie in the 35 bp case, but substantially outperforms Bowtie (11 times faster) when the read length increases to 76 bp and the Hamming distance increases to 3. Moreover, for the 100-bp case with four errors, Bowtie was unable to provide answers because of too many backtracking steps required in the BWT-based algorithm. Hobbes is 24 times faster than the second-best program, mrsFAST in this case. These results suggest that Hobbes outperforms other programs in the Hamming distance case, especially for long reads while allowing many errors.
Table 5.
Results of mapping 250K paired-end reads against HG18
| Read length (Hamming) | 35 bp (two errors) |
76 bp (three errors) |
100 bp (four errors) |
|||
|---|---|---|---|---|---|---|
| Algorithm | Time (h:m) | Mapped pairs (%) | Time (h:m) | Mapped pairs (%) | Time (h:m) | Mapped pairs (%) |
| Bowtie | 0:02 | 84.58 | 0:18 | 80.06 | NA | NA |
| Bowtie* | 0:20 | 84.68 | 0:25 | 80.06 | NA | NA |
| mrsFAST | 0:42 | 84.66 | 0:42 | 80.06 | 0:52 | 83.40 |
| Hobbes | 0:04 | 84.68 | 0:02 | 80.06 | 0:02 | 83.44 |
Mapped pairs: the fraction of read pairs mapped with at least one location satisfying the distance and orientation constraint. Bowtie*: running with-y option. Some entries contain NA because Bowtie does not support >3 mismatches.
Edit distance
Our performance results are summarized in Table 6. The fraction of read pairs that can be aligned to the reference sequence is used as a surrogate of mapping quality. In terms of the fraction of mapped pairs, Hobbes is similar to RaserS2, both of which are significantly better than other programs. In terms of mapping speed, Hobbes is clearly the fastest in all three cases with big margins—22 times faster than BWA, 3 times faster than mrFAST and 15 times faster than RaserS2 in the 100 bp case.
Table 6.
Results of mapping 250K paired-end reads against HG18
| Read length (edit distance) | 35 bp (two errors) |
76 bp (four errors) |
100 bp (six errors) |
|||
|---|---|---|---|---|---|---|
| Algorithm | Time (h:m) | Mapped pairs (%) | Time (h:m) | Mapped pairs (%) | Time (h:m) | Mapped pairs (%) |
| BWAa | 1:02 | 84.93 | 2:15 | 84.45 | 22:48 | 88.14 |
| mrFast-CO | 2:32 | 78.02 | 2:32 | 81.61 | 03:36 | 85.96 |
| RazerS2 | 3:57 | 84.53 | 10:52 | 84.49 | 15:02 | 88.18 |
| Hobbes | 0:26 | 85.35 | 0:21 | 84.60 | 0:50 | 88.37 |
aThe raw number of mapped reads output by BWA are higher with 90.76, 92.60 and 92.66 for 35, 76 and 100 reads, respectively. We removed those mappings that violated the edit distance constraints.
Application in RNA-seq abundance analysis
In RNA-seq applications, it is important to find all mapping positions of a read for quantifying the expression level of a particular gene isoform, due to the occurrence of homologous genes and multiple RNA isoforms originating from the same gene. To show the importance of finding all mappings, we performed a transcript abundance analysis. We used the UCSC genes on the mm9 mouse (NCBI build 37) assembly as target transcripts; both 76 bp paired-end RNA-seq reads were downloaded from Gene Expression Omnibus (Series GSE20846). We compared Bowtie and Hobbes for finding all mappings. The results in Table 7 show that Hobbes was 17–20× faster than Bowtie, and could even map slightly more reads.
Table 7.
Results of mapping 76 bp RNA-seq reads against 55 419 known mouse transcripts, using Hamming distance 3 and a minimum and maximum insert size of 76 bp and 800 bp, respectively
| Reads | SRR047951 (21.8 million) |
SRR047953 (20 million) |
||||
|---|---|---|---|---|---|---|
| Algorithm | Time (h:m) | Reads mapped (%) | Total mappings (millions) | Time (h:m) | Reads mapped (%) | Total mappings (millions) |
| Bowtie | 09:53 | 40.28 | 18.399 | 09:12 | 42.97 | 18.883 |
| Hobbes | 00:35 | 40.29 | 19.157 | 00:27 | 42.99 | 19.393 |
Next, we piped the result of Hobbes directly to eXpress (http://bio.math.berkeley.edu/eXpress), which is a streaming tool for quantifying the abundances of a set of target sequences from sampled subsequences. eXpress estimates the relative abundance by computing the fragments per kilobase per million (FPKM) value. We compared the FPKM values when finding at most k = {1, 10, 100} mappings, with the FPKM when finding all mappings. To quantify their variation, we computed the ratio of the k-mappings FPKM to the all-mapppings FPKM (or its reciprocal). The results are shown in Table 8. We found that among 55 419 target transcripts, the percentage of transcriptions whose ratio was above 1.5 for k = {1, 10, 100} was 42.39, 0.92 and 0.07%, respectively; there was a lot of variability for k = 1, meaning the corresponding FPKM value is a poor estimator. The variability reduced as we increased k to 100. We included the corresponding scatter plots in Supplementary Data.
Table 8.
Transcripts with FPKM ratio above 1.5 and 1.2 on 76 bp RNA-seq reads within Hamming distance 3 against 55 419 known mouse transcripts
| k versus all FPKM ratio | Transcriptions above ratio (%) |
|
|---|---|---|
| 1.5 | 1.2 | |
| k = 1 versus All | 42.39 | 47.12 |
| k = 10 versus All | 00.92 | 01.42 |
| k = 100 versus All | 00.07 | 00.18 |
DISCUSSION
Hobbes efficiently supports Hamming and edit distance while finding all mappings or few mappings per read. Our experiments have shown Hobbes to be just as accurate but significantly faster than current read mapping programs.
Hobbes has a large memory footprint (26 GB in our experiments), but we believe its speed and mapping quality outweigh that drawback, especially considering today's low memory prices. We plan to reduce Hobbes memory requirement, possibly via compression, or by partitioning our index performing the read mapping one partition at a time (mrsFast and mrFast-CO follow this approach).
Given today's trend toward massively multi-core CPUs, we believe that good multithread support is of paramount importance. Both Hobbes' index-construction and read-mapping procedures support multiple threads and scale well (Supplementary Data). Some packages like mrsFast, mrFast-CO and RazerS2 do not support multithreading at all (we could not find this feature in their manuals). Other packages have certain limitations, e.g. in BWA only one of the two steps during read mapping is parallelizable.
We plan to further optimize Hobbes for the edit distance-based mapping, and account for read quality scores.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intel Labs Academic Research Office; National Science Foundation (NSF) (DBI-0846218) in part. Funding for open access charge: NSF (DBI-0846218).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jacob Biesinger and Daniel Newkirk for testing Hobbes and providing valuable feedback. We also thank Knut Reinert and the rest of the SeqAn team for helpful discussions.
REFERENCES
- 1.Burrows M, Wheeler D. 1994. A block sorting lossless data compression algorithm. Technical Report 124, Palo Alto, CA: Digital Equipment Corporation. [Google Scholar]
- 2.Ferragina P, Manzini G. Proceedings of the twelfth annual ACM-SIAM symposium on Discrete algorithms. Society for Industrial and Applied Mathematics; 2001. An experimental study of an opportunistic index; pp. 269–278. [Google Scholar]
- 3.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short dna sequences to the human genome. Genome Biol. 2009;10:r25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. Soap2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 6.Ning Z, Cox AJ, Mullikin JC. Ssaha: a fast search method for large dna databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Ruan J, Durbin R. Mapping short dna sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith A, Xuan Z, Zhang M. Using quality scores and longer reads improves accuracy of solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H, Zhang Z, Zhang M, Ma B, Li M. Zoom! zillions of oligos mapped. Bioinformatics. 2008;24:2431. doi: 10.1093/bioinformatics/btn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rumble S, Lacroute P, Dalca A, Fiume M, Sidow A, Brudno M. Shrimp: accurate mapping of short color-space reads. PLoS Comput. biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weese D, Emde A-K, Rausch T, Döring A, Reinert K. Razers-fast read mapping with sensitivity control. Genome Res. 2009;19:1646–1654. doi: 10.1101/gr.088823.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hach F, Hormozdiari F, Alkan C, Hormozdiari F, Birol I, Eichler EE, Sahinalp SC. mrsfast: a cache-oblivious algorithm for short-read mapping. Nat. Methods. 2010;7:576–577. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. Personalized copy-number and segmental duplication maps using next-generation sequencing. Nat. Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Y, Xu Y, Zhang Q, Tsui K, Yuan Y, Norris C, Jr, Liang S, Liang H. Bm-map: Bayesian mapping of multireads for next-generation sequencing data. Biometrics. 2011;67:1215–1224. doi: 10.1111/j.1541-0420.2011.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung D, Kuan P, Li B, Sanalkumar R, Liang K, Bresnick E, Dewey C, Keleş S. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of chip-seq data. PLoS Comput. Biol. 2011;7:e1002111. doi: 10.1371/journal.pcbi.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newkirk D, Biesinger J, Chon A, Yokomori K, Xie X. Research in Computational Molecular Biology. Springer; 2011. Arem: aligning short reads from chip-sequencing by expectation maximization; pp. 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ukkonen E. Approximae string matching with q-grams and maximal matching. Theor. Comput. Sci. 1992;1:191–211. [Google Scholar]
- 19.Shen AXLK, Torng E. Proceeding of the 27th International Conference on Data Engineering (ICDE) IEEE; 2011. Large scale hamming distance query processing; pp. 553–564. [Google Scholar]
- 20.Bauer MJ, Cox AJ, Evers DJ. Proceeding of the 18th Annual Conference on Intelligent Systems for Molecular Biology, J04. International Society for Computational Biology; 2010. ELANDv2 - fast gapped read mapping for illumina reads. [Google Scholar]
- 21.Burkhardt S, Kärkkäinen J. Better filtering with gapped q-grams. Fundam. Inf. 2002;56:51–70. [Google Scholar]
- 22.Li C, Wang B, Yang X. Proceeding of the 33rd International Conference on Very Large Databases (VLDB) VLDB Endowment; 2007. VGRAM: improving performance of approximate queries on string collections using variable-length grams; pp. 303–314. [Google Scholar]
- 23.Chaudhuri S, Ganti V, Kaushik R. Proceeding of the 22nd International Conference on Data Engineering (ICDE) IEEE; 2006. A primitive operator for similarity joins in data cleaning; pp. 5–15. [Google Scholar]
- 24.Xiao C, Wang W, Lin X. Proceeding of the 34th International Conference on Very Large Databases (VLDB) VLDB Endowment; 2008. Ed-join: an efficient algorithm for similarity joins with edit distance constraints; pp. 933–944. [Google Scholar]
- 25.Collins DW, Jukes TH. Rates of transition and transversion in coding sequences since the human-rodent divergence. Genomics. 1994;20:386–396. doi: 10.1006/geno.1994.1192. [DOI] [PubMed] [Google Scholar]
- 26.Jokinen P, Tarhio J, Ukkonen E. A comparison of approximate string matching algorithms. Softw. Pract. Exper. 1996;26:1439–1458. [Google Scholar]
- 27.Döring A, Weese D, Rausch T, Reinert K. Seqan an efficient, generic c++ library for sequence analysis. BMC Bioinformatics. 2008;9:11. doi: 10.1186/1471-2105-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers G. A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM. 1999;46:395–415. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.