Abstract
Exon arrays are regularly used to analyze differential splicing events. GeneChip Gene 1.0 ST Arrays (gene arrays) manufactured by Affymetrix, Inc. are primarily used to determine expression levels of transcripts, although their basic design is rather similar to GeneChip Exon 1.0 ST Arrays (exon arrays). Here, we show that the newly developed Gene Array Analyzer (GAA), which evolved from our previously published Exon Array Analyzer (EAA), enables economic and user-friendly analysis of alternative splicing events using gene arrays. To demonstrate the applicability of GAA, we profiled alternative splicing events during embryonic heart development. In addition, we found that numerous developmental splicing events are also activated under pathological conditions. We reason that the usage of GAA considerably expands the analysis of gene expression based on gene arrays and supplies an additional level of information without further costs and with only little effort.
INTRODUCTION
Alternative splicing (AS) is a post-transcriptional process based on the joining of exons of a gene in different combinations to generate various isoforms from a single gene. It has been shown that most genes have at least one alternative isoform (1), which can be expressed in a tissue, development or sex-specific manner and fulfill different or even opposing functions (1–3). Traditional microarrays are not capable of detecting alternative spliced isoforms, since their probes target only small regions at the 3′-end of genes (3′ arrays). In contrast, whole-transcript arrays, such as exon arrays from Affymetrix, Inc. (GeneChip Exon 1.0 ST Array), contain probe sets (consisting of up to four probes) representing every exon. Additional probe sets are provided on exon arrays to allow identifications of internal splice sites, retaining introns or putative exons. This approach allows the detection of over 1.4 million expressed sequences.
Recently, GeneChip Gene 1.0 ST Array (gene array) was released by Affymetrix, Inc. This microarray platform was designed to measure gene expressions rather than expressions of single exons. However, gene arrays are very similar to exon arrays in respect to the hybridization protocol and coverage of annotated genes (4,5). The majority of probe sets present on gene arrays is identical to probes of high-quality annotation (core annotation) of exon arrays, but probe sets for putative exons and genes are not integrated into gene arrays (Figure 1A). Additionally, probe sets on gene arrays consist of only two instead of four probes (Figure 1B). Previous studies indicate that both exon and gene arrays correlate well to 3′ arrays in respect to gene expressions [correlation of 0.80 as reported by (7)] and very highly to each other at both gene and exon levels [R = 0.94 and R = 0.91, respectively as reported by (8)]. This implies that gene arrays are also capable of detecting differentially expressed exons, but the coverage of individual exons is limited compared with exon arrays (9). Although gene arrays are more affordable than exon arrays or deep sequencing, their intended usage focuses solely at the gene expression level, which leaves additional information contained in them untouched.
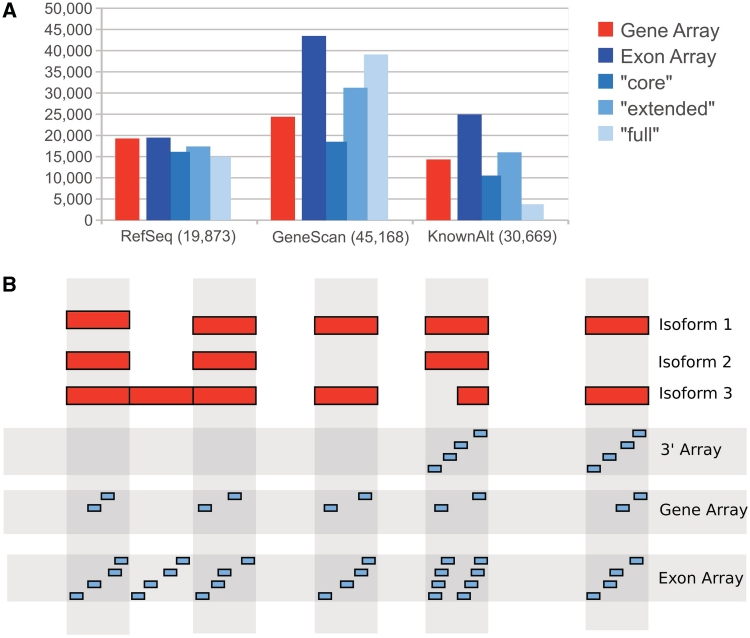
Figure 1.
(A) Probe sets on exon arrays are virtually grouped into three non-overlapping categories, namely ‘core’, ‘extended’ and ‘full’. The ‘core’ set covers well-annotated genes from RefSeq and full-length mRNAs, ‘extended’ cDNA alignments and ‘full’ gene predictions. The number of covered genes on gene and exon array ‘core’ sets are very similar. However, exon arrays have a large number of additional probe sets for hypothetical exons and genes (e.g. from GeneScan) in the ‘extended’ and ‘full’ sets. The numbers shown above were calculated using Galaxy's (6) function ‘Profile Annotations’ on the genomic positions of mouse gene and exon array probe sets. (B) Distribution of probes on 3′, gene and exon arrays. The red boxes represent exons of a hypothetical gene, spliced into three isoforms. 3′ arrays have probes in the 3′ region of a gene to measure gene expressions. For gene and exon arrays, probes distributed across the entire length of the gene are summarized to measure gene expressions.
In a previous study, we introduced the Exon Array Analyzer (EAA) (http://EAA.mpi-bn.mpg.de), an easy-to-use web tool for analyzing exon arrays for differential expressed exons (10). Here, we describe the Gene Array Analyzer (GAA), a versatile tool to analyze GeneChip Array data at both gene and exon levels. To demonstrate the applicability of GAA, we profiled isoform switch events that occur during heart development and cardiomyocyte differentiation of murine embryonic stem (ES) cells. We also used GAA to re-evaluate already existing publicly available datasets, which resulted in the detection of disease-related splicing events to recapitulate the developmental splicing processes.
MATERIALS AND METHODS
Implementation of GAA
The analysis of gene arrays followed a similar approach as our previous work with exon arrays (10). In brief, the Affymetrix Power Tools (http://www.affymetrix.com) were used for normalization, background correction and summarization of raw CEL files uploaded to the GAA server by the user. The ps- and the mps-files for gene arrays were utilized. The ps-file contains a list of probe sets for exon level, and the mps-file contains probe sets grouped into transcript cluster, which represent a gene. All probe set signals of a transcript cluster are used by the Affymetrix Power Tools to estimate the gene expression signal. For the identification of differentially expressed exons, we applied the Splice Index (SI) (11). Gene level normalized intensity (GNI) was calculated for each exon:
| (1) |
where Eij is the exon signal for exon i in sample j. Gj is the gene level signal in j. Using this value, the SI can be calculated as follows:
| (2) |
where A and B are the two samples to compare. This approach has been commonly applied in exon array experiments on the ‘core’ annotation set (12–14). To avoid inflated false positive results produced by the SI and other algorithms (15), we used a set of filters as described by a guideline of Affymetrix (16). Thus, genes that are not expressed in all conditions as well as exons that are not expressed in any condition were filtered out. This eliminates incorrectly identified probe sets with high SI, which are resulting from the background noise (i.e. low signal-to-noise ratio). Cross-hybridizing probe sets and probe sets with an abnormally high SI were discarded, because they may not reflect true expression changes. Furthermore, large differences in gene expression levels can increase the noise resulting in false positives (11). All filters are optional and can be modified by the user.
Since the initial introduction of EAA, we implemented several additional features. Previously, analysis was limited to pairwise comparisons, which we now extended to up to four groups per analysis by implementing limma (17) from the Bioconductor package, which has been originally applied on exon arrays by Shah and Pallas (18). Matching putative splicing events as found by GAA to known splicing events is a critical step to distinguish between false and true positives. For this purpose, we added a table with known splicing events to the result view. To implement this feature, we downloaded the ‘knownAlt’ track from the UCSC genome browser (19). This track contains 132 205 splicing events in human and mouse, which are categorized into nine different event types. In addition, we downloaded the transcript coordinates from the AceView database (20) of human, mouse and rat and identified 211 098 splicing events by utilizing ASTALAVISTA (21). Splicing events from both sources were imported into MySQL tables. Additionally, we added information about tissue-specific splicing events. For this purpose, we analyzed the publicly available tissue dataset from Affymetrix for human, mouse and rat. Each dataset contains 11 tissues with three replicates each. We compared each tissue against all other tissues as a group using EAA. For all 11 × 3 datasets, we used the Robust Multichip Average (RMA) (22) as a preprocessing algorithm and the default settings of the EAA. A probe set (exon) is considered as tissue-specifically (P < 0.01) expressed (SI > 1) or skipped (SI < −1).
mESC culture and differentiation
The clone CM7/1 of J1 mouse embryonic stem cell (mESC) line carrying the neomycin resistance gene driven by cardiac alpha-myosin heavy chain promoter was maintained and differentiated as previously described (23). The selection of cardiomyocytes with 400 µg/ml of G418 (GIBCO) was initiated on 9 days after the initiation of differentiation.
Animals
Hearts [E17.5, P1 and adult (2-month old)] were isolated from individual C57BL/6 mice. All animal experiments in this study were approved by the local animal care committee.
Microarray experiment
Total RNA was prepared with TRIzol Reagent (Invitrogen). The quality of extracted RNA was assessed by the Agilent Bioanalyzer (Agilent Technologies). Three hundred nanograms of total RNA from each sample was processed for hybridization onto GeneChip®Mouse Gene 1.0 ST Array (Affymetrix) or GeneChip®Mouse Exon 1.0 ST Array according to the manufacturer's protocol and scanned. Raw and normalized data have been submitted to GEO and are available under series GSE33183.
AltAnalyze
For a comparison to the result of AltAnalyze, we downloaded the most recent release (2.0.4) from http://www.altanalyze.org/. To reduce complexity, we excluded EB from the analysis and compared ES to CB only. We used the ENSEMBL release 55, since GAA is based on this version. To retrieve all probe sets (not just significant ones), we set ‘Minimum alternative exon score’ and ‘Max MiDAS/normalized intensity P-value’ to 1. All other parameters were left as default settings.
Gene Ontology analysis
We used PantherDB (http://www.pantherdb.org/) to identify up- and downregulated Gene Ontology terms. All genes and exons, which were significantly different between any condition (F-statistic P < 0.001), were used.
Hierarchical clustering
The normalized expression files were loaded onto MeV (24) to filter for the 25% of genes with highest standard deviations. The same software was used for hierarchical clustering of samples by using Pearson's correlation with average linkage clustering. The dendogram was generated by TreeGraph2 (25).
RESULTS
Gene Array Analyzer
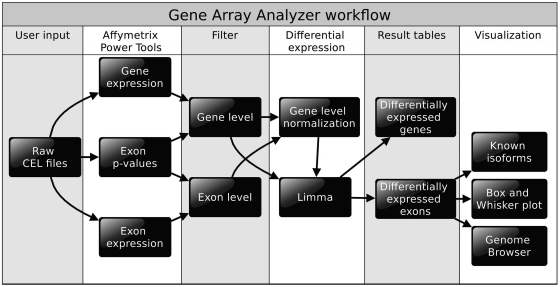
The reliability of gene arrays compared to exon arrays has been studied at the gene (7) and exon levels (9), suggesting that gene arrays have the potential to profile exon expression changes (8). Inspired by these studies, we reasoned that gene arrays might be utilized to analyze alternative splicing events with existing algorithms. We therefore reconfigured our web server for exon array analysis (10) to match requirements of gene arrays and named this new tool GAA. To use GAA, the user only has to provide raw CEL files. GAA is capable of performing a complete analysis, including pre-processing (background correction, normalization and summarization), filtering, gene expression analysis, alternative splicing analysis and visualization of spliced genes (Figure 2). Unlike the former version of EAA, which was only able to perform a pairwise comparisons, up to four groups per analysis can be processed (Figure 3A). All groups can be compared to each other individually by setting up a contrast matrix. GAA allows graphical representation of known isoforms within its built-in genome browser to evaluate expressed isoforms, which is a critical step to distinguish between false and true positives. Moreover, known splicing events from two different sources were integrated into GAA to assess whether a differentially expressed probe set may correspond to known splicing events (Figure 3D). We also implemented exon array-derived datasets from 11 different normal tissues containing differentially spliced exons to serve as a reference for potential splicing events. The main features of GAA are shown in Figure 3. A more detailed description can be found in the ‘Materials and Methods’ section and in the online help section of GAA (http://GAA.mpi-bn.mpg.de/help.php).
Figure 2.
Work flow of GAA. After uploading the raw CEL files to the GAA server, preprocessing is initiated. The Affymetrix Power Tools are utilized for background correction, normalization and summarization at the gene and exon level. A P-value for each exon (probe set) is estimated to determine whether an exon is expressed above background levels. In the next steps, filters are applied to eliminate false positives. GNI are passed to limma for calculation of the SI and statistics. Results are imported into MySQL tables for visualization.
Figure 3.
In the first two steps of an analysis, the user has to select the species and upload CEL files to the server (data not shown). (A) Afterwards, names for the analysis and groups as well as a password to access the result are needed to be provided by the user. Each uploaded CEL file needs to be assigned to its group by selecting a box. The groups are used to set up comparisons. After processing by GAA has finished, the user will receive a link to access the result. (B) On the result page, thresholds for SI and P-value can be chosen to filter at the exon or gene levels. It is also possible to search for certain genes, e.g by Gene Symbol. A table with all exons or genes meeting the thresholds will be shown on the same page (data not shown). Each entry is linked to a page with more details, including links to other databases, SI of all exons of the gene and probe set sequences. (C) This page also displays a box plot of GNIs of all exons of a gene, here shown on the example of Add3 which has been validated by reverse transcription polymerase chain reaction (RT–PCR) (see text). Not shown here is the genome browser, which is also displayed on the same page (see Figures 4B and 5B for examples). (D) A table shows the SI, P-value and known splicing events of all probe sets. (E) By clicking on a probe set ID in the genome browser, exon sequences are shown. They can be used directly to design primers. Screen shots are modified to fit the page. Additional information on how to use GAA can be found in the online help section at http://gaa.mpi-bn.mpg.de/help.php.
Alternative splicing during heart development
Heart is the earliest organ in mammals to acquire functionality in order to support survival. Although the heart is a vital organ, very few studies have been published up until recently to identify genome-wide alternative splicing events (26,27). To gain further information about alternative splicing during heart development, we performed gene array experiments using hearts from embryonic day 17.5 (E17.5), post-natal day 1 (P1), and 2-month-old male mice and compared these three developmental stages with each other at the gene and exon levels. Analysis was conducted using GAA as described in the ‘Materials and Methods’ section. We identified 2521 differentially expressed genes (DEGs) and 3452 probe sets [termed ‘differentially expressed exon (DEE)’ hereafter] as significantly regulated (F-statistic P < 0.001). All significant DEGs and DEEs are listed in Supplementary Table S1, and the full result can be accessed at GAA online.
Next, we performed Gene Ontology analysis through the PANTHER software (28). As reported by others (27), significantly enriched terms are not identical at the gene and exon levels but do overlap with each other. We found enrichments for system development, and helicase activity at the exon level, whereas oxidoreductase activity and receptor binding activity at the gene level. In agreement with our data set, highly significant terms in both groups are metabolic process, developmental process, cytoskeletal protein binding and structural constituent of cytoskeleton, which are known to be important for the development of heart (Supplementary Table S2).
Alternative splicing during cardiomyocyte differentiation
Heart is made up of cardiomyocytes, fibroblasts, endothelial and smooth muscle cells. To obtain a pure source of cardiomyocytes, we differentiated J1 mouse ES cells (strain 129S4/Jae; ATCC# SCRC-1010) to cardiomyocytes and eliminated all non-cardiomyocyte cells using a construct consisting of the cardiac alpha-myosin heavy chain promoter driving the neomycin resistance gene (23). Gene arrays were hybridized with RNA obtained from undifferentiated ES cells (ES) (Day 0 before the induction of differentiation), embryonic bodies (EB) (Day 7 after the induction of differentiation) and cardiac bodies (CB) (14 days after the induction of differentiation with spontaneous beating, which is a characteristic of cardiomyocytes). Data were analyzed with GAA as described in the ‘Materials and Methods’ section. The P-value of the F-test was used to identify genes that are differentially expressed between any of the three conditions. GAA identified 1 852 DEG and a relatively low number of 166 DEE within the threshold P < 0.001 (Supplementary Table S1 and at GAA online).
The Gene Ontology analysis during differentiation of cardiomyocytes shows that significant terms at the exon level represent a subset of the terms at the gene level. The much higher number of DEG compared with DEE might explain this observation. Terms found at both levels represent metabolic process, protein binding and transcription factor activity, etc. One of the most significant biological process at the gene level was developmental process. Other highly enriched terms were heart development and muscle organ development, which are expected terms during differentiation of cardiomyocytes (Supplementary Table S2).
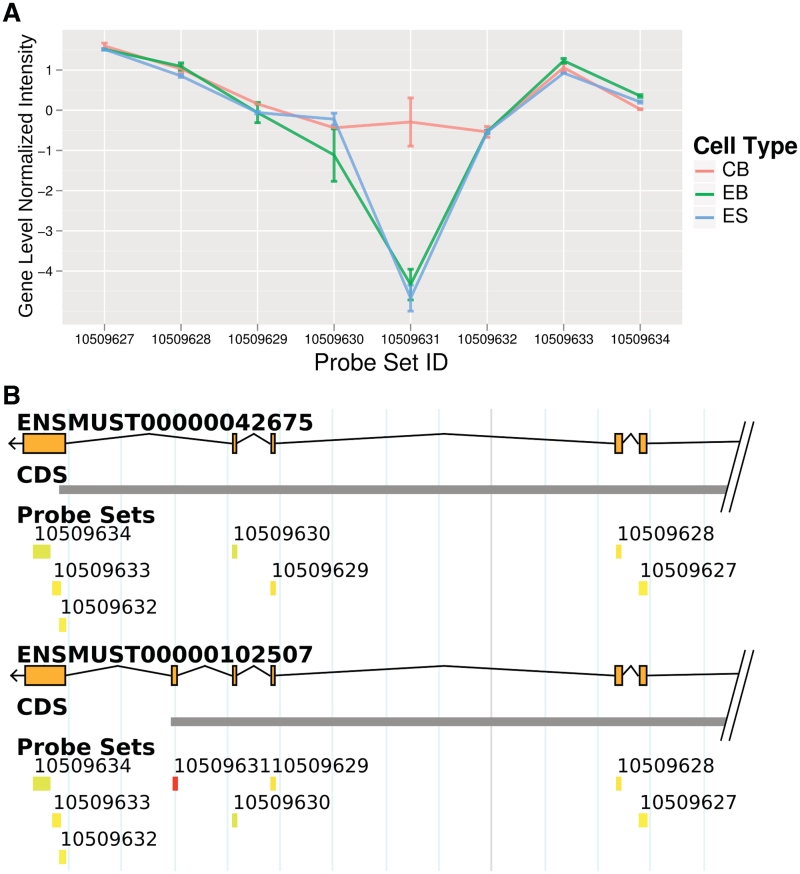
To demonstrate the usability of GAA, we selected a set of genes with the highest difference at the exon level between CB and ES as well as EB. The probe set with the largest difference (SI = 4.38, Figure 4A) was Capzb. As shown in the GAA's genome browser, several isoforms of this gene are known (only two out of six isoforms are shown in Figure 4B). The differentially expressed probe set (probe set ID ‘10509631’) indicates that the longer isoform of Capzb1 was expressed in CB while the corresponding exon for this longer isoform was skipped in ES and EB, thus, generating Capzb2 (29). The knownAlt track and AceView also identified this splicing event as indicated by the GAA-generated expression table. In addition, the GAA detected tissue specificity of this splicing event using the exon array dataset. The significantly higher expression values of this probe set were recorded for heart and muscle compared with nine other tissues. This finding is in agreement with the result in Ref. (29), which reported that Capzb1 is predominately expressed in muscle and cardiac tissue while the shorter form is present in non-muscle tissues. Another study found these two isoforms have distinct functions and cannot replace each other (30).
Figure 4.
Splicing during cardiomyocyte differentiation. (A) The probe set ‘10509631’ of Capzb shows one of the highest SI during cardiomyocyte differentiation (CB compared to ES and EB). In contrast, the flanking probe sets in all three cell lines show a similar GNI indicating that they are expressed equally (in relation to the individual gene expression). (B) The integrated genome browser revealed that ‘10509631’ is targeting an exon, which is skipped in the upper and included in the lower isoform. Literature search revealed that the longer isoform is known as cardiac and muscle specific (see text). Both genome maps display a region of Capzb.
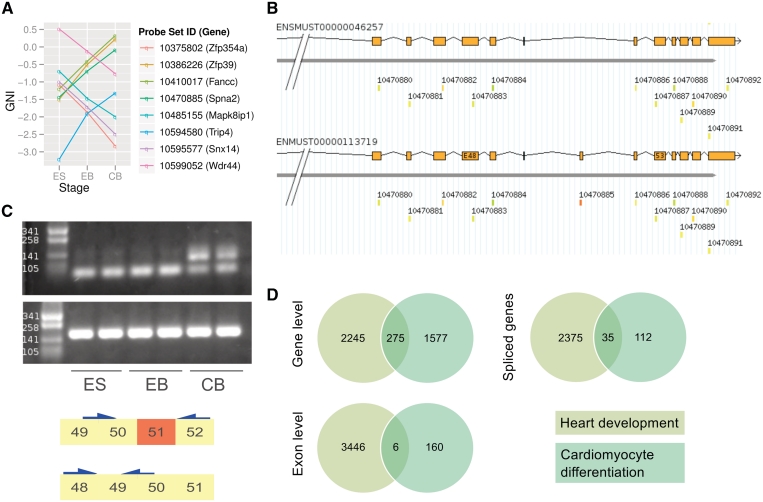
Next, we reason that there should be a set of genes whose expressions do not change but create different isoforms with altered expression patterns during cardiomyocyte differentiation, in which we termed this phenomenon as ‘isoform switch’. To screen for such genes, the following criteria were used: exons with increasing or decreasing expression levels (SI > 0.5, P < 0.05) of unregulated genes with opposite thresholds (gene level fold-change < 0.5, P > 0.05). As a result, eight genes were selected (Figure 5A). To provide an evidence for such isoform switched genes, we validated the known cassette exon of Spna2 (Spectrin alpha 2) by reverse transcription polymerase chain reaction (RT–PCR) experiment (Figure 5B and C).
Figure 5.
(A) Plot represents GNI of eight selected probe sets, which are up and downregulated at the exon level during cardiomyocyte differentiation but are expressed equally on the gene level (see text). (B) One such gene is Spectrin alpha 2 (Spna2), which is known to be spliced into many isoforms. The parts of the 3′-end of two isoforms is shown above. During cardiomyocyte differentiation, probe set ‘10470885’ is differentially expressed while the expression level of the gene does not change. (C) The RT-PCR validation of the cassette Exon 51 (according to ENSMUST00000113719) in ES cells compared with CB (upper). Additional primers were used to measure the global gene expression of Spna2 (lower). (D) Venn diagrams show the number of DEG, DEE and spliced genes during heart development and differentiation of cardiomyocytes. A gene or exon was considered as regulated if any of the three conditions in the two datasets differs significantly (F-statistic P < 0.001).
Comparison between heart development and cardiomyocyte differentiation
The comparison of heart development and cardiomyocyte differentiation using the same thresholds as described above revealed an overlap of regulated genes and exons (Figure 5D). The number of DEE derived from the cardiomyocyte differentiation dataset was remarkably smaller compared with the heart development dataset. To further investigate the reasons for the high discrepancy of DEE, we searched for genes that are involved in ‘RNA splicing’ (by Gene Ontology term GO:0008380). Interestingly, we found only three genes that are regulated during cardiomyocyte differentiation, but 34 splicing factors were identified during development of the heart (P=1 and P=0.04725, respectively, by right-tailed Fisher's exact test). Hierarchical clustering of these 37 genes shows the large difference among different samples (Supplementary Figure S1).
Comparison to exon arrays
Previously, we described the identification of eight heart-specific splicing events using EAA and exon arrays (10). For the current analysis, gene array data from the same set of tissues were employed (http://www.affymetrix.com) and used for a direct comparison between exon and gene arrays. Independently validated splicing events were compared to the result of the gene array analysis and are listed in Supplementary Table S3. In the case of Idh3b, gene array analysis indicated a wrong splicing event due to insufficient probe set coverage (Supplementary Figure S2). However, in half of the observed cases, both types of arrays identified identical splicing events.
The origin of samples which were used in the Affymetrix tissue dataset has not been published. For a systematic comparison between gene and arrays, we performed exon arrays with the same samples as we used for cardiomyocyte differentiation. First, we measured the correlation at the gene and exon levels (Supplementary Figure S3). As previously reported by others (8), we detected a very high correlation at the gene level (of R = 0.91) and a slightly lower correlation at the exon level (R = 0.87). To identify DEG and DEE, we analyzed the exon arrays with EAA using its default parameters. We found 1,014 DEGs and 769 DEEs using the same cut-off as for gene arrays (F-statistic P < 0.001). The identified genes and exons can be found in Supplementary Table S4. Next, we asked how many DEGs and DEEs were detected on both arrays. We identified 762 significantly regulated genes by both GAA and EAA, which corresponds to 76% of all DEG on exon arrays. For a comparison at the exon level, we mapped the significantly regulated gene array probe sets to ENSEMBL exons. Of these 89 unique ENSEMBL exons, 14 were identified as DEE by exon array probe sets as well (Supplementary Table S5 for a comparison). We performed RT–PCR experiments for nine splicing events and were able to validate all events. To demonstrate that gene arrays are capable of detecting splicing events that were not found with exon arrays, we randomly chose 14 probe sets that show no significant regulation on exon arrays. Of these, two splicing events were validated as true positive results. Results of RT–PCR experiments can be found in Supplementary Figure S4 and the used primer pairs in Supplementary Table S6.
Comparison to other software products
Since the idea of using gene arrays for the analysis of alternative splicing events is new, there are only a very limited number of other products that can be directly compared to GAA. FIRMAGene, a modification of FIRMA to gene arrays, represents an alternative approach, although it is an algorithm not a software product (9). Since there is no ‘gold standard’ dataset, a direct comparison between FIRMAGene and the use of Splice Indices employed by GAA is not possible.
While we were building GAA, a software work flow became available that can be used to analyze gene arrays as well as exon arrays at the gene and exon levels (31). The work flow consists of two products: ‘AltAnalyze’ for pre-processing and statistic analysis of gene arrays; and DomainGraph for visualization of downstream analysis (e.g. miRNA predicted binding sites, pathway analysis and protein domain information). Compared to this work flow, which requires a manual installation to the work station of the users, GAA users do not need to worry about the computational power of analysis because all calculations are carried out through server machines, which should minimize the requirement for high-end, powerful computers at the user side. Furthermore, since GAA is completely browser based, no set up on the user side is required, which avoids time-consuming operations, such as downloading annotation files and other relevant and required files for processing of gene array data. Another point is that AltAnalyze is a stand-alone application that provides the confidentiality of experimental data. To cope with confidentiality concerns, the uploaded CEL files are password protected and are kept at the server for 1 week, which can be extended through request by users, and will be eliminated thereafter. Moreover, users can use pseudo-sample names to further increase the confidentiality. Unlike AltAnalyze and DomainGraph, GAA provides mRNA sequences of exons upstream and downstream of the identified differentially expressed exon in a new window (Figure 3E). This feature allows a user to simply copy and paste sequences to a primer designing program, such as ‘Primer3’ (32), to obtain a set of primers for the validation by RT–PCR experiments.
To compare the result of AltAnalyze with GAA, we analyzed our gene arrays as described in the ‘Materials and Methods’ section. Since AltAnalyze uses SI > 1 and P < 0.05 as default thresholds, we also applied these values for the GAA result. Under these thresholds, we found 1707 regulated probe sets using GAA and 830 using AltAnalyze. Of which, 312 were found with both tools. To evaluate the true positive rate of both tools, we checked which probe sets match to known splicing events. In all, 17% of the probe sets detected as differentially expressed with both tools and 15% of the probe sets detected with either tool match to known splicing events. We also compared the result to our seven validated RT–PCR experimental results, which show differences between ES and CB. Two of these DEEs were also identified by AltAnalyze, further three show regulation but no significant P-value. The other three DEEs appear not in the result of AltAnalyze. A direct comparison of the results can be found in Supplementary Table S7.
Aberrant alternative splicing events under pathological conditions
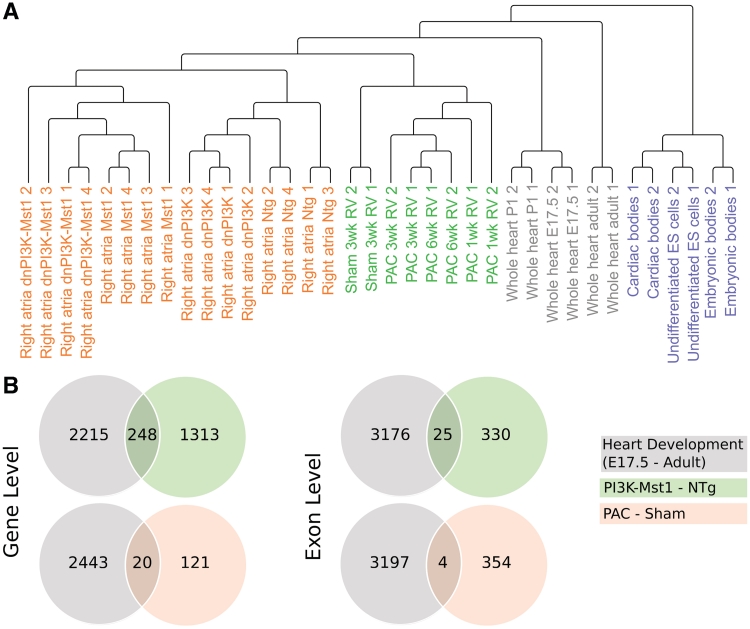
The major advantage of GAA is that it only requires Affymetrix CEL files. In other words, it is possible to re-evaluate previously published gene array data, which were analyzed at the gene level but not at the exon level. Hence, the information about changes in alternative splicing events can be obtained without additional experiments. As a proof-of-principle for such an approach, we re-analyzed public datasets dealing with various pathological conditions of the heart using GAA and compared the results to the dataset of heart development (Figure 6A).
Figure 6.
Gene and exon expression changes under pathological conditions. (A) Hierarchical clustering of gene arrays of different studies. The data set from cell culture experiments are clearly separated from those derived from whole hearts. In addition, hearts at early developmental stages are distant from pathological conditions and adult hearts. (B) Venn diagrams of regulated genes under three different studies. Only genes that are significantly regulated (P < 0.001 in both comparisons) and show the same trend of differentially expression (fold change <−0.5 or >0.5 in the compared conditions) are counted.
Phosphoinositide 3-kinase (PI3K) is essential for the heart development and plays an important role for the induction of physiological cardiac hypertrophy (33). Pretorius et al. (34) studied whether loss of PI3K makes the heart susceptible to atrial fibrillation (AF) in a mouse model of dilated cardiomyopathy (DCM). To evaluate molecular changes, the authors analyzed transcriptomes of the left and right atria by gene arrays. Essentially, the authors found that a reduction of PI3K is correlated with a reduction of heart weight, although the heart functions normally. However, combination of PI3K loss and DCM leads to a more severe phenotype than DCM alone. We obtained the raw CEL files from the Gene Expression Omnibus (GSE12420) and analyzed them with GAA using the default settings. We found 565 DEEs and 2285 DEGs between Mst1 (mammalian sterile 20-like kinase 1 overexpression as a model for DCM) and PI3K knock-out and Mst1-PI3K (Mst1 overexpression and PI3K knock-out) compared to the NTg (non-transgenic) mice (Supplementary Table S8).
Next, we used our previously published gene array data (GSE30428) of right ventricular hypertrophy induced by using pulmonary artery clipping (PAC) (35). In this data set, we found 105 DEGs and 419 DEEs using the same thresholds as above (P-value of the F-test < 0.001) (Supplementary Table S8).
The comparison of the results of the above two studies with our study of heart development identified 268 overlapping genes among the three groups, but only 29 probes showing similar expression patterns at the exon level (Figure 6B).
DISCUSSION
In our study, we describe a simple and efficient way to analyze splicing events using the widely applied gene arrays. Gene arrays were originally designed to measure genome-wide expression changes. However, their probe design also allows for the analysis of changes at the exon level to identify alternative splicing events. Gene arrays have limitations due to the lower number of probes per probe set, less probe sets per gene and no probes for putative genes in comparison to exon arrays, but they are much cheaper to use. Furthermore, numerous datasets from gene arrays are available, which have not yet been exploited to study differential splicing events.
The applicability of GAA was demonstrated by analyzing datasets from heart development and cardiomyocyte differentiation. We were able to identify DEGs and DEEs in both datasets and illustrated how the graphical output of GAA helps to recognize different isoforms. Gene arrays are not limited to a priori known splicing events, which may lead to the detection of de novo isoforms. To support such novel discoveries, GAA provides the built-in genome browser and the inclusion of known splicing events to easily separate known and novel splicing isoforms.
Our own data (Supplementary Figure S3) and others' (7–9) demonstrated that gene arrays provide a comparable signal for exon expression as exon arrays. To demonstrate this potential, we analyzed similar data sets by using both types of arrays and compared the outcome on eight validated examples (Supplementary Table S3). As expected, gene arrays were able to identify most splicing events, although one splicing event was falsely recognized because of the lower probe set coverage. To further evaluate the performance of gene arrays compared to exon arrays, we analyzed the same data set of cardiomyocyte differentiation with exon arrays. We compared the number of DEGs and DEEs identified by both array types. At the gene level, we found less DEG with exon arrays compared to gene arrays. The reasons for this difference have been discussed elsewhere (7). At the exon level, we found 14 DEEs with both arrays and validated nine splicing events by RT–PCR experiments (Supplementary Figure S4). In addition, we performed RT–PCR experiments for further 14 DEEs. Of which, two showed the expected bands. This demonstrates that splicing events that could not be detected by exon arrays could be identified and validated by gene arrays.
AltAnalyze is a stand-alone software package to analyze different types of microarrays, including gene arrays, for alternative splicing. We compared the result of GAA with AltAnalyze side-by-side (Supplementary Table S6). Regarding known splicing events, both tools discovered the same proportion. However GAA found 48% more DEEs than AltAnalyze using the same thresholds. Five out of seven validated splicing events between ES and CB were not significant or not found with AltAnalyze. The difference between both tools originates from different annotations and filters. Furthermore, AltAnalyze uses ‘constitutive exons’ instead of all core exons to estimate the gene expression, which influences the calculation of the SI. Since we cannot test all putative spliced exons, it is not possible for us to compare comprehensively with other tools. However, we showed by RT–PCR validations that GAA identifies splicing events reliably and we believe that GAA outperforms AltAnalyze in respect to usability.
In the comparison between heart development and cardiomyocyte differentiation, we made an interesting observation: although 275 genes showed statistically significant differences among both related conditions (Figure 5D), only very limited number of exons (six exons) matched both processes. One might speculate that alternative splicing is a very common process by which genes respond to different conditions. This hypothesis is also quoted frequently to explain discrepancies between transcriptomics and proteomics datasets (36). To further investigate the discrepancy, we searched for regulated splicing factors and found a significant enrichment during heart development, but not during cardiomyocyte differentiation (Supplementary Figure S1). One of the strongly regulated splicing factors in the adult heart is A2bp1 (7.36 fold upregulated), which encodes the Fox-1 protein. Fox-1 is known to be expressed in the adult mouse heart (37) and involved in the post-natally development of the heart (27). Other regulated splicing factors are the serine/arginine-rich family (Snrnp27, Sfrs2, Srsf5 and other) and the heterogeneous nuclear ribonucleoproteins (Hnrnpa1, Hnrnpk and other) of which both can act as splicing activator and silencer by binding to short cis-motifs on the pre-mRNA [reviewed in (38)]. These regulated splicing factors in adult hearts may have caused significantly more splicing events, which cannot be observed during cardiomyocyte differentiation.
In addition, we found genes that did not show differences of expression levels during differentiation of cardiomyocytes but underwent alternative splicing, in which we named this phenomenon to be ‘isoform switch’. The gene ‘Spna2’ is an example of such phenomenon. In our study, we identified an isoform switch (inclusion of Exon 51 upon the differentiation of ES cells into beating cardiomyocytes), which was validated by an RT–PCR experiment (Figure 5C). The spliced exon lies in the spectrin/alpha-actinin domain (IPR018159; 47-2,315aa) at position 2217–2237aa, on the last of 20 repeats. A previous study demonstrated that homozygous mice appear healthy and morphologically normal when Exon 25∼27 of Spna2, which lays downstream of the spectrin/alpha-actinin domain (1154–1189aa, which corresponds to repeat 10) and named CCC (calpain, caspase, calmodulin) domain, are deleted in the mouse (39). However, in rat hearts, an insertion of 20 aa exists next to repeat 10 called ‘alpha II-SH3i’, which is important for intracellular targeting of Connexin 43 to gap junctions and suspected to be a potential target for stress signaling pathways (40). Inclusion or exclusion of exon 51 might affect localization of Spna2 encoded ‘alpha II-spectrin’ at the Z disc and the plasma membrane of myofibrils (41), and thereby biological properties of cardiomyocytes. It will be of interest to study functional consequences of differences in the distribution of isoforms during tissue development. Application of GAA might serve as a starting point for such approaches using existing data sets.
We obtained CEL files from studies of pathological conditions of dilated cardiomyopathy (34) and right ventricular hypertrophy (35), which have been analyzed at the gene but not at the exon level. Since these experiments used different parts of the heart and have been conducted at different laboratories with animals of different genetic backgrounds, a direct comparison between all experiments needs careful evaluation (Figure 6A shows the similarity between all samples). Yet, comparison of datasets at the gene level revealed that some genes showed the same expression pattern during the heart development and under pathological conditions. However, this correlation was somewhat lost when the comparison was made at the exon level (Figure 6B). In contrary to the popular belief that there is a re-initiation of embryonic gene expressions under pathological conditions (42,43), such trend does hold at the gene level but not so well at the exon level. This indicates that developmental alternative splicing events are not activated to full extent under pathological conditions. Hence, the activation of the fetal gene program under stress conditions is incomplete in respect to specific isoforms and represents a distinct state that can be distinguished from the regular developmental program. These findings further underline that changes in the transcriptome originate from gene expressions, but a care must be taken at the exon level by taking into account for alternative splicing events.
Up until recently, high-throughput transcriptome studies mostly concentrated on changes in gene expression. This trend has changed dramatically with the introduction of RNA-seq using deep sequencing technology, which uncovered that over 93% of genes have isoforms (1) indicating that alternative splicing events are more common than one anticipated. Although RNA-seq technology is improving constantly, results cannot be analyzed without proper bioinformatical analysis, which imposes a hurdle for experimental biologists. GAA is an easy to use tool that allows experimental biologists with limited bioinformatical skills to detect new splicing isoforms. GAA is a one-stop-shop for analysis of gene arrays to which a user can upload CEL files of gene arrays. Through a click of the mouse, a list of analytical methods can be selected. DEEs are indicated by box-and-whisker plot (Figure 3C) and by the genome viewer (Figure 5B) to identify locations of individual exons in the genome. We reason that GAA is a valuable tool for biologists and adds another layer of analysis to gene array without additional costs. The web interface of GAA is easy to use, requires no set up and is freely available at http://GAA.mpi-bn.mpg.de.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4, Supplementary Tables 1–8.
FUNDING
Excellence Cluster Cardio-Pulmonary System (ECCPS); LOEWE Center for Cell and Gene Therapy (to S.U.); by fellowships of the International Max Planck Research School for Heart and Lung Research (IMPRS-HLR) (to K.J. and P.D.G.); Max-Planck-Society; DFG (Br1416); Excellence Initiative ‘Cardiopulmonary System’ (to T.B.); University of Giessen-Marburg Lung Center (UGMLC); Cell and Gene Therapy Center (CGT) supported by the HMWK. Funding for open access charge: Max-Plank-Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Petra Uchida for her valuable advice and comments on this article, and Silvia Thomas for help with gene array experiments. S.U. conceived of the project. P.G. developed GAA. P.G. and D.J. performed computational analysis. M.T. performed gene array experiments. M.T., P.D.-G. and K.J. performed biological experiments. K.gK. operated cardiac hypertrophy mice and provided samples. T.B. provided general guidance. S.U., P.G., M.T. and T.B. drafted the manuscript. All authors read and approved the final manuscript.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2009;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stetefeld J, Ruegg M. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 4. Affymetrix Inc. (2009) GeneChip Exon Array System for Human, Mouse, and Rat. www.affymetrix.com.
- 5. Affymetrix Inc. (2007) GeneChip Gene 1.0 ST Array System for Human, Mouse and Rat. www.affymetrix.com.
- 6.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson MD, Speed TP. A comparison of Affymetrix gene expression arrays. BMC Bioinformatics. 2007;8:449. doi: 10.1186/1471-2105-8-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha KC, Coulombe-Huntington J, Majewski J. Comparison of Affymetrix Gene Array with the Exon Array shows potential application for detection of transcript isoform variation. BMC Genomics. 2009;10:519. doi: 10.1186/1471-2164-10-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson MD, Speed TP. Differential splicing using whole-transcript microarrays. BMC Bioinformatics. 2009;10:156. doi: 10.1186/1471-2105-10-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gellert P, Uchida S, Braun T. Exon Array Analyzer: a web interface for Affymetrix exon array analysis. Bioinformatics. 2009;25:3323–3324. doi: 10.1093/bioinformatics/btp577. [DOI] [PubMed] [Google Scholar]
- 11.Clark T, Schweitzer A, Chen T, Staples M, Lu G, Wang H, Williams A, Blume J. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Revil T, Gaffney D, Dias C, Majewski J, Jerome-Majewska LA. Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics. 2010;11:399. doi: 10.1186/1471-2164-11-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorsen K, Mansilla F, Schepeler T, ster B, Rasmussen MH, Dyrskjt L, Karni R, Akerman M, Krainer AR, Laurberg S, et al. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002998. M110.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bemmo A, Benovoy D, Kwan T, Gaffney DJ, Jensen RV, Majewski J. Gene expression and isoform variation analysis using Affymetrix Exon Arrays. BMC Genomics. 2008;9:529. doi: 10.1186/1471-2164-9-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Affymetrix Inc. (2008) Identifying and validating alternative splicing events. www.affymetrix.com. [Google Scholar]
- 17.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 18.Shah SH, Pallas JA. Identifying differential exon splicing using linear models and correlation coefficients. BMC Bioinformatics. 2009;10:26. doi: 10.1186/1471-2105-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):S12.1–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foissac S, Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007;35:W297–W299. doi: 10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 23.Zweigerdt R, Burg M, Willbold E, Abts H, Ruediger M. Generation of confluent cardiomyocyte monolayers derived from embryonic stem cells in suspension: a cell source for new therapies and screening strategies. Cytotherapy. 2003;5:399–413. doi: 10.1080/14653240310003062. [DOI] [PubMed] [Google Scholar]
- 24.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 25.Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schafer D, Korshunova YO, Schroer T, Cooper J. Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. Cell Biol. 1994;127:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart MC, Cooper J. Vertebrate isoforms of actin capping protein beta have distinct functions in vivo. J. Cell Biol. 1999;147:128798. doi: 10.1083/jcb.147.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755–W762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 33.McMullen JR, Shioi T, Zhang L, Tarnavski O, Sherwood MC, Kang PM, Izumo S. Phosphoinositide 3-kinase(p110alpha) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc. Natl Acad. Sci. USA. 2003;100:12355–12360. doi: 10.1073/pnas.1934654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pretorius L, Du X-J, Woodcock EA, Kiriazis H, Lin RCY, Marasco S, Medcalf RL, Ming Z, Head GA, Tan JW, et al. Reduced phosphoinositide 3-kinase (p110alpha) activation increases the susceptibility to atrial fibrillation. Am. J. Pathol. 2009;175:998–1009. doi: 10.2353/ajpath.2009.090126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreymborg KG, Uchida S, Gellert P, Schneider A, Boettger T, Voswinckel R, Wietelmann A, Szibor M, Weissmann N, Ghofrani AH, et al. Identification of right heart-enriched genes in a murine model of chronic outflow tract obstruction. J. Mol. Cell Cardiol. 2010;49:598–605. doi: 10.1016/j.yjmcc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Irmler M, Hartl D, Schmidt,T, Schuchhardt J, Lach C, Meyer HE, Hrab de Angelis M, Klose J, Beckers J. An approach to handling and interpretation of ambiguous data in transcriptome and proteome comparisons. Proteomics. 2008;8:1165–1169. doi: 10.1002/pmic.200700741. [DOI] [PubMed] [Google Scholar]
- 37.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Meary F, Metral S, Ferreira C, Eladari D, Colin Y, Lecomte M-C, Nicolas G. A mutant alphaII-spectrin designed to resist calpain and caspase cleavage questions the functional importance of this process in vivo. J. Biol. Chem. 2007;282:14226–14237. doi: 10.1074/jbc.M700028200. [DOI] [PubMed] [Google Scholar]
- 40.Ursitti JA, Petrich BG, Lee PC, Resneck WG, Ye X, Yang J, Randall WR, Bloch RJ, Wang Y. Role of an alternatively spliced form of alphaII-spectrin in localization of connexin 43 in cardiomyocytes and regulation by stress-activated protein kinase. J. Mol. Cell Cardiol. 2007;42:572–581. doi: 10.1016/j.yjmcc.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett PM, Baines AJ, Lecomte M-C, Maggs AM, Pinder JC. Not just a plasma membrane protein: in cardiac muscle cells alpha-II spectrin also shows a close association with myofibrils. J. Muscle Res. Cell Motif. 2004;25:119–126. doi: 10.1023/b:jure.0000035892.77399.51. [DOI] [PubMed] [Google Scholar]
- 42.Creemers EE, Wilde A, Pinto YM. Heart failure: advances through genomics. Nat. Rev. Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 43.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.