Abstract
RIG-I is a cytosolic receptor for non-self RNA that mediates immune responses against viral infections through IFNα/β production. In an attempt to identify novel tools that modulate IFNα/β production, we used SELEX technology to screen RNA aptamers that specifically target RIG-I protein. Most of the selected RIG-I aptamers contained polyU motifs in the second half regions that played critical roles in the activation of RIG-I-mediated IFNβ production. Unlike other known ligands, RIG-I aptamer bound and activated RIG-I in a 5′-triphosphate-independent manner. The helicase and RD domain of RIG-I were used for aptamer binding, but intact RIG-I protein was required to exert aptamer-mediated signaling activation. Furthermore, replication of NDV, VSV and influenza virus in infected host cells was efficiently blocked by pre- or post-treatment with RIG-I aptamer. Based on these data, we propose that RIG-I aptamer has strong potential to be an antiviral agent that specifically boosts the RIG-I-dependent signaling cascade.
INTRODUCTION
Infection by both known and newly emerging RNA viruses, such as hepatitis C virus (HCV), Ebola virus, influenza virus and SARS-CoV, are a severe threat to public health. RNA viruses cause injury to the infected organs and disrupt normal functioning, leading to high levels of morbidity and mortality (1–3). RNA viruses exhibit genomic hyper-variation, mainly caused by low fidelity of the virus-derived RNA-dependent RNA polymerase activity (4,5). Instability of the RNA viral genome allows viruses to escape from virus-specific drugs and vaccines. Because the development of stable vaccines for RNA viruses is a challenging task, alternative tools that modulate the immune response against a viral infection are of great interest (6–8). Among them, IFN-based therapy shows a strong potential for HCV clearance by stimulation of the innate and adaptive immune system of the infected host, although IFN resistance is often reported in HCV-infected patients. The concept of IFN-based therapy for virus eradication prompted the exploration of therapeutic agents that target innate immune systems of the infected host (9). In this regard, synthetic agonists for Toll-like receptors (TLR) have been successfully developed as potent IFN-inducers, and their potential to control viral replication and infection-related diseases has been extensively studied (6,8,10).
Early detection of the virus in the infected host relies on specific receptors that recognize virus-derived molecular patterns, such as double stranded RNA (dsRNA) or 5′-triphosphate-single stranded RNA (ssRNA) (11). TLR3, TLR7, TLR8 and TLR9 localize in the membrane of the endosomal compartment and sense non-self molecular patterns that are derived from the viral particles. In contrast, retinoic acid-inducible genes I (RIG-I) and melanoma differentiation-associated gene five (MDA-5) detect molecular patterns derived from replicating RNA viruses in the cytoplasm (12). Upon recognition of the intracellular non-self RNA, both RIG-I and MDA5 proteins are recruited to the mitochondria, via association with Cardif (also called IPS-1, VISA or MAVS), and trigger IFNβ production and signaling through the activation of IRF3 and NFκB (13). Although RIG-I and MDA5 utilize the same signaling pathways for IFNβ production, they exhibit differential preferences toward RNA substrates, and therefore detect different types of viruses. It has been suggested that an RNA structure with a 5′-triphosphate moiety can be found during the replication of ssRNA viruses, such as HCV, Newcastle disease virus (NDV), respiratory syncytial virus (RSV) and influenza virus, and that this structure is selectively recognized by RIG-I (14–17). Meanwhile, MDA5 preferentially detects long stretches of dsRNA structures, which are mainly produced by replicating Encephalomyocarditis virus (EMCV) or Reovirus (16,18,19). The significance of viral recognition and signaling cascades mediated by cytosolic RIG-I and MDA5 was well demonstrated in vivo using RIG-I or MDA5 knockout mice. RIG-I−/− mice exhibit defects in the immune response against NDV, Sendai virus (SeV) and vesicular stomatitis virus (VSV) infection, whereas MDA5−/− mice exhibit specific defects against EMCV infection (20).
Aptamers are single-stranded nucleic acid molecules that are selected with SELEX (systematic evolution of ligands by exponential enrichment) technology based on their affinities and specificities to target proteins (21,22). Due to their rapid selection process, high stability and relatively low immunogenicity, aptamers have become a favorite choice among molecular detection tools, replacing the use of antibodies (23). Furthermore, depending on the modes of action, aptamers are capable of modulating activities of target molecules, which facilitates the development of aptamers for therapeutic purposes (24–26). Most aptamers with anti-viral activity have been developed against virus-specific proteins, such as Tat of HIV or NS3 of HCV, and less effort has focused on aptamers that target the innate immune response against viral infection (22,27–29). In an attempt to develop aptamers that specifically control RIG-I-mediated anti-viral responses in the infected host cells, we screened RNA oligonucleotides specific for RIG-I using advanced-SELEX technology.
MATERIALS AND METHODS
Cells, virus and reagents
HepG2 cells were maintained in minimum essential medium (MEM) containing 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) and penicillin-streptomycin (Gibco). Huh7 cells and A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS and antibiotics. NDV-GFP, VSV-GFP and Influenza A (PR8 strain)-GFP were kindly provided by Dr Peter Palese (Mount Sinai School of Medicine, USA) and by Dr Adolfo Garcia-Sastre (Mount Sinai School of Medicine, USA), respectively. For transfection of DNA plasmids and RNA oligonucleotides including aptamers, Lipofectamine 2000 (Invitrogen) was used. IFNβ and polyI:C were purchased from R&D Systems and Amersham Biosciences, respectively. For synthetic 5′-triphosphate-dsRNA, both 3P-G and AS G24 were separately synthesized and annealed, following the method as described (30). For secreted TNFα and RANTES, human TNFα/TNFSF1A ELISA Development kit (R&D System) and human RANTES ELISA kit (Thermo scientific) was used, respectively.
Plasmids
The pIRES-Luc (Stratagene) and pNFκB-Luc (Stratagene) plasmids were purchased, and the pPRD-III-I-Luc plasmid was provided by Dr Katherine A. Fitzgerald (University of Massachusetts). pEF-BOS-Flag-RIG-I was provided by Dr Takashi Fujita (Institute for Virus Research, Kyoto University). The construction of pEF-BOS-Flag-Helicase+RD and pEF-BOS-Flag-CARD plasmids was described previously (31). pEF-BOS-Flag-ΔRD was sub-cloned using following primers: 5′-GCTCTAGATAAGCCACCATGGAT-3′, 5′-CCATCGA TTCATCTTGCTCTTGCTCTTCCTCTGCC-3′. pcDNA3-Flag-MAVS was kindly provided by Dr Zhijian J. Chen (University of Texas Southwestern Medical Center), and was subsequently sub-cloned into the XhoI/XbaI site of pCS2+MT plasmid.
Expression and purification of human RIG-I protein
A DNA fragment encoding human RIG-I (1–925 amino acids) was sub cloned into an expression vector derived from the pFastBac plasmid (Invitrogen). The vector was designed to express RIG-I as a fusion protein containing (His)10-glutathion-S-transferase (GST) at the N-terminus. Recombinant protein was produced in the Sf9 insect cells based on the Baculovirus expression vector system. Harvested cells were resuspended and lysed by sonication in lysis buffer consisting of 50 mM Tris (pH 7.5), 100 mM NaCl and 1 mM dithiothreitol. The supernatant of cell lysates was loaded onto a column packed with GST binding resin (Novagen) and eluted with the same buffer containing 10 mM reduced glutathione. To purify Flag-RIG-I, Flag-Helicase+RD (218–925 amino acids) and Flag-CARD (1–217 amino acids) proteins from the BL21(DE3)-pLsys (Novagen) bacteria, human RIG-I gene was sub cloned into an expression vector derived from the pProEX HTa (Invitrogen) or pET-22b CPD plasmid. The bacteria cells were cultured in LB broth and induced for over-night at 18°C with 100 µM IPTG, and cells were resuspended and lysed by sonication in lysis buffer consisting of 50 mM Tris (pH 7.5), 1 M NaCl and 4 mM β-mercaptoethanol. The supernatant was loaded onto a column packed with Cobalt resin (Novagen) and eluted with 20 mM Tris (pH 7.5), 200 mM NaCl, 4 mM β-mercaptoethanol and 150 mM Imidazole. The (His)10-GST-tag and CPD-(His)10-tag were cleaved by TEV protease or phytate, and then removed from RIG-I, Helicase+RD, and CARD by using Cobalt resine. Subsequently, HiTrap Q and HiLoad 26/60 Superdex 200 columns (GE Healthcare) were used for further purification of proteins. The following primers were used for cloning of RIG-I and their mutants: Flag-RIG-I, 5′-GAAGGCCTAATGGACTACAAAGACG-3′ and 5′-CCGCTCGAGTCATTTGGACATTT CTGC-3’; Flag-Helicase+RD, 5′-CGGCATATGGACTACAAAGACG-3′ and 5′-CCGGTC GACTTTGGACATTTCTGCTGG-3′; Flag-CARD, 5′-CGGCATATGGATTATAAGGAT GAT-3′ and 5′-CCGGTCGACTGGATCTTCTTGGTAG-3′.
Aptamer screening (SELEX) and synthesis
A synthetic DNA library pool of 5′-GCGCAAGCGTGCTGGGCC-(N40)-CATAACCC AGAGGTCGAT-3′ was amplified by PCR using the following primers: 5′-GGTAATACG ACTCACTATAGGGAGAGCGCAAGCGTGCTGGG-3′; 5′-GGGGGGATCCATCGACCT CTGGGTTATG-3′. To prepare RNA oligonucleotides, the PCR product was in vitro transcribed using a Durascribe® T7 Transcription Kit containing 2′-F-UTP and 2′-F-CTP (Epicentre Biotechnologies) at 37°C for 3 h, followed by purification with G25-sephadex column. At the first SELEX cycle, 1 nmol of the RNA oligonucleotide pool was incubated with 200 pmol of biotinylated human RIG-I protein for 30 min in 1× PBS (with 1 mM MgCl2). Oligonucleotide bound to RIG-I was harvested using biotin-streptavidin system (Sigma), reverse transcribed and then PCR amplified. Concentration of the RNA oligonucleotide pool and RIG-I protein were gradually reduced at the next round of aptamer selection. After 13 cycles of SELEX, sequences of the selected clones were analyzed using automated DNA sequencing. To remove the 5′-triphosphate, 20 µg of in vitro-transcribed RNA transcripts was incubated with 30 U of alkaline phosphate (NEB) and 40 U of RNase inhibitor (Promega) at 37°C for 1 h (14).
Luciferase assay
Cells were transfected with pPRD-III-I-, pISRE-, or pNKκB-Luc together with pRL-CMV for 36 h. For stimulation, cells were transfected with cognate ligands by Lipofectamine 2000. Using a dual-luciferase assay kit (Promega), firefly luciferase activity was measured and normalized with the value of renilla activity, and the ‘Rel. activity (fold)’ relative to the mock-treated sample was used.
Real-time RT-PCR
Total RNA (1 µg) was reverse transcribed using the Improm-II reverse transcription system (Promega). To detect NDV-GFP and VSV-GFP, random oligomers were used for reverse transcription. For PCR, SYBR premix Ex-Taq (Takara) was used and the ΔCt values were obtained using One-step™ Real-time PCR system (Applied Biosystem). Data were normalized against β-actin. The following primers were used: IFNβ, 5′-TGCTCT CCTGTTGTGCTTCTCC-3′ and 5′-CATCTCATAGATGGTCAATGCGG-3′; RANTES, 5′-ATGAAGGTCTCCAAAGAG-3′ and 5′-GCTCATCTCCAAAGAG-3′; TNFα, 5′-CACCATCAGCCGCATC-3′ and 5′-CAGGGCAATGATCCCA-3′; GFP, 5′-TTGATGGCAGGCCTCTTGC-3′ and 5′-GGAGGATGTTGGACGCATT-3′; β-actin, 5′-TCATGAAGTGTGACGTTGACATCCGT-3′ and 5′-CCTAGAAGCATTTGCGGTGCA CGATG-3′.
RNA interference
Cells were transfected with the appropriate siRNA using Lipofectamine 2000. The following sequences were used: siControl, 5′-GAAUUUGCACGAAAACGCC-3′; siRIG-I, 5′-GAG GUGCAGUAUAUUCAGG-3′; siMDA5, Qiagen validated siRNA (S103649037). siControl and siRIG-I were synthesized by the custom design service of Qiagen.
Western blotting and immunofluorescence staining
For western blot analysis, antibodies against GFP (Santacruz), RIG-I (ALEXIS Biochemicals), MDA5 (ProSci Incorporated), Flag (M2, Sigma), c-Myc (Roche), GAPDH (Chemicon) and β-actin (Santacruz) were used. Immunoreactive signals were developed using a LAS4000 luminescent image analyzer (Fujifilm). To detect intracellular localization of IRF3, cells on coverglass were stimulated with polyI:C (1.5 µg/ml) or aptamers (75 pmol) for 4 h, fixed with 4% paraformaldehyde, and then permeabilized in 0.2% TritonX-100 solution. Cells were incubated with anti-IRF3 antibody (Santacruz) in 1× PBS containing 0.05% Tween20 at room temperature for 2 h. Cells were then incubated with anti-Rabbit IgG conjugated with Alexa Fluor 488 (Invitrogen) and Hoechst 33258 (Sigma). Slides were mounted and then analyzed using an Olympus fluorescence microscope.
In vitro binding assay
For radioactive-labeled aptamer production, aptamers were in vitro transcribed using Durascribe® T7 Transcription Kit with the mixture of 25 µM α32P-GTP and 2.5 mM GTP at 37°C for 3 hours. For end-labeling, 500 pmole of AP-treated aptamer was incubated with T4 polynucleotide kinase (20 U, New England Biolabs, Beverly, MA, USA) and 100 pmol of γ-32P-ATP for 30 min. The labeled aptamers were subsequently purified with CHROMA spin30 column (Clontech). For binding assay, recombinant protein (10 µg) or total lysate (1 mg) from HEK293 cell was incubated with 400 pmol of the labeled aptamer, 2 µg of Flag antibody and Protein G plus/Protein A-Agarose bead (Calbiochem) at 4°C for 1 hour. The 32P-labeled-aptamers bound to RIG-I fraction was quantitated using liquid scintillation analyzer (LSA).
RESULTS
In vitro selection of RNA aptamers against RIG-I
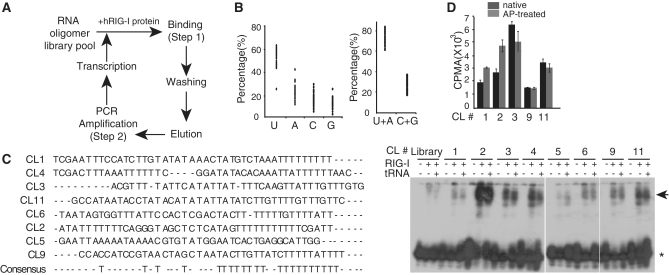
We screened RIG-I aptamers from a library pool of ∼1 × 1014 random RNA oligonucleotides using a purified full-length human RIG-I protein (Figure 1A). RNA oligonucleotides bound to RIG-I were harvested, reverse transcribed and PCR amplified and then transcribed in vitro to generate RNA olibonucleotide pools for the next round of selection (Supplementary Figure S1). After 13 cycles of selection and amplification, 45 independent clones of RIG-I aptamers with unique sequences were obtained. Sequence analysis of the 45 clones indicated that the sequences of the selected RIG-I aptamers contained high U content (average 51.9%), with a repeated polyUn motif in the second half of the sequence (Figure 1B). Among individual aptamer clones, lengths of the repeated polyUn varied between n = 2 to n = 11 (Figure 1C). We chose eight independent RIG-I aptamer clones at random and examined for their in vitro binding affinity to purified RIG-I protein. For this purpose, individual RIG-I aptamers were end-labeled with 32P and were incubated with purified full-length RIG-I proteins in the absence or presence of tRNA as a competitor of non-specific binding (Figure 1C). Despite differences in their binding potentials, all of the tested RIG-I aptamers exhibited strong RIG-I binding ability compared to the control RNA (library). We attempted to measure in vitro binding affinity of the selected aptamers by estimating bound and free forms of labeled aptamer under equilibrium conditions using different amounts of RIG-I protein. For this purpose, we picked CL9, which showed relatively weak binding compared to the other clones, hence provides lower bound value of the selected aptamer. In vitro Bmax and Kd value of CL9 were 0.29 fmol/mg and 37.2 nM, respectively (Supplementary Figure S2).
Figure 1.
SELEX-based RIG-I aptamer screening. (A) Schematic diagram depicting the RIG-I specific RNA aptamer screening process. RNA nucleotides with specific binding to the purified human recombinant RIG-I protein were selected with multiple rounds of the binding, washing, elution, PCR amplification, and RNA transcription, starting with the library pool of the RNA oligomers (see ‘Materials and Methods’ section). (B) Nucleotide usages found in the 45 selected RIG-I RNA aptamers are shown. (C) Left, Sequences of the RIG-I aptamer used in this study. Right, RNA-EMSA using 32P-labeled RIG-I aptamers and purified human RIG-I protein. Arrowhead indicates aptamer-RIG-I complex. Asterisk, free aptamer. (D) The level of 32P-labeled-aptamers (native or AP-treated) bound to RIG-I protein in vitro were quantitated using liquid scintillation analyzer (LSA). Data represent mean values ± standard deviation (±SD), from three independent experiments.
RNA aptamers were prepared in vitro using T7 RNA polymerase, and as a result, the 5′-triphosphate moiety in the first nucleotide position remained (32). Because the 5′-triphosphate moiety in ssRNA is known to be recognized by RIG-I (33,34), it is possible that the 5′-triphosphate in the RNA aptamer directly acts as a ligand for RIG-I protein. To test this possibility, the in vitro binding affinity of native and alkaline phosphatase (AP)-treated aptamers to RIG-I were directly compared (Figure 1D). For every aptamer tested, the binding of aptamers to RIG-I protein was not significantly altered by the presence of the 5′-triphosphate moiety, indicating that the selected aptamer sequences governs specific binding to RIG-I protein.
Stimulation of the RIG-I-dependent IFNβ production by RIG-I RNA aptamers
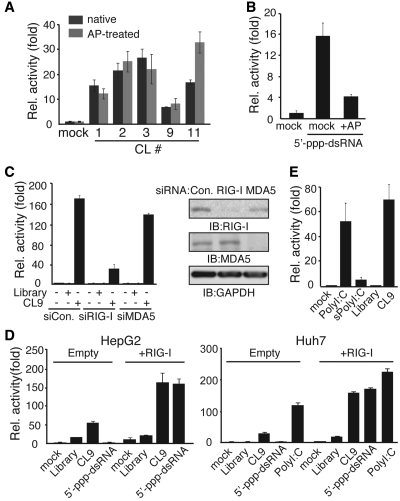
To determine whether the RIG-I aptamers were able to modulate RIG-I-mediated IFNβ production, we stimulated HepG2 cells with native or AP-treated RNA aptamers and measured IFNβ promoter activity using the PRD-III-I-Luc reporter (35). HepG2 cells express a fair amount of endogenous RIG-I protein and are therefore suitable for testing the regulatory effects of RIG-I aptamers (36,37). Every tested aptamer elicited stimulatory effects on the IFNβ promoter (PRDIII-I) (Figure 2A), and there were no significant differences between native and AP-treated aptamers, except for CL11. Efficacy of 5′-triphosphate removal by AP was separately tested using chemically synthesized dsRNA with 5′-triphosphate (Figure 2B). Reporter activity induced by 5′-triphosphate-dsRNA has been significantly reduced by AP treatment, indicating that AP efficiently removes the 5′-triphosphate moiety. To examine whether the regulatory effect of the screened aptamers depends RIG-I for their function, cells were transfected with RIG-I or MDA5 siRNA before aptamer treatment. The ability of RIG-I aptamer, CL9, to stimulate the IFNβ-driven promoter activity was significantly impaired when RIG-I, but not MDA5, expression was reduced (Figure 2C). Functional knock-down of MDA5 was separately confirmed in the polyI:C stimulated cells (Supplementary Figure S3). Signaling activity of the RIG-I aptamer was then compared to those of the conventional ligands of RIG-I, 5′-triphosphate-dsRNA or polyI:C (Figure 2D). Stimulating cells with 5′-triphosphate-dsRNA induced IFNβ promoter activity, only when RIG-I was overexpressed in the HepG2 or Huh 7 human hepatoma cells. In contrast, CL9 exhibited moderate levels of the IFNβ promoter activity in the HepG2 and Huh7 cells, and this activity was further enhanced by RIG-I overexpression (Figure 2D). Depending on its length, polyI:C was reported to activate RIG-I and MDA5 differentially: Long (1–10 kb) strands of polyI:C bind to MDA5, while short (<1 kb) strands of polyI:C preferentially activate RIG-I (18). CL9 exhibited similar levels of signaling activity relative to long forms of polyI:C, and >10-fold higher activity than RNaseIII-digested, short form of polyI:C (Figure 2E and Supplementary Figure S4).
Figure 2.
RIG-I aptamers strongly activate RIG-mediated signaling activity. After transfection with PRD-III-I-Luc and pRL-CMV for 36 h, HepG2 or Huh7 cells were stimulated with (A) 75 pmol of RIG-I aptamers, (B) 100 pmol of 5′-triphosphate-dsRNA for 6 h. (C) Thirty-six hours after transfection with indicated siRNAs, HepG2 cells were stimulated with AP-treated aptamers (75 pmol) for 6 h. Endogenous RIG-I and MDA5 were detected in the same cell lysates used for luciferase assay. (D) Similar to (A), except that cells were co-transfected with Empty (pEF-BOS) or RIG-I (pEF-BOS-Flag-RIG-I) with luciferase vectors, and then stimulated with 100 pmol of indicated activators for 6 h. (E) HepG2 cells were stimulated with equal amounts (1.5 µg/ml) of polyI:C, RNaseIII-treated short polyI:C, AP-treated RNA library, or AP-treated CL9 for 6 h before luciferase assay. Data from three independent experiments, mean ± SD.
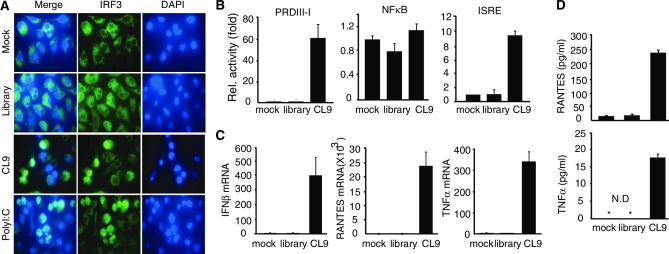
We next examined whether RIG-I aptamer activates signaling pathways for IFNβ production. When treated, CL9 induced nuclear translocation of IRF3, a transcription factor that occupies the IFNβ promoter (Figure 3A), and stimulated STAT1-mediated, but not NFκB-mediated, trans-activation activity (Figure 3B). As a result, mRNA expression of IFNβ, RANTES and TNFα was dramatically enhanced by CL9 (Figure 3C). Protein levels of RANTES and TNFα were separately confirmed in the CL9-stimulated condition (Figure 3D). These results indicate that RNA aptamers with specific binding potential to RIG-I protein stimulate IFNβ-mediated signaling pathways in a RIG-I-dependent, yet 5′-triphosphate-RNA-independent manner.
Figure 3.
RIG-I aptamer activates signaling pathways for IFNβ produciton. (A) Nuclear translocation of IRF3 by RIG-I aptamer. The stimulated cells were stained with an IRF3 antibody (green) and Hoechst 33258 (nucleus). (B) HepG2 cells were transfected with PRD-III-I-Luc, NFκB-Luc, or ISRE-Luc reporter, and stimulated with 75 pmol of AP-treated aptamers for 8 h. (C and D) HepG2 cells were stimulated with AP-treated CL9 (75 pmol) for 12 h. The expression of IFNβ, RANTES, and TNFα gene was measured by qRT-PCR in mRNA level (C), and by ELISA assay in protein level (D). Data from three independent experiments, mean ± SD.
PolyU motif of the aptamer CL9 is critical to activate RIG-I-mediated signaling pathways
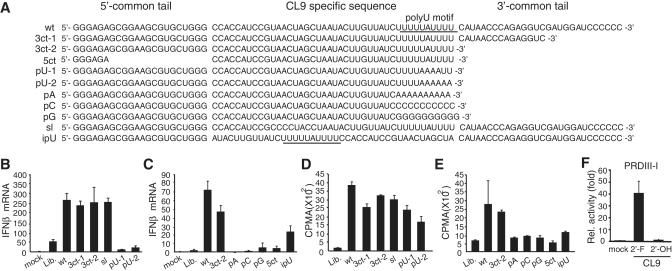
To identify the key regulatory feature that is responsible for RIG-I activation, we constructed various CL9 mutants and measured their effect on the expression of IFNβ mRNA (Figure 4A). Every RNA oligonucleotide selected for the RIG-I aptamer harbored common 5′ and 3′ tails that were used for reverse transcription and amplification step of SELEX screening (see ‘Materials and Methods’ section). Therefore we first evaluated the contribution of the 5′ and 3′ common tails. Deletion mutations in the 3′ common tail that flank polyU motif in the CL9 (3ct-1, 3ct-2) did not alter stimulatory effect on IFNβ production (Figure 4B), while additional deletion of the 5′ common tail was deteriorative (Figure 4C). Base substitutions in the predicted stem-loop structure (sl; Supplementary Figure S5) did not have any effect on the IFNβ expression. In contrast, a long stretch of polyU motif was critical for its activity to produce IFNβ, as partial substitution of consecutive ‘U's to ‘A's (pU-1, pU-2) almost completely abolished its activity (Figure 4B). Furthermore, complete substitution of polyU motif to poly A, C or G (pA, pC, pG) impaired its activity to induce IFNβ mRNA expression (Figure 4C). Interestingly, relative position of polyU motif within aptamer was critical, as ipU mutant which contains same nucleotide composition but internal polyU motif induces less IFNβ mRNA expression, compare to wt (Figure 4C). These results clearly indicate that a polyU motif within aptamer sequence plays critical roles in the activation of RIG-I-mediated IFNβ production.
Figure 4.
Unique sequence features of CL9 for RIG-I activation. (A) Sequence of the CL9 mutants used in this study. 3ct, deletion mutants in the 3′-common tail region; 5ct, a deletion mutant in the 5′-common tail region; pU, mutants with point mutations in the polyU motifs; pA, pC, pG, mutants with poly A- C- or Gs in the polyU motif, respectively; sl, a mutant with substitutions in the predicted internal stem-loop; ipU, a mutant with internal polyU motif. (B and C) Effect of various CL9 mutants on IFNβ production. HepG2 cells were stimulated with 75 pmol of the indicated CL9 mutants for 4 h, and IFNβ mRNA was measured by qRT-PCR. Mock, Lipofectamine alone. Lib., library control. (D and E) Binding of CL9 mutants to the purified RIG-I protein, in vitro. The 32P-end labeled-aptamers bound to RIG-I protein in vitro were quantitated using LSA method. (F) CL9 was prepared by in vitro transcription using 2′-F-UTP/CTP (2′-F) or 2′-OH-UTP/CTP (2′-OH). Signaling activity was measured by luciferase assay in the pPRD-III-I-luc transfected HepG2 cells. Representative data of three independent experiments are shown, mean ± SD.
To investigate whether signaling activity of the selected aptamer is depends on its binding ability to RIG-I protein, we then evaluated the binding potential of the mutant aptamers to the purified RIG-I protein. In general, signaling activity of the mutant aptamers to produce IFNβ positively correlates with their in vitro RIG-I binding, except pU-1 and pU-2 mutants (Figure 4D and E). Although pU-1 and pU-2 mutants fail to activate IFNβ production, both of them exhibited comparable levels of RIG-I binding. These data suggest that RIG-I aptamer might not act as a conventional ligand for RIG-I, but function to stabilize the conformation of RIG-I in an active state. Finally, we examined the IFNβ production activity of CL9 synthesized with 2′-OH-UTP and 2′-OH-CTP (Figure 4F). Signaling activity of CL9 was completely abolished when conventional nucleotides were used instead of 2′-F-UTP and 2′-F-CTP, suggesting that 2′-F-containing CL9 has a unique structural feature for RIG-I activation.
RNA aptamer CL9 requires full-length RIG-I for its stimulatory effect
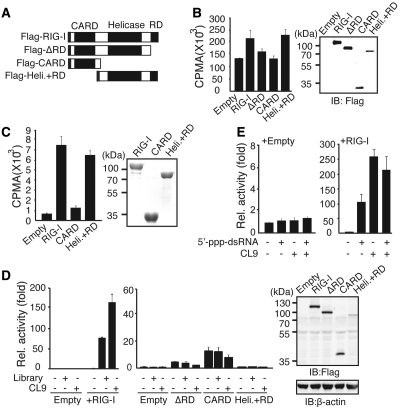
We next examined the binding domains of RIG-I to aptamer CL9. For this purpose, each mutant form of RIG-I was overexpressed in HEK293 cells, and 32P-labeled CL9 binding to the immunoprecipitated RIG-I proteins was measured (Figure 5A and B). CL9 was present in the immunoprecipitated Flag-RIG-I complex, but this binding was significantly diminished in the immunocomplex with Flag-ΔRD or Flag-CARD. In contrast, CL9 was found in the immunocomplex with Flag-Helicase+RD, which contains a helicase domain as well as a carboxyl-terminal RD domain. Direct binding between CL9 and mutant forms of RIG-I was separately confirmed using bacterially purified recombinant proteins in vitro (Figure 5C).
Figure 5.
CL9 binds to the RD-containing Helicase domain of RIG-I. (A) Schematic diagram of the RIG-I mutants used in this study. (B) Left, AP-treated 32P-CL9 was mixed with the indicated proteins that were over-expressed in HEK293 cells. Right, The expression level of the transfected protein was measured by western blot assay. (C) Left, Wild-type or mutant forms of RIG-I protein were purified from bacteria and incubated with AP-treated 32P-CL9 in vitro. Right, The expression of each protein was measured by Coomassie blue staining (D) Left, Huh7 cells were transfected with various RIG-I mutant plasmids as indicated, along with PRD-III-I-Luc and pRL-CMV, and luciferase activity was assayed after stimulation for 6 h. Right, Ectopic expression of RIG-I mutants was measured using an anti-Flag antibody. (E) Similar to (D), except cells were stimulated with 5′-triphosphate-dsRNA (100 pmol) and/or AP-treated CL9 (100 pmol) for 6 h. Data from more than three independent experiments, mean ± SD.
Although our data indicate that CL9 binds directly to the C-terminal RD and helicase domain of RIG-I, aptamer binding itself was not sufficient to activate RIG-I-mediated signaling activity, as none of the RIG-I mutants was able to transmit aptamer-specific signaling activity (Figure 5D). These data indicate that CL9 requires an intact form of RIG-I to elicit its signaling activity. Because CL9 functions in a 5′-triphosphate-independent manner, we wondered whether providing a 5′-triphosphate-dsRNA moiety in trans can further enhance signaling activity of CL9. To test this idea, the signaling activity of CL9 in the absence or presence of 5′-triphosphate-dsRNA was examined in the RIG-I overexpressed Huh7 cells (Figure 5E). However, no additive effect was observed, indicating that CL9 binding was sufficient to provide conformational change of RIG-I for full activity.
RIG-I specific RNA aptamer CL9 efficiently blocks viral replication
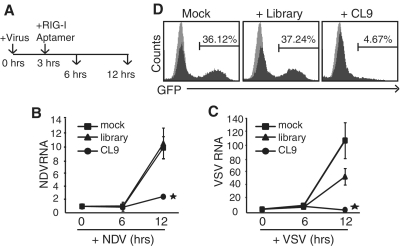
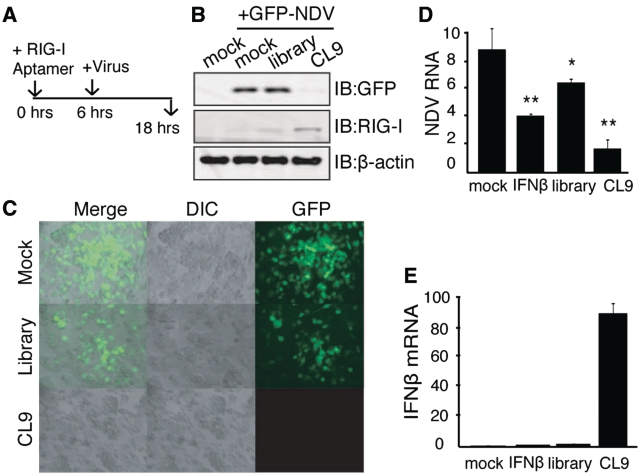
RIG-I-dependent activation of IFNβ expression is a key step to trigger antiviral responses against foreign RNA viruses, such as influenza virus, NDV, VSV and HCV (20). Therefore, we evaluated whether RIG-I aptamer has the potential to inhibit the replication of infected RNA viruses. To do this, HepG2 cells were infected with either NDV-GFP, or VSV-GFP, which both contain a gene coding Green Fluorescent Protein (GFP) in the viral genome (38) (Supplementary Figure S6). To evaluate its therapeutic potential, cells were treated with CL9 3 h after virus infection, and viral replication was assayed 3 or 9 h later (Figure 6A). In contrast to the RNA library pools used as negative control, CL9 treatment efficiently blocked replication of NDV and VSV (Figure 6B and C). Similar effect on the repression of influenza A (PR8 strain)-GFP replication was also demonstrated in the cells treated with CL9 (Figure 6D). We then examined whether pre-treating cells with CL9 prior to viral infection had any preventative effects (Figure 7A). NDV replication was almost completely blocked when cells were treated with CL9 6 h before infection, visualized by fluorescence of GFP (Figure 7B and C) or PCR with virus specific primers (Figure 7D). Efficient induction of IFNβ mRNA by CL9 was separately verified (Figure 7E). These results suggest that RIG-I aptamer has strong potential to inhibit viral replication, via activation of RIG-I and IFNβ production.
Figure 6.
Post-treatment of CL9 blocks viral replication. (A) Experimental scheme: HepG2 cells were infected with virus for 3 h before aptamer (75 pmol) treatment and incubated for an additional 3 or 9 h before the assay. Presence of the viral fragment specific for NDV (B) or VSV (C) was analyzed by qRT-PCR. Mean (±SD) values of three independent samples are shown (Student t-test, *P < 0.05). (D) A549 cells were infected with Influenza A (H1N1, PR8)-GFP virus for 3 h before aptamer treatment and incubated for additional 9 h. The fraction of virus-infected cells was measured by FACS analysis (gray: mock-infection, black: virus-infection).
Figure 7.
Pre-treatment of CL9 inhibited viral replication. (A) Experimental scheme: Prior to virus infection, HepG2 cells were pre-treated with 75 pmol of aptamers for 6 h. Viral replication was assayed after 12 h of treatment by immunoblot against anti-GFP (B) or GFP fluorescence (C). (D and E) Same experimental scheme as (A), except RNA for NDV (D) and IFNβ (E) were analyzed: Student t-test, **P < 0.005, *P < 0.05, relative to the mock-treated sample. Data from more than three independent experiments.
DISCUSSION
RIG-I is a DExH/D RNA helicase that senses intracellular non-self RNA structures derived from infected RNA viruses. In this article, we report the identification of RIG-I-specific RNA aptamers with anti-viral potential. The selected RIG-I aptamer behaves as a strong agonist for RIG-I, as determined by its tendency to activate RIG-I-mediated signaling pathways and its dependence on the expression of RIG-I. As a result, RIG-I aptamer efficiently blocked replication of RNA virus in infected cells, suggesting it has a strong potential for therapeutic applications.
Originally identified as a cytosolic receptor for non-self RNA, RIG-I monitors unique RNA structures in the cytoplasm. It binds dsRNA with 5′-triphosphate moiety via carboxyl-terminal RD domain of RIG-I (14,39,40). Double-stranded RNA of intermediate length (between 300 and 2000 bp) and blunt-ended short dsRNA (23 ∼ 30 bp) are also known to be recognized by RIG-I, although they lack 5′-triphosphate moiety (12,41,42). RIG-I tends to have a preference for dsRNA, as ssRNA with 5′-triphosphate moiety fails to activate RIG-I (30). In contrast to the conventional ligands of RIG-I, the selected RIG-I aptamers seem to utilize distinct binding strategies: the 5′-triphosphate moiety of the aptamer was not required for its binding to RIG-I, yet the helicase and C-terminal RD domains of RIG-I were essential for its binding. Because it has been reported that self-templated ‘copy-back’ activity of T7 RNA polymerase generates double-stranded RNA (34), we wondered whether in vitro transcribed RIG-I aptamer forms a dsRNA structure. When we evaluated the size of the transcribed RIG-I on a denaturing polyacrylamide gel, in vitro transcribed RIG-I aptamer migrated to the position that matches the size of the single-stranded RIG-I aptamer (Supplementary Figure S7). In addition, its stimulating potential of RIG-I-mediated signaling activity was strictly dependent on the polyU motifs and its position within aptamer (Figure 4), rather than its physical binding to RIG-I, suggesting that the unique feature derived from the selected aptamer plays an important role in the activation of the RIG-I. It is noteworthy to mention that unanchored K63-poly-Ub chains have been reported to act as a potent ligand of RIG-I, thereby suggesting that there are another class of RIG-I ligand that functions non-conventional manner (43).
In this study, we showed that RIG-I aptamer binds to the RD-containing helicase domain of RIG-I and stimulates RIG-I-dependent signal in a 5′-triphosphate-RNA-independent manner. Specifically, the polyU motif found near the 3′ side of RIG-I aptamer, CL9, was essential for the activation of the RIG-I. This polyU motif is somewhat reminiscent of the polyU/UC in the RNA of replicating HCV, which has been identified as a strong ligand for RIG-I (44,45). However, a couple of differences exist between these two motifs. First, polyAG/A showed comparable activity to polyU/UC of HCV genome in the production of IFNβ, but the replacement of U with polyA of CL9 almost completely blocked IFNβ expression. Second, the RIG-I activating signal of the polyU/UC in the HCV genome was diminished when 2′-F-uridine was used instead of 2′-OH-uridine. However, in vitro-synthesized CL9 with 2′-F-uridine and 2′-F-cytosine exhibited much stronger activity over transcripts generated with 2′-OH-uridine and 2′-OH-cytosine. It is noteworthy to mention that RNA aptamers synthesized with 2′-F-uridine and 2′-F-cytosine exhibits increased RNA stability compared to 2′-OH-uridine and 2′-OH-cytosine, due to the resistance to nuclease digestion (46). And finally, polyU/UC of HCV is reported to lose its activity after PAGE purification (44,45), but in vitro-synthesized CL9 keeps its activity after the same purification procedure. In all, these data indicate that RIG-I activation by RIG-I aptamer is quite distinct that of the polyU/UC sequence derived from the HCV RNA genome.
We propose that RIG-I aptamers can be potential tools based on their ability to enhance IFNβ production and block viral replication. Due to the immune-cell-restricted expression patterns of Toll-like receptor (TLR) family members and their localization on the surface membrane, reagents that target these proteins have been actively screened for the treatment of viral infection or inflammation-related diseases (47,48). In contrast, RIG-I is mainly expressed in non-immune cells and localizes in the cytoplasm, presenting technical difficulties associated with intracellular delivery of the targeting agents. Furthermore, RIG-I is mainly expressed in fibroblasts, epithelial cells, hepatocytes, and conventional dendritic cells (cDCs) (20), all of which express very low levels of TLRs, suggesting that reagents that target TLRs may not be ideal for systemic application. Instead, local application of RIG-I aptamers to the infected sites may present an attractive alternative. For example, local injection of RIG-I aptamers in olfactory or airway epithelial cells may be effective for treatment of human metapneumovirus (HMPV) or respiratory syncytial virus (RSV) infection, which are both recognized by RIG-I and are known to cause bronchiolitis and even pneumonia (15,49). In summary, we report RIG-I RNA aptamers have a strong tendency to stimulate IFNβ expression and block viral replication. Further studies to increase the efficacy and stability of RIG-I aptamers, as well as the development of cellular delivery tools, are necessary.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7, Supplementary Methods and Supplementary References (18,50–52).
FUNDING
Funding for open access charge: POSCO Research Fund, the Korea Healthcare Technology R&D Project (Ministry for Health, Welfare & Family Affairs; A080084, A084584 to J.Y.Y.); Bio R&D Program (NRF, Korea; 2011-0018331 to J.Y.Y.); Regional Core Research Program (Korean Ministry of Education, Science and Technology, to J.Y.Y.); Global Research Program of National Research Foundation of Korea (grant number: K20815000001 to B.H.O.) and EPB center (grant number: 20110001019 to B.H.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Aptamer Core Technology Unit of POSTECH for technical support.
REFERENCES
- 1.Chen J, Subbarao K. The immunobiology of SARS*. Annu. Rev. Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 2.Doherty PC, Turner SJ, Webby RG, Thomas PG. Influenza and the challenge for immunology. Nat. Immunol. 2006;7:449–455. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 3.Racanelli V, Rehermann B. Hepatitis C virus infection: when silence is deception. Trends Immunol. 2003;24:456–464. doi: 10.1016/s1471-4906(03)00178-9. [DOI] [PubMed] [Google Scholar]
- 4.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 5.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 6.Averett DR, Fletcher SP, Li W, Webber SE, Appleman JR. The pharmacology of endosomal TLR agonists in viral disease. Biochem. Soc. Trans. 2007;35:1468–1472. doi: 10.1042/BST0351468. [DOI] [PubMed] [Google Scholar]
- 7.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman ES, Smith RE, Renaud RC., Jr From the analyst's couch: TLR-targeted therapeutics. Nat. Rev. Drug Discov. 2005;4:879–880. doi: 10.1038/nrd1880. [DOI] [PubMed] [Google Scholar]
- 9.De Francesco R, Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- 10.Ishii KJ, Uematsu S, Akira S. ‘Toll’ gates for future immunotherapy. Curr. Pharm. Des. 2006;12:4135–4142. doi: 10.2174/138161206778743484. [DOI] [PubMed] [Google Scholar]
- 11.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann. NY Acad. Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 14.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 15.Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 2007;81:1401–1411. doi: 10.1128/JVI.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl Acad. Sci. USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 21.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 22.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 23.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl Acad. Sci. USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 26.Sullenger BA, Lee TC, Smith CA, Ungers GE, Gilboa E. Expression of chimeric tRNA-driven antisense transcripts renders NIH 3T3 cells highly resistant to Moloney murine leukemia virus replication. Mol. Cell. Biol. 1990;10:6512–6523. doi: 10.1128/mcb.10.12.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browning CM, Cagnon L, Good PD, Rossi J, Engelke DR, Markovitz DM. Potent inhibition of human immunodeficiency virus type 1 (HIV-1) gene expression and virus production by an HIV-2 tat activation-response RNA decoy. J. Virol. 1999;73:5191–5195. doi: 10.1128/jvi.73.6.5191-5195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda K, Vishnuvardhan D, Sekiya S, Hwang J, Kakiuchi N, Taira K, Shimotohno K, Kumar PK, Nishikawa S. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur. J. Biochem. 2000;267:3685–3694. doi: 10.1046/j.1432-1327.2000.01400.x. [DOI] [PubMed] [Google Scholar]
- 29.Kumar PK, Machida K, Urvil PT, Kakiuchi N, Vishnuvardhan D, Shimotohno K, Taira K, Nishikawa S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology. 1997;237:270–282. doi: 10.1006/viro.1997.8773. [DOI] [PubMed] [Google Scholar]
- 30.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MJ, Yoo JY. Active caspase-1-mediated secretion of retinoic acid inducible gene-I. J. Immunol. 2008;181:7324–7331. doi: 10.4049/jimmunol.181.10.7324. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 33.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5’-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl Acad. Sci. USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 36.Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 37.Preiss S, Thompson A, Chen X, Rodgers S, Markovska V, Desmond P, Visvanathan K, Li K, Locarnini S, Revill P. Characterization of the innate immune signalling pathways in hepatocyte cell lines. J. Viral Hepat. 2008;15:888–900. doi: 10.1111/j.1365-2893.2008.01001.x. [DOI] [PubMed] [Google Scholar]
- 38.Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 2003;77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 40.Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. Cytosolic 5’-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 42.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan W, Craven RC, Qiu Q, Wilson CB, Wills JW, Golovine S, Wang JF. Isolation of virus-neutralizing RNAs from a large pool of random sequences. Proc. Natl Acad. Sci. USA. 1995;92:11509–11513. doi: 10.1073/pnas.92.25.11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat. Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 48.Zuany-Amorim C, Hastewell J, Walker C. Toll-like receptors as potential therapeutic targets for multiple diseases. Nat. Rev. Drug Discov. 2002;1:797–807. doi: 10.1038/nrd914. [DOI] [PubMed] [Google Scholar]
- 49.Liao S, Bao X, Liu T, Lai S, Li K, Garofalo RP, Casola A. Role of retinoic acid inducible gene-I in human metapneumovirus-induced cellular signalling. J. Gen. Virol. 2008;89:1978–1986. doi: 10.1099/vir.0.2008/000778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B. Generation of species cross-reactive aptamers using “toggle” SELEX. Mol. Ther. 2001;4:567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- 51.Fitzwater T, Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- 52.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.