Abstract
Ribosomal proteins are essential to life. While the functions of ribosomal protein-encoding genes (RPGs) are highly conserved, the evolution of their regulatory mechanisms is remarkably dynamic. In Saccharomyces cerevisiae, RPGs are unusual in that they are commonly present as two highly similar gene copies and in that they are over-represented among intron-containing genes. To investigate the role of introns in the regulation of RPG expression, we constructed 16 S. cerevisiae strains with precise deletions of RPG introns. We found that several yeast introns function to repress rather than to increase steady-state mRNA levels. Among these, the RPS9A and RPS9B introns were required for cross-regulation of the two paralogous gene copies, which is consistent with the duplication of an autoregulatory circuit. To test for similar intron function in animals, we performed an experimental test and comparative analyses for autoregulation among distantly related animal RPS9 orthologs. Overexpression of an exogenous RpS9 copy in Drosophila melanogaster S2 cells induced alternative splicing and degradation of the endogenous copy by nonsense-mediated decay (NMD). Also, analysis of expressed sequence tag data from distantly related animals, including Homo sapiens and Ciona intestinalis, revealed diverse alternatively-spliced RPS9 isoforms predicted to elicit NMD. We propose that multiple forms of splicing regulation among RPS9 orthologs from various eukaryotes operate analogously to translational repression of the alpha operon by S4, the distant prokaryotic ortholog. Thus, RPS9 orthologs appear to have independently evolved variations on a fundamental autoregulatory circuit.
Author Summary
Eukaryotic genes are littered with non-coding intervening sequences, or introns, that must be precisely excised from a messenger RNA before it can be properly translated into protein. Despite their ubiquity, the evolution and function of introns remain poorly understood. Consequently, we cannot accurately predict the functions of individual introns in any organism. In this manuscript, we used a combination of comparative genomics and experimental tests to identify functional introns. First, we looked for signatures of selection to identify important introns in the model yeast Saccharomyces cerevisiae, which focused our attention on the introns of ribosomal protein genes. We then genetically deleted these introns to assess their function. Unlike mammalian introns, we found that yeast introns were not required for high levels of gene expression. Instead, particular introns (we focus on those within genes encoding ribosomal protein S9) were required to fine-tune gene expression through autoregulation. Surprisingly, animal orthologs of these genes also use introns to autoregulate through multiple forms of alternative splicing. We speculate that the introns of ribosomal protein genes, in particular, readily evolve means for autoregulation to meet the demanding requirements of ribosomal protein genes to maintain tight control of gene expression.
Introduction
The evolution and function of spliceosomal introns are among the largest unsolved mysteries of eukaryotic genomes. Pronounced differences in intron evolution between lineages and between introns within the same lineage provide insight into 1) the selective and mutational forces governing intron evolution and 2) the potential roles of introns in gene function. Here we study the case of ribosomal protein genes (RPGs) in the model yeast Saccharomyces cerevisiae. RPGs are highly over-represented among intron-containing genes (69% of RPGs contain introns compared to ∼5% of non-RPGs), which has been suggested to reflect ongoing selection for introns that provide one or more functions in gene expression [1], [2]. However, two major facets of this hypothesis — the action of selection (intron evolution), and the source of this selection (intron function) — remain unknown. First, biased intron loss has not been specifically tested within hemiascomycetous yeasts (S. cerevisiae and relatives). And second, the effect of intron loss on RPG expression remains uncertain.
RPG expression is remarkable both in terms of synthesis rate and control [3]; thus, RPG introns may function to promote these aspects of gene expression. One proposal predicts that RPG introns function to promote high levels of expression. Consistent with this view, intron-containing genes, including RPGs, produce some of the highest transcript and protein abundances in S. cerevisiae [4]. However, the requirement for introns to enhance RPG expression has not been directly tested.
In addition to the above, two other proposals predict that RPG introns function by providing an opportunity for splicing regulation. One possibility is that introns provide rapid regulation in response to environmental stress, as suggested by splicing inhibition of RPG pre-mRNAs in response to amino acid starvation [5]. Another possibility is that introns provide an opportunity to fine-tune gene expression through autoregulation. For example, negative feedback control of RPL30 and RPS14B expression is achieved through the binding of their respective protein products to RNA structures within their own unspliced transcripts, thereby regulating splicing [6], [7]. Interestingly, nearly all the ribosomal proteins of Escherichia coli are regulated by key ribosomal proteins in an analogous manner; for example, bacterial S4 directly binds its own mRNA to repress the translation of itself and three other RPGs [8], [9]. Given that the majority of S. cerevisiae RPGs contain introns, intron-dependent autoregulation may be more common than previously appreciated.
We report the first direct tests of both the action and the source of selection on RPG introns. First, we used comparative genomics to show that RPG introns have been preferentially retained following whole genome duplication (WGD), indicating ongoing selection for retention of RPG introns. Second, we generated S. cerevisiae strains harboring precise deletions of 16 RPG introns to distinguish between selective hypotheses. We found that RPG introns generally reduce gene expression, suggesting that RPG introns allow for splicing regulation rather than promoting high levels of expression. In particular, we identified intron-dependent cross-regulation between the RPS9A and RPS9B genes, which both encode ribosomal protein S9 (S9). Finally, overexpression of RpS9 in D. melanogaster S2 cells, and analysis of available EST sequences, suggest that autoregulation of RPS9 orthologs may involve different forms of splicing regulation between species, but also appears to be widespread across disparate lineages.
Results
Yeast ribosomal protein genes have resisted recent intron loss
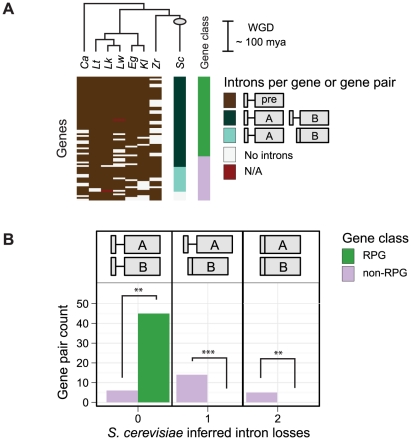
Introns are over-represented in the RPGs of both Candida albicans and S. cerevisiae [1], [2]. While this shared over-representation may reflect selection pressure to maintain RPG introns prior to the divergence of these two species from a common ancestor, it may also reflect the action of selection in more recent history, since their divergence from a common ancestor. This distinction is important, since selection pressure to maintain RPG introns in more recent history is more likely to be relevant to the biology of S. cerevisiae. To determine if RPGs have resisted intron loss compared to other genes since the divergence of C. albicans and S. cerevisiae (∼200–800 million years ago [10]), we assessed the fates of S. cerevisiae introns in paralogs (a.k.a. gene pairs) that were duplicated ∼100 million years ago by whole-genome duplication (WGD) [11]. To determine the fates of introns after genome duplication, we took advantage of the well-annotated genome of S. cerevisiae, which has been exhaustively searched for introns [12], [13]. With these annotations, we identified 121 intron-containing genes among 554 WGD-derived gene pairs obtained from Yeast Gene Order Browser [14]. Assuming that intron loss has largely dominated intron evolution in hemiascomycetous yeast species [15], we inferred intron loss if one of the WGD-derived gene copies had fewer introns than the other. Using this criterion, we calculated the number of apparent intron losses in RPG pairs compared to all other gene pairs. Strikingly, this simple accounting revealed that 16 of 23 non-RPG pairs have a gene with fewer introns than its copy, whereas none of the 46 RPG pairs did. Nonetheless, this analysis ignores intron losses that occurred independently in both gene copies and assumes that intron gain did not occur.
To better assess whether WGD-derived RPG pairs have been biased for either intron gain or loss (including losses in both gene copies), we reconstructed the hypothetical intron distribution of the pre-WGD ancestor that existed prior to the WGD event. For each of the 554 S. cerevisiae duplicated gene pairs, we assigned the presence or absence of an intron in the hypothetical pre-WGD ancestral ortholog based on intron annotations and predictions from the genomes of the pre-WGD (so-called protoploid) species (C. albicans, Lachancea waltii, L. thermotolerans, L. kluyveri, Eremothecium gossypii, Kluyveromyces lactis, and Zygosaccharomyces rouxii) and the genomes of the post-WGD species (Vanderwaltozyma polyspora, Naumovia castellii, C. glabrata, and S. bayanus). A complete list of intron predictions and annotations can be found in Table S1. Our analysis revealed 73 intron-containing genes that were likely present in the pre-WGD ancestor from which the duplicated gene pairs in S. cerevisiae were descended (Figure 1A). Based on this hypothetical intron distribution of the pre-WGD ancestor, we inferred the number of S. cerevisiae WGD-derived gene pairs that have gained or lost an intron for each post-WGD gene pair (Figure 1B). From this improved analysis, we identified 5 S. cerevisiae non-RPG pairs that appear to have independently lost introns from both gene copies after gene duplication. This was in addition to 14 non-RPG pairs in which one of two introns were lost (Figure 1B, right and middle columns, respectively). Once again, we inferred no intron losses in S. cerevisiae RPG pairs (Figure 1B, left column). Thus, RPG introns appear to have been biased against loss in the lineage leading to S. cerevisiae during the last ∼100 million years.
Figure 1. Biased intron loss in hemiascomycetous yeasts after the recent whole-genome duplication event.
A) The dendogram (top) illustrates the assumed topology of the phylogenetic relationships used to infer intron-containing genes present in the pre-WGD ancestor prior to the WGD event (based on [63]; not to scale); the estimated time of the WGD event is indicated (gray circle and scale bar) [11]. A heatmap (bottom) illustrates the number of introns in S. cerevisiae gene pairs and their orthologs (rows) by species (columns). Genes containing an intron in pre-WGD species (brown tiles) were used to infer the intron-containing genes present in the pre-WGD ancestor (see Methods). Among the 95 S. cerevisiae gene pairs derived from an intron-containing gene in the pre-WGD ancestor, those with an intron in both gene copies (dark blue-green) were inferred to have no intron losses. S. cerevisiae gene pairs with an intron in only one of two gene copies (light blue-green) or no intron in either gene copy (white) were inferred to have had one or two intron losses, respectively. Missing genes (red) are indicated. RPGs (green) and other functional gene classes (purple) are indicated (right-most column). See Table S1 for intron predictions and annotations. Ca = C. albicans, Lw = L. waltii, Lt = L. thermotolerans, Lk = L. kluyveri, Eg = E. gossypii, Kl = K. lactis, Zr = Z. rouxii, Sc = S. cerevisiae. B) A histogram counts the number of inferred intron losses for each S. cerevisiae gene pair that descended from an intron-containing pre-WGD ortholog. Intron losses from RPGs (green) are compared to other functional gene classes (purple). Asterisks indicate statistical significance values p<0.01 (**), and 0.001 (***); exact binomial test.
Next, we asked whether intron gains contributed to the bias for introns in S. cerevisiae RPGs. For a given S. cerevisiae gene, we inferred that an intron was gained if introns were absent in both the pre-WGD ancestor and the majority of post-WGD orthologous gene pairs. Using this criterion, we did not infer intron gains in any of the S. cerevisiae RPGs. On the other hand, two introns in non-RPGs (i.e. USV1 and BMH2) have possibly been gained in the S. cerevisiae lineage (Table S1); however, since both of these introns are located in the 5′ UTR and are not well annotated in other species, it is therefore difficult to be confident of this conclusion. Taken together, the bias for introns in S. cerevisiae RPG pairs appears to have been dominated not by intron gains in RPGs, but by intron losses in non-RPGs.
Introns repress ribosomal protein gene expression
Having found a bias against RPG intron loss, we sought to determine if RPG introns have a function in gene expression. To mimic the effect of RPG intron loss, we created S. cerevisiae mutant intron deletion strains (henceforth denoted as Δi). Each Δi mutant was created with a precise deletion of a single RPG intron, such that only an intronless copy of the gene remained at the endogenous locus (See Methods).
Because RPGs are among the most highly expressed genes in the genome, we tested the model that introns are required in cis for high levels of gene expression by assessing the expression profiles of 16 Δi mutants compared to a wild-type strain. We also considered the possibility that Δi mutations may affect other genes in trans, in particular, the WGD-derived gene copies of RPG pairs. To measure changes in expression of the gene from which an intron was deleted (in addition to 124 RPG and 911 non-RPG features) we used custom splicing-sensitive microarrays designed to detect pre-, mature, and total mRNA species (using intron, junction, and exon probes, respectively [16]). To assess the effect of Δi mutation on gene expression, we plotted the expression change for the intronless gene (Figure 2A, red lines) compared to all the other genes on the microarray (Figure 2A, boxplots). Thus, the most significant expression changes lie outside the whiskers of the boxplot and are, by definition, statistical outliers. Intron deletion mutations, as assessed by microarray, typically had only modest effects on gene expression (Figure 2A, compare red lines to boxplots). Nonetheless, these effects were biased toward increased expression of the intronless gene (14 out of 16), rather than decreased expression (Figure 2A “up” and “down,” respectively). Moreover, the four most substantial expression changes increased the expression of the intronless gene (Figure 2A “outlier”). These data suggest that yeast introns are generally not required for the high expression levels of RPGs. Further, only a few genes showed substantial increases in expression, which suggests that splicing may be more inefficient for these genes than most other RPGs.
Figure 2. RPG intron deletions reveal gene-specific effects on steady-state mRNA levels.
A–C) Microarray expression data for 16 RPG Δi mutants compared to a common wild-type strain. In each panel, the change in expression due to intron deletion is shown for either the intronless gene (red lines) or its paralogous gene copy (blue lines) compared to all other changes detected by microarray (boxplots). The effect of intron deletion is shown for each Δi mutant on A) the expression of the intronless gene copy, B) the expression of the paralogous gene copy, and C) the Intron Accumulation Index of the paralogous gene copy. Microarray data are expressed as the normalized log2 transformed probe intensity for exon features averaged from at least two replicate microarrays. Whiskers represent 1.5 times the interquartile range. D) RT-qPCR quantification of RPS9A (red circles) and RPS9B (blue triangles) expression changes for each Δi mutant relative to wild-type (columns). RPS9A and RPS9B values were divided by SCR1 values to obtain ratios controlled for variations in cDNA quantity. Log2 transformed ratios are plotted relative to wild-type (based on the mean of three biological replicates). Each of three biological replicates is shown as a point and the mean as a dash. E) The effect of intron deletion on the total number of transcripts encoding S9. Stacked barplots illustrate the percent of RPS9A (red bars) and RPS9B (blue bars) transcripts calculated for each Δi mutant. For a wild-type strain (first column), the percent of RPS9A and RPS9B transcripts encoding S9 were estimated from published RNA-seq data [19]. Changes in RPS9A and RPS9B transcript numbers for each Δi mutant (columns) were calculated by multiplying wild-type percentages by relative expression changes determined by qPCR.
We also sought to determine if any of the deleted introns were required for splicing regulation. As controls, we deleted the introns of RPS14A and RPS14B, as it has been known for some time that S14 binds to the RPS14B intron (but not the RPS14A intron) to inhibit splicing and to cause rapid degradation [7], [17]. As expected, deletion of the RPS14B intron led to a substantial increase in its expression compared to the other genes on the microarray (Figure 2A “outlier”), whereas deletion of the RPS14A intron had little effect on expression (Figure 2A “down”). Thus, our microarrays have the sensitivity required to detect the derepression of RPS14B expression. An unexpected and novel finding is the substantial effect that Δi mutations have on the expression of the two gene copies encoding ribosomal protein S9 (hereafter referred to as S9). Our microarray experiments revealed that RPS9A and RPS9B Δi mutations increased the expression of the intronless genes (Figure 2A “outlier”) and also decreased the expression of the wild-type gene copies (Figure 2B). We hypothesized that the decreased expression of the wild-type RPS9A and RPS9B genes was caused by decreased splicing efficiency due to negative feedback. Therefore, we tested whether Δi mutations caused an increase in the ratio of pre-mRNA to total mRNA of the wild-type gene copies by calculating the Intron Accumulation Index of these genes, which is a measure of inefficient splicing [18]. Of all the mutants tested by microarray, only RPS9A and RPS9B showed substantial increases in the Intron Accumulation Index compared to the other intron containing genes on the array (Figure 2C, compare blue lines to boxplots). Taken together, these data suggest that the RPS9A and RPS9B genes require introns to repress their own expression. Further, derepression of RPS9A resulted in increased repression of RPS9B through splicing inhibition (and vice versa), suggesting that these genes cross-regulate.
Our custom microarray platform is precise; however, it lacks control probe sets needed for highly accurate quantification. As such, our microarrays “compress” fold-changes compared to equivalent determination by qPCR. To validate our most surprising observations, we assessed RPS9A and RPS9B expression by RT-qPCR. Importantly, we designed at least one qPCR primer to the 3′UTR in an effort to maximize specificity and to minimize artifacts caused by primer cross-hybridization to the other gene copy. As expected, qPCR measurements validated our microarray results for both RPS9A and RPS9B genes in the rps9bΔi and rps9bΔi mutants (Figure 2D, second and third columns). In the case of the rps9aΔi mutant, Δi mutation was associated with a substantial increase (>4-fold of wild-type) in RPS9A expression and a modest decrease (<2-fold of wild-type) in RPS9B expression (Figure 2D, second column). Conversely, in the rps9bΔi mutant, Δi mutation was associated with a modest increase (<2-fold of wild-type) in RPS9B expression and a substantial decrease (>8-fold of wild-type) in RPS9A expression (Figure 2D, third column).
Having validated the surprising effects of deleting the RPS9A and RPS9B introns, we hypothesized that the genes reciprocally cross-regulate through a shared negative feedback circuit. We made two strong predictions from this hypothesis: 1) deletion of both the RPS9A and RPS9B introns should eliminate cross-regulation, and therefore, derepress both gene copies and 2) the wild-type gene copy should compensate for a derepressed copy by an equal and opposite number of transcripts. First, to determine if repression of RPS9A expression in the rps9bΔi mutant required the RPS9A intron (and vice versa), we created a double rps9a/bΔi mutant and tested the effect on expression by RT-qPCR. As predicted, both RPS9A and RPS9B were derepressed in the rps9a/bΔi mutant (Figure 2D, fourth column). Second, we sought to determine if changes in the number of RPS9A transcripts were compensated by a nearly equal and opposite change in number of RPS9B transcripts. We first estimated the percent of transcripts encoding S9 contributed by the RPS9A and RPS9B genes (6% and 94%, respectively) from a published RNA-seq data set from a wild-type strain [19]. In order to calculate the number of transcripts in each Δi mutant, we then simply multiplied the percent of transcripts encoding S9 (as determined by RNA-seq) by the relative change in expression (as determined by qPCR) for each Δi mutant. As predicted for the rps9aΔi mutant, a substantial relative increase in RPS9A expression mutant was nearly equally compensated by a modest relative decrease in RPS9B expression, such that the total number of transcripts encoding S9 was nearly unchanged (Figure 2D and 2E, second column). In the rps9bΔi mutant, however, a modest relative increase in RPS9B expression mutant was only partially compensated at the expense of nearly all RPS9A transcripts (Figure 2D and 2E, second column). In this case, it appears that RPS9A defied our prediction and presumably because its contribution to the total number of S9 transcripts was limiting. Lastly, deletion of both introns increased the total number of transcripts encoding S9 to 170% of wild-type levels (Figure 2E, fourth column). Taken together, these data suggest that the RPS9A and RPS9B genes reciprocally cross-regulate by a common intron-dependent mechanism. Further, the large relative effects detected for RPS9A compared to RPS9B may simply reflect the large difference in expression level between the two gene copies.
Drosophila RpS9 autoregulates through alternative splicing and NMD
Reminiscent of the cross-regulation between S. cerevisiae RPS9A and RPS9B genes, several metazoan RPGs have been shown to autoregulate through alternative splicing coupled to NMD (so-called “Regulated Unproductive Splicing and Translation” or RUST): a process in which the synthesis of productively-spliced mRNA is repressed in favor of unproductive mRNA isoforms encoding premature termination codons (PTC+) [20]–[23] (reviewed in [24]). While this process is conserved between distantly related eukaryotes, there is no known overlap between the genes regulated by RUST in yeast and metazoans to facilitate mechanistic comparisons. Intriguingly, an alternatively-spliced RpS9 PTC+ mRNA isoform was recently identified in Drosophila melanogaster [25]. Thus, we considered the possibility that other RPS9 orthologs autoregulate in a manner analogous to RPS9A and RPS9B cross-regulation.
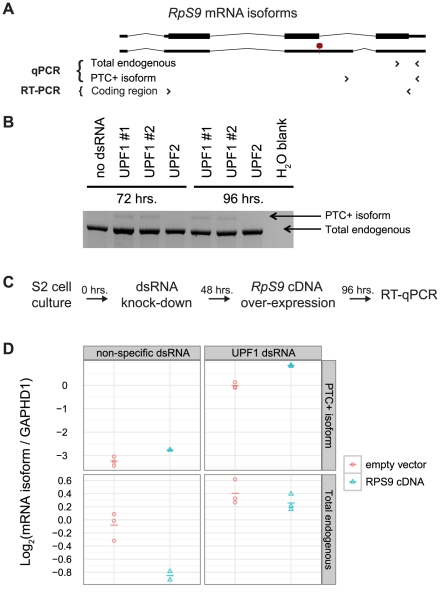
We hypothesized that D. melanogaster RpS9 expression is regulated in response to excess protein production by alternative splicing coupled to NMD. Therefore, we predicted that increased RpS9 expression would result in increased abundance of the PTC+ mRNA isoform. To test this hypothesis, we measured the affect of exogenous RpS9 overexpression and NMD inhibition on alternative splicing of RpS9 messages using RT-qPCR primer sets specific to endogenous RpS9 mRNA isoforms (Figure 3A). We first verified that the previously identified RpS9 PTC+ isoform in S2 cells was degraded by NMD through RT-PCR amplification of RpS9 transcripts from S2 cells incubated with either of two dsRNAs targeting Upf1 (Figure 3B). To then test the effect of increased RpS9 expression on the abundance of the PTC+ mRNA isoform, we exogenously overexpressed a cDNA copy of RpS9 (Figure 3C). In S2 cells overexpressing RpS9, we detected an increase in the abundance of the PTC-containing mRNA isoform (Figure 3D, top panels, compare red and blue points) and a decrease in the total RpS9 expression as compared to the empty vector control (Figure 3D, bottom left panel, compare red and blue points). As expected, we observed a UPF1-dependent decrease in total endogenous RpS9 abundance in response to increased RpS9 expression (Figure 3D, compare bottom left and right panels, blue points). Taken together, these results suggests that Drosophila RpS9 autoregulates by RUST, in which excess expression shifts the balance of alternative splicing from the synthesis of productively spliced messages towards the synthesis of unproductive RpS9 PTC+ messages that are selectively degraded by NMD.
Figure 3. D. melanogaster RpS9 is autoregulated by alternative splicing coupled to NMD.
A) Illustration of RpS9 mRNA isoforms assessed by PCR (the PTC+ isoform is indicated by a red octagon). Primers sets (arrows) were designed to amplify multiple or specific RpS9 mRNA isoforms (RT-PCR and qPCR primers, respectively). B) RT-PCR validation of the RpS9 PTC+ mRNA isoform degraded by NMD. C) Experimental design used to assess the affect of UPF1 knock-down on the abundance of RpS9 mRNA isoforms. D) RT-qPCR determination of RpS9 PTC+ mRNA isoform abundance (top panels) and total endogenous RpS9 mRNA abundance (bottom panels) in S2 cells transfected with a plasmid constitutively expressing an RpS9 cDNA (red circles) or an empty vector control (blue circles). The affect of UPF1 knock-down (via incubation with dsRNA) on each RpS9 mRNA isoform (right panels) is compared to a non-specific dsRNA control (left panels). RpS9 mRNA isoform abundance values were divided by GAPDH1 mRNA abundance values to obtain ratios internally controlled for variations in cDNA quantity. Log2 transformed ratios for each of three biological replicates is shown as a point and the mean as a dash.
Diverse forms of RPS9 alternative splicing are associated with structured and conserved RNA sequences
We hypothesized that RpS9 autoregulation had an important function and would thus be conserved in other animals. Further, we hypothesized that conserved RNA structures were involved in the cross-regulation of RPS9A and RPS9B in S. cerevisiae and the autoregulation of RpS9 in D. melanogaster, because E. coli S4 (the bacterial ortholog), requires an RNA structure to autoregulate by translational repression. Therefore, we predicted that RPS9 orthologs would be associated with alternatively-spliced mRNA isoforms, conserved RNA structures, and PTCs. To identify such messages, we summarized expressed sequence tags (ESTs) data from diverse animals. Indeed, EST coverage extends outside exons and into introns, which support the existence of rare unspliced or alternatively-spliced transcripts (<5% maximum coverage) (Figure 4, gray bars). To identify ESTs that specifically support alternative splice site usage or cassette exon inclusion, we mapped putative EST exon-exon junctions that spanned both 5′ GT and 3′ AG splice sites (Figure 4, blue and red bars, respectively). With the exception of Petromyzon marinus, ESTs from various vertebrates (e.g. H. sapiens, Rattus norvegicus, Xenopus tropicalis, Danio rerio, and Oryzias latipes) reveal cassette exons that introduce PTCs from the last canonical intron (Figure 4 and Figure S1). P. marinus and D. melanogaster ESTs, on the other hand, reveal alternative 5′ splice sites that also introduce PTCs from a homologous intron (Figure 4 and Figure S1). Most intriguingly, Ciona intestinalis ESTs also support alternative 5′ splice site usage, but in a non-homologous intron compared to those of other animals (Figure 4). Thus, our surveys of animal ESTs suggest that animal RPS9 orthologs are often alternatively-spliced to utilize RUST. Further, the conservation of alternatively-spliced cassette exons within the last intron among distantly related vertebrates (e.g. ∼400 million years between humans and fish [10]) suggest that these isoforms are functional.
Figure 4. Diverse alternatively spliced RPS9 isoforms encode PTC+ exons associated with high nucleotide conservation and predicted RNA structures.
Summaries of ESTs, predicted RNA structures, and sequence conservation from animal RPS9 orthologs (H. sapiens, X. tropicalis, O. latipes, D. melanogaster, C. intestinalis, and S. cerevisiae) are presented along a dendogram illustrating their phylogenetic relationships (not to scale). For each species, histograms summarize EST coverage (gray bars) and inferred splice junctions with both 5′ GT (blue bars) and 3′ AG splice sites (red bars). Dashed lines separate the lower 5% and upper 95% histogram values; EST coverage is labeled on the y-axis. Two gene models (below each histogram) are plotted to scale (black line; 1 kb) representing either the major isoform (top gene model) or a spliced PTC+ EST (bottom gene model) for each species (an “unspliced” pre-mRNA is modeled for S. cerevisiae in lieu of an EST). The major isoform sequence is annotated as coding (thick black lines) or UTR (thin black lines) and interrupted by GT-AG introns (angled black lines). The first PTC (red line and octagon) in the representative PTC+ EST sequence (thin lines) is indicated. Below the two gene models, PhastCons scores (black bars), and RNAz predictions (green lines) indicate regions associated with high nucleotide conservation and statistically significant (P>0.9) RNA structure predictions, respectively (X. tropicalis not shown; C. intestinalis not applicable). PhastCons scores and RNAz predictions were based on MultiZ alignments obtained from the UCSC Genome Browser where available (see Methods).
Also consistent with function, PTC positions in RPS9 orthologs were associated with high nucleotide conservation (Figure 4). To determine if RPS9 orthologs were also associated with thermodynamically-stable and structurally-conserved RNA structures, we screened the gene bodies of RPS9 orthologs for statistically significant RNA structures using RNAz [26] on alignments obtained from the UCSC Genome Browser [27]. In order to examine both intronic and exonic sequences, we obtained sets of nucleotide alignments from closely-related groups of organisms: mammals, drosophilids, teleosts, and hemiascomycetous yeasts. Scanning RPS9 ortholog alignments in 400 bp windows, we identified predicted RNA structures (P>0.9), specifically within the last intron of mammalian, drosophilid, and teleost RPS9 orthologs, each overlapping with PTC positions (Figure 4, green lines and red octagons). Similarly, sequence alignments of RPS9 orthologs from hemiascomycetous yeasts also revealed predicted RNA structures specifically within the single yeast intron, which if unspliced, would introduce a PTC (Figure 4). Due to the lack of sequences similar to the C. intestinalis RPS9 gene corresponding to the PTC in its third intron, we did not test this region for conserved elements and predicted RNA structures. In any case, these data indicate the potential for autoregulation among distantly related RPS9 orthologs through the use of different forms of alternative splicing, perhaps through structured RNA elements.
Discussion
The complex evolutionary history of introns immediately raises three fundamental questions. First, why do introns persist? Second, what functions of introns promote their selection and persistence? Third, are intron functions general across species, or have they acquired different functions in different organisms? Our study sheds light onto these questions.
Biased intron loss may reflect selection for functional introns
The genes of S. cerevisiae, and hemiascomycetous yeasts in general, contain very few introns compared to other eukaryotes [28], which is generally attributed to uncommonly high rates of intron loss within this lineage [15]. Previous observations that S. cerevisiae introns are biased for RPGs [1], [2], [29], [30] and other highly expressed genes [4] have been cited as evidence that many S. cerevisiae RPG introns have one or more functions. Intriguingly, similar biases are also observed in the intron-poor genomes of Encephalitozoon cuniculi [31], [32] and the nucleomorph of Guillardia theta [33], suggesting that the bias against RPG intron loss is not limited to yeasts. By measuring the rates of intron loss among recently-duplicated genes, we confirm that an ongoing bias against RPG intron loss is apparent in the lineage leading to S. cerevisiae (Figure 1). Thus, the few remaining introns in S. cerevisiae may reflect biases in 1) the mechanisms of intron loss and/or 2) selection to keep important introns. In addition to previously-proposed functions of RPG introns (see below), several lines of evidence suggest that the conservation of RPG introns is not merely a function of mutation rates. Reverse transcription-mediated intron loss is expected to preferentially remove 3′ end biased introns from highly-expressed genes [34]. First, intron biases for RPGs run counter to the expectation for intron loss among highly-expressed genes, since these transcripts would be more likely to be reverse transcribed (as discussed in [32]). Second, the majority of S. cerevisiae intron losses observed here are not 3′ end biased; in fact, several introns were lost from the 5′ UTR (e.g. GBP1, NHP6A and ARF1). Lastly, at least 21 RPG introns that are present in both the Lachantea and Saccharomyces clades appear to have been lost from Z. rouxii, indicating that species-specific RPG intron losses can occur, but have not done so in the lineage leading to S. cerevisiae (Table S1). Biased intron loss, therefore, may reflect species-specific selective pressure to retain functional introns.
Intron function in the absence of alternative spicing
In many eukaryotes, the presence of large numbers of introns permit alternative splicing, which can be used to increase protein diversity [35]. However, the simple gene architectures of S. cerevisiae provide limited opportunity for the generation of multiple protein isoforms through alternative splicing (although a few instances have been described [36], [37]). Instead, S. cerevisiae RPG introns have been proposed to confer other functions, such as transcriptional enhancement [4] and splicing regulation. We tested these two hypotheses directly by deleting introns from 16 S. cerevisiae genes and assessing the effect on gene expression by microarray.
Unlike intronless copies of some mammalian genes [38], the expression of many RPGs were unaffected or even increased by deleting introns (Figure 2A). Thus, the persistence of these introns may be due to selection for other intron functions, such as splicing regulation, perhaps in response to amino acid starvation [16]. Alternatively, our splicing microarray platform may not provide the sensitivity needed to confidently identify subtle, but potentially important, changes in expression levels. Nonetheless, we did observe large increases in gene expression for RPS14B, which is known to autoregulate through splicing inhibition [7]. Thus, it seems likely that this intron and the RPS9A and RPS9B introns are under additional selection pressure to maintain homeostasis of protein levels. Consistent with this view, regions within the RPS9A and RPS9B introns are highly conserved (Figure 4 and Figure S2), which strongly suggests that mutations within these introns have been detrimental to fitness during natural history. Therefore, the strong bias against RPG intron loss (see above) may reflect ongoing selection for splicing regulation.
Evolution of the RPS9 autoregulatory circuit
The propensity for RNA-binding proteins to utilize alternative splicing for the purpose of autoregulation has long been noted [39] and, in the case of RNA-binding proteins, is remarkably common [40]–[42]. To our knowledge, however, regulation at the level of splicing between organisms as evolutionarily distant as S. cerevisiae and humans is exceedingly rare. While autoregulation of RPGs by alternative splicing is common and can be conserved as distantly as worms and humans [20], [21], we find no evidence that other yeast RPGs (i.e. RPL30 and RPS14) are regulated by splicing in both yeast and mammals (Figure S3). Interestingly, S9 orthologs in bacteria (and possibly archaea) are among a small class of RPGs that autoregulate by translational repression [43], [44]. Thus, an intriguing notion is that S9 autoregulation is of particular importance to life or particularly likely to evolve. Presumably, autoregulation of S9 production would benefit the cell by reducing waste [3] and by preventing potentially harmful interactions with low-affinity targets [45].
Cross-regulation, such as between the RPS9A and RPS9B genes, has also been observed between multiple sets of paralogous splicing regulators, including hnRNPL and hnRNPLL [46], as well as PTB, nPTB and ROD1 [47]. We speculate that these genes exemplify a straightforward principle of gene duplication and evolution: upon gene duplication, autoregulation would inherently become cross-regulation. As the paralogs diverge in abundance and/or protein function, this cross-regulation could become asymmetric (Figure 5A). In theory, such asymmetric cross-regulation among RPG pairs may allow differential expression of functionally-distinct ribosomal proteins to produce a “ribosome code” [48]. What distinct functions are provided by RPS9A and RPS9B gene products remain to be seen. Interestingly, the RPS9A and RPS9B genes encode S9 proteins that differ primarily within a small C-terminal acidic patch that may be required for proper ribosomal disassociation [49]. Mutational analyses of these three differing amino acids are needed to definitively test whether S. cerevisiae utilizes differential expression of RPS9A and RPS9B genes to exploit functional differences in the proteins they encode.
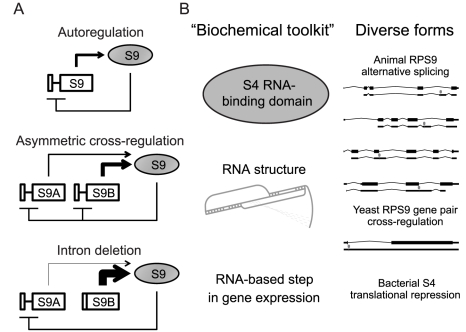
Figure 5. Hypothetical evolution of RPS9 autoregulation.
A) Hypothetical evolution of the RPS9 autoregulatory circuit after duplication and divergence. Autoregulation of pre-WGD RPS9 (top) is conserved between post-WGD gene copies despite divergence in expression levels to produce asymmetrical cross-regulation (middle). In S. cerevisiae, RPS9A and RPS9B intron deletions shift the burden of autoregulation onto the other intron-containing gene copy (bottom). B) A theoretical “biochemical toolkit,” which minimally requires an S4 RNA-binding domain and a suitable RNA binding site to perturb an essential step in gene expression (left), could potentially produce the many forms of splicing regulation observed in yeasts and animal RPS9 orthologs (right).
How does excess S9 regulate the splicing of the RPS9 orthologs? One possibility is that S9 binds its own mRNA like bacterial S4. A strong paradigm has been set by S14 and L30 in yeast and S26 and S13 in animals, in which these ribosomal proteins bind RNA structures present in their introns [6], [7], [22], [23]. It seems likely that S9 might operate under the same paradigm. Intuitively, conserved RNA structures within the introns of RPS9 orthologs make for likely targets for S9 binding (Figure 4). However, we do not observe obvious similarities between these predicted structures and the E. coli S4 regulatory site, which forms a double pseudoknot [43]. E. coli S4 can also bind and regulate a Bacillis subtilis mRNA that contains a dissimilar pseudoknot structure [50]. Intriguingly, the conserved RNA structures in RPS9A and RPS9B also appear to have the potential to form a pseudoknot (Figure S4). Thus, it seems plausible that the putative RNA structures within yeast and animal introns may yet be binding sites for S9 despite considerable structural divergence. This, however, is mere speculation and in vitro binding assays are needed to determine if ribosomal protein S9 directly regulates its own expression in S. cerevisiae and other eukaryotes. If auto- and cross-regulation were indeed directly mediated by ribosomal protein S9 binding, then comparative biochemical studies using proteins and RNA sequences from different species could provide mechanistic detail to describe how S9 mediates the different forms of alternative splicing.
Why are there so many forms of splicing regulation among RPS9 orthologs? One possibility is that particular aspects of these forms are ancient and conserved, while others have evolved independently in different lineages. For example, the genetic circuits that specify the development of diverse animal forms (e.g. eyes and limbs) exemplify deep homology, where recent evolutionary innovations overlay a shared “genetic toolkit” [51]. By analogy, genetic circuits themselves (in this case, autoregulation) may share a common “biochemical toolkit” comprised of highly conserved biochemical processes (e.g. RNA∶protein interactions), while independently evolving elaborations on these basic circuits. Thus, translational inhibition of the alpha-operon by S4-binding may represent just one of many possible forms of regulation accessible to the highly conserved S4 RNA-binding domain proteins found throughout cellular life. Alternative splicing in animals and regulated splicing in S. cerevisiae may be different elaborations on this autoregulatory circuit, perhaps mediated by different RNA structures within introns (Figure 5B). Thus, we propose that the highly-conserved function of ribosomal protein S9 (and RNA-binding proteins in general) is one part of a biochemical toolkit that is frequently used and reused, as the fundamental autoregulatory circuit is maintained, elaborated and reinvented.
Methods
Intron gain and loss analysis
To estimate the propensity for intron loss among RPGs and non-RPGs, we compared annotated S. cerevisiae intron-containing genes and WGD-derived gene pairs. S. cerevisiae intron annotations were obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/) on 7/20/2011. WGD-derived gene pairs (as inferred from genomic synteny; a.k.a. “Ohnologs”) were obtained from the Yeast Gene Order Browser (http://wolfe.gen.tcd.ie/ygob/) [14]. Because introns are commonly identified by gaps in BLAST-based homology searches, intron-containing genes with short first exons are commonly misannotated. To identify annotated introns upstream of an annotated gene, custom scripts written in R (http://www.r-project.org/) were used to scan 800 bp upstream and 100 bp downstream of the ORF start site with a regular expression that recognizes >90% of S. cerevisiae introns by identifying the most common splice sites to minimize false positive matches. The regular expression matches sequences meeting the following criteria in order: 1) any one of the 4 most common of 5′ SSs, 2) an S1 length of at least 30, 3) any one of the 5 most common branchsites, 4) an S2 length between 1 and 50, and 5) any one of the 3 most common 3′ SS trinucleotides, which was formalized as: “(gtatgt|gtacgt|gtaagt|gtatga).{30,}?(tactaac|gactaac|aactaac|tgctaac|cactaac).{1,50}?[tca]ag”. The pre-WGD ancestor was inferred to contain an intron if the majority of available outgroup pre-WGD species orthologs (C. albicans, L. waltii, L. thermotolerans, L. kluyveri, E. gossypii, K. lactis) and 1) the Z. rouxii ortholog had an intron or 2) the majority of post-WGD species intron (S. cerevisiae, S. bayanus, C. glabrata, N. castellii and V. polyspora) gene pairs had at least one intron. In this manner, we distinguished independent intron gains and losses in Z. rouxii from intron gains and losses immediately after the WGD event.
S. cerevisiae strain construction
Intron deletion mutants were generated by a replacement strategy similar to a previously-described method for intron deletion [52]. Briefly, a PCR product amplified from the plasmid pJPS1232 (generously provided by J. Staley, University of Chicago), which contains the CORE construct [53] fused to the I-SceI endonuclease site, using gene-specific primers containing exon 1 and exon 2 sequences that allow integration and subsequent intron deletion via homologous recombination. Transformed diploid cells (yAP047) were incubated for 4 h at 30°C in the presence of 2% galactose to induce I-SceI endonuclease expression and precise deletion of the CORE cassette. Sporulated haploid cells were confirmed to harbor intron deletions by PCR. To ensure that a precise intron deletion was obtained without any additional mutations, the region surrounding the newly-created exon-exon junction (at least 100 bp) was PCR amplified and sequenced. Strains described in Figure 2 were also confirmed for Δi mutation by decreased microarray intron probe signal. Gene specific primers used for mutagenesis are detailed in Table S2.
D. melanogaster S2 cell RNAi and transfection
Routine passaging of S2 cell cultures and RNAi depletion was performed as described [54] with the following modifications. Briefly, 1 µg/ml of dsRNA was incubated with 3.5E5 cells in 350 µl of media in 24-well plates. After 48 h incubation with dsRNA, cells were transfected with 0.2 µg plasmid with Effectene (Quigen) according to manufacturer's instructions. Cells were harvested with 1 ml TRIzol (Invitrogen) for analysis by RT-qPCR (see below). Primers used to generate PCR products used for dsRNA synthesis (Promega RiboMAX) are described in Table S2.
RNA isolation and cDNA synthesis
To analyze the expression of genes in S. cerevisiae intron deletion strains, 15 ml cultures of mutant and wild-type yeast were grown in parallel at 30°C in rich medium supplemented with 2% glucose to an optical density between A600 = 0.5 and 0.7. For microarray hybridization, RNA was isolated by acid-phenol extraction and converted to cDNA as described [16]. A similar protocol was performed for qPCR applications with the following modifications. After RNA isolation, 2 µg of DNase-treated RNA was random primed in a 40 µl reaction containing 1 µg dN9 primer, 50 mM TrisHCl (pH 8.4), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM dNTPs, and 5 ng murine Moloney leukemia virus (M-MLV) RT. Primers were hybridized at 60°C for 7 min prior to the addition of enzyme, and then incubated with enzyme at 42°C for at least 2 h. Prepared cDNA was diluted at least 10-fold before use in qPCR. Similarly, to analyze D. melanogaster S2 cell, 725 µl cultures of S2 cells (UCSF cell culture facility) were grown in 24-well plates at 25°C in Schneider's Drosophila Medium (Gibco) supplemented with 10% fetal bovine serum (UCSF) to a count of ∼5E6 cells. RNA was extracted with 1 ml TriZol (Invitrogen) according to manufacturer's instructions. After RNA isolation, cDNA was prepared with SuperScript III (Invitogen) and random priming according to manufacturer's instructions. Prepared cDNA was diluted at least 10-fold before use in qPCR.
Microarray analysis
Splicing-sensitive microarrays were constructed and performed as described [16]. In each experiment, a wild-type strain derived from the same parent as the intron deletion mutant strain was used as a reference. Data was analyzed using the R Bioconductor packages marray() and limma() [55] in a custom pipeline based on the Goulphar program [56]. Microarray data used in this study are available in the Gene Expression Omnibus at NCBI (GSE35541).
Quantitative PCR
Quantitative PCR primers (Table S2) were designed using Primer3 [57] and S. cerevisiae or D. melanogaster genomic sequence obtained from the UCSC Genome Browser (SacCer1 or dm3, respectively) [58], [59]. Serial dilutions of DNA ranging from 100 to 0.16 ng of the genomic DNA were used to obtain calibration curves, measure primer efficiencies, and ensure that quantification was in a linear dynamic range. Primer sets yielding multiple amplification products or calibration curves with R-squared values of <0.96 were excluded. For each qPCR sample, diluted cDNA was amplified in 25 µl volume reactions containing 250 µM dNTPs, 1× (NH4)2SO4 buffer (Fermentas), 0.5 µM primer, 1.5 mM MgCl2, 1.25 Units Dynazyme II (Finnzymes), and Sybr Green I fluorescent dye (Sigma). Fluorescence was measured on a BioRad Opticon machine using standard cycling conditions (3 min at 95°C, 40 cycles of 15 s at 95°C, 30 s at 55°C, and 15 s at 72°C). Biological replicate qPCR values were determined as the median of technical replicates. For each of 3 biological replicates, target gene values (e.g. RPS9A) were divided by reference gene values (e.g. SCR1) before log transformation. Plots were generated using the R package ggplot2() [60].
Assessment of alternative splicing among animal RPS9 orthologs
To assess EST coverage and splicing, we obtained genomic coordinates corresponding to GenBank ESTs from the UCSC Genome Browser ‘spliced EST’ track, which span at least one canonical intron of at least 32 bases [61]. Custom R scripts were used to calculate EST coverage per genomic nucleotide position, and identify all exon-exon junctions that span putative GT/AG splice site [62]. The following genome assemblies were used in the analysis: Branchiostoma floridae (braFlo1); Ciona intestinalis (Ci2); Danio rerio (danRer7); Drosophila melanogaster (dm3), Oryzias latipes (oryLat2); Petromyzon marinus (petMar1); Xenopus tropicalis (xenTro2); Rattus norvegicus (rn4); Mus musculus (mm9); Homo sapiens (hg19).
Supporting Information
Comparison of spliced isoforms by EST analysis of RPS9 orthologs from 10 animals. EST summaries of RPS9 orthologs from 10 animal species illustrated as in Figure 4. Genes are plotted to scale (black line; 1 kb).
(PDF)
Conserved intronic regions among yeast RPS9 orthologs. Nucleotide alignment of genes encoding ribosomal protein S9 from L. waltii, L. thermotolerans, L. kluyveri, E. gossypii, K. lactis, Z. rouxii, S. cerevisiae, S. bayanus, C. glabrata, N. castellii and V. polyspora. Note that all genes contain an intron with regions of identical sequences (black shading). Positions of compensatory base pair changes supporting RNA stems shown in Figure S4 are highlighted in purple.
(PDF)
EST analysis of mammalian RPL30 and RPS14 does not reveal conserved alternatively-spliced isoforms. EST summaries of RPL30 and RPS14 orthologs from human, mouse, and rat illustrated as in Figure 4. Genes are plotted to scale (black line; 1 kb).
(PDF)
Putative RNA structure within the RPS9A and RPS9B introns. Conserved elements in the RPS9A and RPS9B introns are associated with a putative pseudoknot structure near the 5′ splice site. A) An illustration of the S. cerevisiae RPS9A gene model and nucleotide conservation among closely related yeasts (from the UCSC Genome Browser). Putative RNA stems (predicted by the RNAz program [26]) that overlap with conserved regions are indicated (numbered 1–4). B) Illustration of a putative H-H type pseudoknot (predicted by the IPknot program [64]) based on nucleotide alignment of pre- and post-WGD yeasts (Figure S2). Positions of compensatory base pair changes that support the pseudoknot stems are indicated in purple. Pseudoknot illustration was created with PseudoViewer v3.0 (http://pseudoviewer.inha.ac.kr/).
(PDF)
Intron annotations and predictions.
(PDF)
Primers.
(PDF)
Acknowledgments
We thank Jon Staley for the very generous gift of S. cerevisiae strains, procedures, and advice required for the generation of intron deletion strains. We thank Jeff Farrell and Hansong Ma for their excellent advice concerning S2 cell culture and dsRNA knockdowns. We also thank Sandy Johnson, Jason Huff, Imke Listerman, Scott Roy, and the Guthrie group for their helpful discussions in the preparation of this manuscript and throughout the course of this work.
Footnotes
The authors have declared that no competing interests exist.
Research support was provided by NIH grant GM21119. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bon E, Casaregola S, Blandin G, Llorente B, Neuvéglise C, et al. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitrovich QM, Tuch BB, Guthrie C, Johnson AD. Computational and experimental approaches double the number of known introns in the pathogenic yeast Candida albicans. Genome Res. 2007;17:492–502. doi: 10.1101/gr.6111907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 4.Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW. Introns regulate RNA and protein abundance in yeast. Genetics. 2006;174:511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabeva MD, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J Biol Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- 7.Fewell SW, Woolford JL., Jr Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol Cell Biol. 1999;19:826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean D, Nomura M. Feedback regulation of ribosomal protein gene expression in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3590–3594. doi: 10.1073/pnas.77.6.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 10.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 11.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 12.Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci USA. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Hesselberth JR, Fields S. Genome-wide identification of spliced introns using a tiling microarray. Genome Res. 2007;17:503–509. doi: 10.1101/gr.6049107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stajich JE, Dietrich FS, Roy SW. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Paulovich AG, Woolford JL., Jr Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol Cell Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 19.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Res. 2005;33:5965–5977. doi: 10.1093/nar/gki905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov AV, Malygin AA, Karpova GG. Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim Biophys Acta. 2005;1727:134–140. doi: 10.1016/j.bbaexp.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35:6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lareau LF, Brooks AN, Soergel DAW, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 25.Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffares DC, Mourier T, Penny D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Ares M, Jr, Grate L, Pauling MH. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spingola M, Grate L, Haussler D, Ares M. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 32.Lee RCH, Gill EE, Roy SW, Fast NM. Constrained intron structures in a microsporidian. Mol Biol Evol. 2010;27:1979–1982. doi: 10.1093/molbev/msq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- 34.Fink GR. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- 35.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grund SE, Fischer T, Cabal GG, Antúnez O, Pérez-Ortín JE, et al. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol. 2008;182:897–910. doi: 10.1083/jcb.200803098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juneau K, Nislow C, Davis RW. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics. 2009;183:185–194. doi: 10.1534/genetics.109.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattox W, Ryner L, Baker BS. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992;267:19023–19026. [PubMed] [Google Scholar]
- 40.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 41.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang CK, Draper DE. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- 44.Sano K, Taguchi A, Furumoto H, Uda T, Itoh T. Cloning, sequencing, and characterization of ribosomal protein and RNA polymerase genes from the region analogous to the alpha-operon of escherichia coli in halophilic archaea, halobacterium halobium. Biochem Biophys Res Commun. 1999;264:24–28. doi: 10.1006/bbrc.1999.1480. [DOI] [PubMed] [Google Scholar]
- 45.Von Hippel PH, Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, et al. Autoregulation of gene expression. Quantitative evaluation of the expression and function of the bacteriophage T4 gene 32 (single-stranded DNA binding) protein system. J Mol Biol. 1982;162:795–818. doi: 10.1016/0022-2836(82)90548-4. [DOI] [PubMed] [Google Scholar]
- 46.Rossbach O, Hung L-H, Schreiner S, Grishina I, Heiner M, et al. Auto- and Cross-Regulation of the hnRNP L Proteins by Alternative Splicing. Mol Cell Biol. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pnueli L, Arava Y. Genome-wide polysomal analysis of a yeast strain with mutated ribosomal protein S9. BMC Genomics. 2007;8:285. doi: 10.1186/1471-2164-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grundy FJ, Henkin TM. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991;173:4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 52.Parenteau J, Durand M, Véronneau S, Lacombe A-A, Morin G, et al. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell. 2008;19:1932–1941. doi: 10.1091/mbc.E07-12-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- 55.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemoine S, Combes F, Servant N, Le Crom S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics. 2006;7:467. doi: 10.1186/1471-2105-7-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 58.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 59.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 60.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer New York; 2009. [Google Scholar]
- 61.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R Development Core Team. Development Core Team, R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 63.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 64.Sato K, Kato Y, Hamada M, Akutsu T, Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:i85–93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of spliced isoforms by EST analysis of RPS9 orthologs from 10 animals. EST summaries of RPS9 orthologs from 10 animal species illustrated as in Figure 4. Genes are plotted to scale (black line; 1 kb).
(PDF)
Conserved intronic regions among yeast RPS9 orthologs. Nucleotide alignment of genes encoding ribosomal protein S9 from L. waltii, L. thermotolerans, L. kluyveri, E. gossypii, K. lactis, Z. rouxii, S. cerevisiae, S. bayanus, C. glabrata, N. castellii and V. polyspora. Note that all genes contain an intron with regions of identical sequences (black shading). Positions of compensatory base pair changes supporting RNA stems shown in Figure S4 are highlighted in purple.
(PDF)
EST analysis of mammalian RPL30 and RPS14 does not reveal conserved alternatively-spliced isoforms. EST summaries of RPL30 and RPS14 orthologs from human, mouse, and rat illustrated as in Figure 4. Genes are plotted to scale (black line; 1 kb).
(PDF)
Putative RNA structure within the RPS9A and RPS9B introns. Conserved elements in the RPS9A and RPS9B introns are associated with a putative pseudoknot structure near the 5′ splice site. A) An illustration of the S. cerevisiae RPS9A gene model and nucleotide conservation among closely related yeasts (from the UCSC Genome Browser). Putative RNA stems (predicted by the RNAz program [26]) that overlap with conserved regions are indicated (numbered 1–4). B) Illustration of a putative H-H type pseudoknot (predicted by the IPknot program [64]) based on nucleotide alignment of pre- and post-WGD yeasts (Figure S2). Positions of compensatory base pair changes that support the pseudoknot stems are indicated in purple. Pseudoknot illustration was created with PseudoViewer v3.0 (http://pseudoviewer.inha.ac.kr/).
(PDF)
Intron annotations and predictions.
(PDF)
Primers.
(PDF)