Abstract
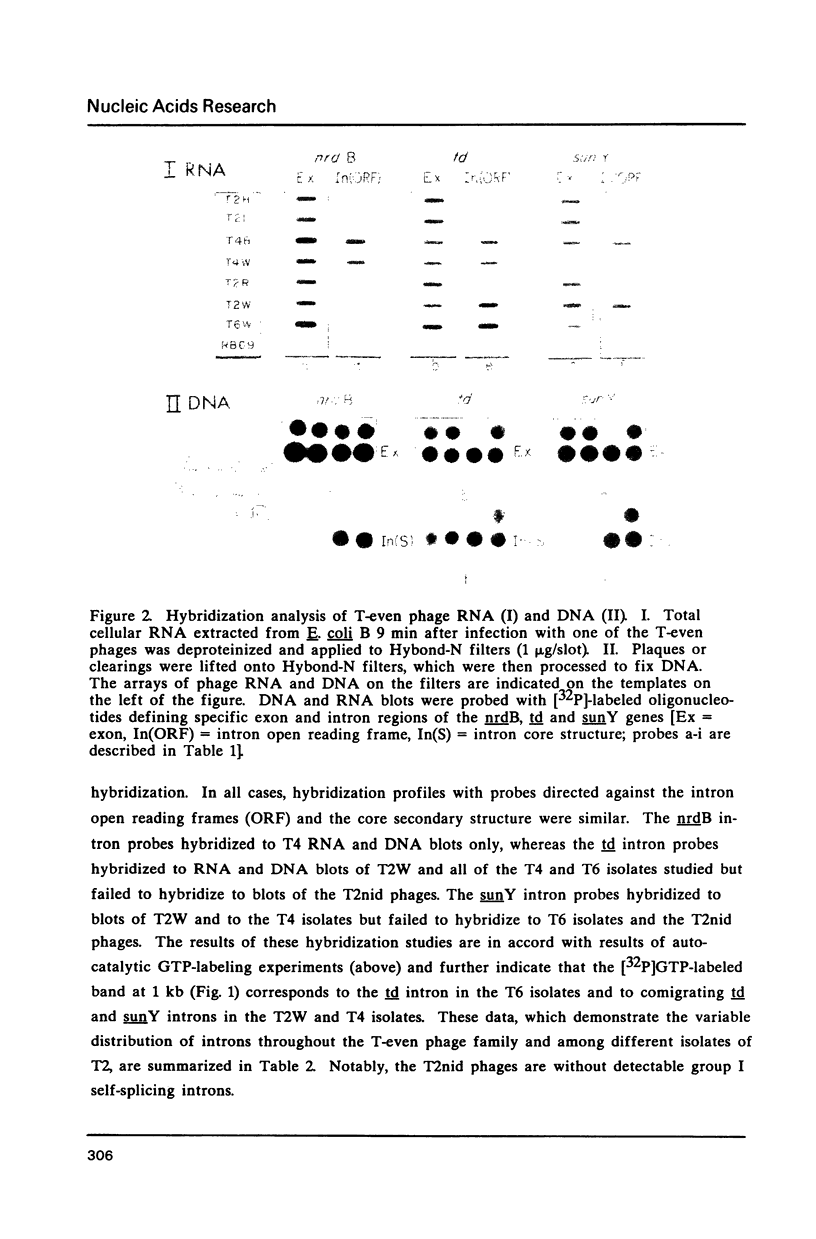
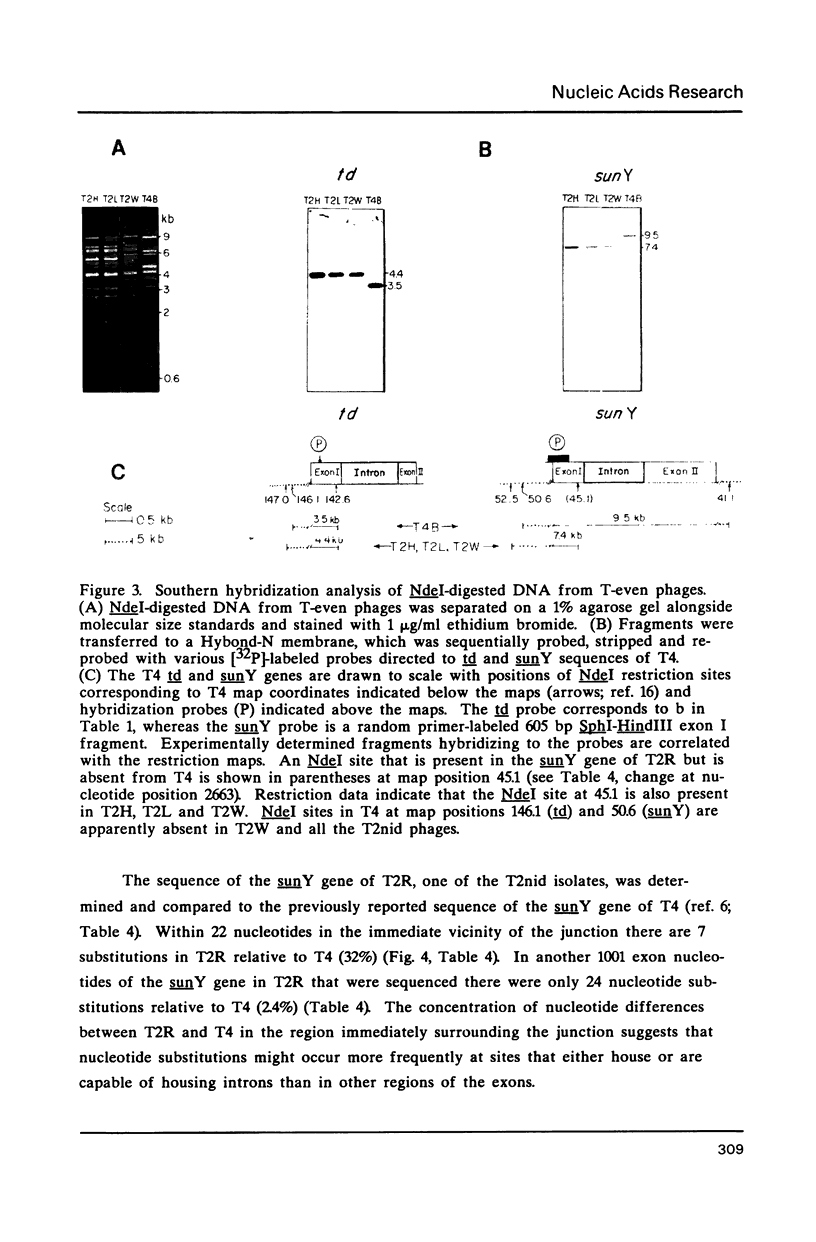
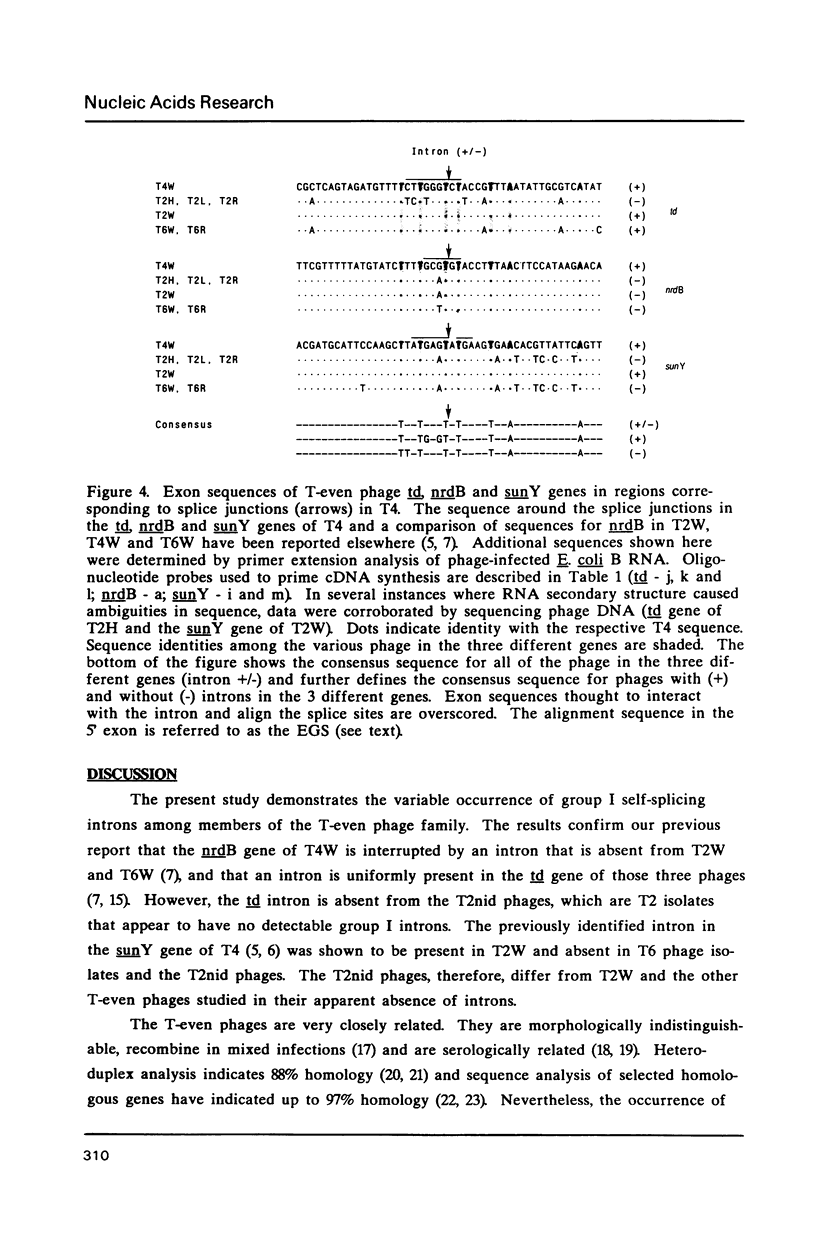
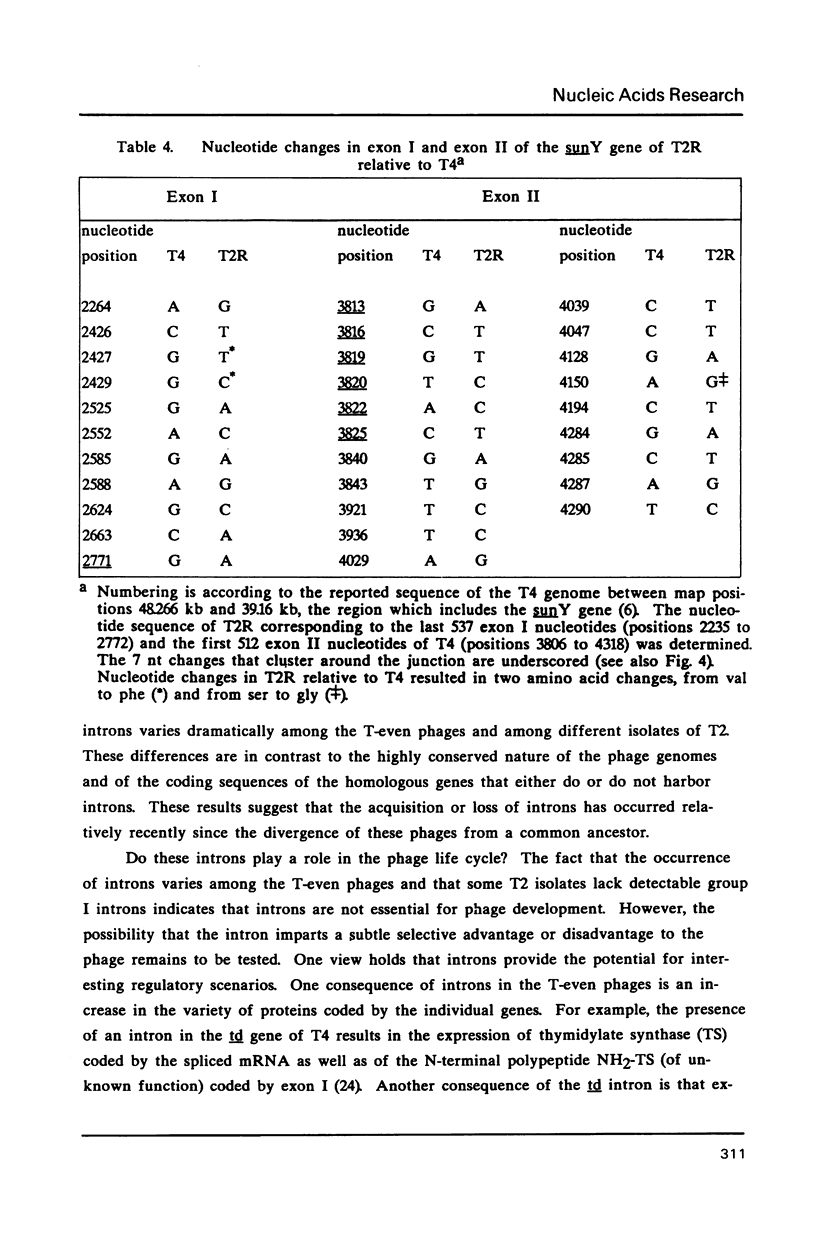
Group I self-splicing introns are present in the td, nrdB and sunY genes of bacteriophage T4. We previously reported that whereas the td intron is present in T2, T4 and T6, the nrdB intron is present in T4 only. These studies, which argue in favor of introns as mobile genetic elements, have been extended by defining the distribution of all three T4 introns in a more comprehensive collection of T2, T4 and T6 isolates. The three major findings are as follows: First, all three introns are inconsistently distributed throughout the T-even phage family. Second, different T2 isolates have different intron complements, with T2H and T2L having no detectable introns. Third, the intron open reading frames are inherited or lost as a unit with their respective flanking intron core elements. Furthermore, exon sequences flanking sites where introns are inserted in the T4 td, sunY and nrdB genes were determined for all the different T-even isolates studied. Six of eighteen residues surrounding the junction sequences are identical. In contrast, a comprehensive comparison of exon sequences in intron plus and intron minus variants of the sunY gene indicate that sequence changes are concentrated around the site of intron occurrence. This apparent paradox may be resolved by hypothesizing that the recombination events responsible for intron acquisition or loss require a consensus sequence, while these same events result in sequence heterogeneity around the site.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baylor M. B. The distribution of conditional lethals and host range mutants in the genetic map of bacteriophage T2H. Virology. 1977 Dec;83(2):380–389. doi: 10.1016/0042-6822(77)90182-9. [DOI] [PubMed] [Google Scholar]
- Belfort M., Pedersen-Lane J., Ehrenman K., Chu F. K., Maley G. F., Maley F., McPheeters D. S., Gold L. RNA splicing and in vivo expression of the intron-containing td gene of bacteriophage T4. Gene. 1986;41(1):93–102. doi: 10.1016/0378-1119(86)90271-4. [DOI] [PubMed] [Google Scholar]
- Belfort M., Pedersen-Lane J., West D., Ehrenman K., Maley G., Chu F., Maley F. Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level. Cell. 1985 Jun;41(2):375–382. doi: 10.1016/s0092-8674(85)80010-6. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley F., Martinez J., Maley G. F. Interrupted thymidylate synthase gene of bacteriophages T2 and T6 and other potential self-splicing introns in the T-even bacteriophages. J Bacteriol. 1987 Sep;169(9):4368–4375. doi: 10.1128/jb.169.9.4368-4375.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., Maley F., Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci U S A. 1984 May;81(10):3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., West D. K., Belfort M., Maley F. Characterization of the intron in the phage T4 thymidylate synthase gene and evidence for its self-excision from the primary transcript. Cell. 1986 Apr 25;45(2):157–166. doi: 10.1016/0092-8674(86)90379-x. [DOI] [PubMed] [Google Scholar]
- Colleaux L., d'Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986 Feb 28;44(4):521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- Cowie D. B., Avery R. J., Champe S. P. DNA homology among the T-even bacteriophages. Virology. 1971 Jul;45(1):30–37. doi: 10.1016/0042-6822(71)90109-7. [DOI] [PubMed] [Google Scholar]
- Demerec M, Fano U. Bacteriophage-Resistant Mutants in Escherichia Coli. Genetics. 1945 Mar;30(2):119–136. doi: 10.1093/genetics/30.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenman K., Pedersen-Lane J., West D., Herman R., Maley F., Belfort M. Processing of phage T4 td-encoded RNA is analogous to the eukaryotic group I splicing pathway. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5875–5879. doi: 10.1073/pnas.83.16.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott J. M., Shub D. A., Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell. 1986 Oct 10;47(1):81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- Gram H., Rüger W. The alpha-glucosyltransferases of bacteriophages T2, T4 and T6. A comparison of their primary structures. Mol Gen Genet. 1986 Mar;202(3):467–470. doi: 10.1007/BF00333278. [DOI] [PubMed] [Google Scholar]
- Hall D. H., Povinelli C. M., Ehrenman K., Pedersen-Lane J., Chu F., Belfort M. Two domains for splicing in the intron of the phage T4 thymidylate synthase (td) gene established by nondirected mutagenesis. Cell. 1987 Jan 16;48(1):63–71. doi: 10.1016/0092-8674(87)90356-4. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Bonen L., de Haan M., van der Horst G., Grivell L. A. Two intron sequences in yeast mitochondrial COX1 gene: homology among URF-containing introns and strain-dependent variation in flanking exons. Cell. 1983 Feb;32(2):379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985 Jun;41(2):383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- LANNI F., LANNI Y. T. Antigenic structure of bacteriophage. Cold Spring Harb Symp Quant Biol. 1953;18:159–168. doi: 10.1101/sqb.1953.018.01.026. [DOI] [PubMed] [Google Scholar]
- Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985 Jun;41(2):395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Genetic exchanges between bacteriophage T4 and filamentous fungi? Cell. 1986 Aug 1;46(3):323–323. doi: 10.1016/0092-8674(86)90651-3. [DOI] [PubMed] [Google Scholar]
- Mota E. M., Collins R. A. Independent evolution of structural and coding regions in a Neurospora mitochondrial intron. Nature. 1988 Apr 14;332(6165):654–656. doi: 10.1038/332654a0. [DOI] [PubMed] [Google Scholar]
- Paddock G. V., Abelson J. Nucleotide sequence determination of bacteriophage T2 and T6 species I ribonucleic acids. J Biol Chem. 1975 Jun 10;250(11):4207–4219. [PubMed] [Google Scholar]
- Pedersen-Lane J., Belfort M. Variable occurrence of the nrdB intron in the T-even phages suggests intron mobility. Science. 1987 Jul 10;237(4811):182–184. doi: 10.1126/science.3037701. [DOI] [PubMed] [Google Scholar]
- Shub D. A., Gott J. M., Xu M. Q., Lang B. F., Michel F., Tomaschewski J., Pedersen-Lane J., Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg B. M., Hahne S., Mathews C. Z., Mathews C. K., Rand K. N., Gait M. J. The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J. 1986 Aug;5(8):2031–2036. doi: 10.1002/j.1460-2075.1986.tb04460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L., Ingold A., Karlok M., Nielsen H., Engberg J. Phylogenetic evidence for the acquisition of ribosomal RNA introns subsequent to the divergence of some of the major Tetrahymena groups. EMBO J. 1986 Dec 20;5(13):3625–3630. doi: 10.1002/j.1460-2075.1986.tb04691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaschewski J., Rüger W. Nucleotide sequence and primary structures of gene products coded for by the T4 genome between map positions 48.266 kb and 39.166 kb. Nucleic Acids Res. 1987 Apr 24;15(8):3632–3633. doi: 10.1093/nar/15.8.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West D. K., Belfort M., Maley G. F., Maley F. Cloning and expression of an intron-deleted phage T4 td gene. J Biol Chem. 1986 Oct 15;261(29):13446–13450. [PubMed] [Google Scholar]
- Zimmer M., Welser F., Oraler G., Wolf K. Distribution of mitochondrial introns in the species Schizosaccharomyces pombe and the origin of the group II intron in the gene encoding apocytochrome b. Curr Genet. 1987;12(5):329–336. doi: 10.1007/BF00405755. [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell. 1985 Apr;40(4):887–895. doi: 10.1016/0092-8674(85)90348-4. [DOI] [PubMed] [Google Scholar]





