Abstract
During the human B cell (Bc) recall response, rapid cell division results in multiple Bc subpopulations. The TLR-9 agonist CpG oligodeoxynucleotide, combined with cytokines, causes Bc activation and division in vitro and increased CD27 surface expression in a sub-population of Bc. We hypothesized that the proliferating CD27lo subpopulation, which has a lower frequency of antibody-secreting cells (ASC) than CD27hi plasmablasts, provides alternative functions such as cytokine secretion, costimulation, or antigen presentation. We performed genome-wide transcriptional analysis of CpG activated Bc sorted into undivided, proliferating CD27lo and proliferating CD27hi subpopulations. Our data supported an alternative hypothesis, that CD27lo cells are a transient pre-plasmablast population, expressing genes associated with Bc receptor editing. Undivided cells had an active transcriptional program of non-ASC B cell functions, including cytokine secretion and costimulation, suggesting a link between innate and adaptive Bc responses. Transcriptome analysis suggested a gene regulatory network for CD27lo and CD27hi Bc differentiation.
Introduction
Several groups have suggested a “division of labor” between populations of re-activated memory B cell (mBc). Some of the proposed binary classifications include: antibody versus cytokine producing Bc1,2, effector versus regulatory Bc (Reviewed in3), plasma cells versus resting mBc4, and central versus effector mBc1. Such classification schemata generally focus on the terminal functions of activated naïve Bc5,6, rather than the early differentiation of re-activated mBc. However, it is not clear that activated and dividing mBc can be partitioned into only 2 categories: antibody secretion and another function, or how adjuvants like oligodeoxynucleotides (CpG), may alter such a balance.
Among the biological activities of activated IgG class-switched, mBc are division, antibody secretion, cytokine secretion, and antigen presentation (reviewed in7). In several in vitro Bc activation systems, 30–50% of activated Bc become antibody secreting cells (ASC) by the third cell division8,9. In vivo, such activation takes place at the T cell zone-lymphoid follicle border, where Bc undergo co-stimulation with T follicular helper cells (TFh). Activated mBc present antigen and provide reciprocal cytokine stimulation to T cells10,11.
Bc activation and proliferation can also be induced by CpG acting through Toll-like receptor 9 (TLR-9). TLR-9 agonists improve production of antibody by Bc responding to vaccine12, and are in clinical trials as vaccine adjuvants13. We have previously shown that a combination of CpG2008 ODN and cytokines (IL-2, IL-10, IL-15, and BAFF) can induce in vitro mBc differentiation into CD138+ plasma cells9. Previously, a division of labor was postulated between CpG-activated Bc, including a partition into antibody and cytokine-secreting cells14. However, these studies have not examined the potential signaling networks and transcriptome patterns in the subsets of CpG-activated mBc. Such an analysis is important in understanding the differences between TFh and TLR-9 activated mBc and the antibody response generated by these two pathways, especially with respect to Bc responses to TLR-9 adjuvanated vaccines15,16.
Here we have used transcriptome analysis to characterize differences between CpG ODN activated, class switched, human mBc in three phenotypic categories: non-dividing, activated dividing, and activated-dividing-antibody secreting Bc. Our group and others have previously described increased surface expression of CD27 as correlating with division-dependent antibody secretion in CpG stimulated CD27+ class switched mBc9,17. CD27hi cells have a higher frequency of antibody-secreting cells than CD27lo cells18. For this reason, we hypothesized that CD27lo cells provide other germinal center Bc functions such as cytokine production, antigen presentation or co-stimulation for T cells. Vaccine adjuvants such as CpG may alter the balance between these states.
To this end, we describe significant differences between gene expression patterns of CpG-activated CD27hi IgG secreting versus CD27lo non-secreting mBc. These transcription patterns suggest two gene expression pathways in proliferating CpG activated mBc, with upregulation of antibody production pathways in the CD27hi subset, and upregulation of NF-κB activation pathways in the CD27lo subset preceding differentiation into a plasmablast phenotype. Gene expression patterns in undivided cells showed surprisingly active transcription of antigen presentation, cytokine secretion, and co-stimulation genes.
Results
CpG-stimulated human memory B cells are CD27 heterogeneous
To investigate a potential division of labor between the CD27hi and CD27lo cell subsets, we tested the hypothesis that CD27lo cells function in antigen presentation, co-stimulation or cytokine production. CpG activated human mBc exhibit considerable heterogeneity as they divide and differentiate, especially with respect to IgG secretion and secretion rates. In vitro CpG ODN stimulation of IgG class-switched, CD27+ human peripheral blood mBc leads to development of a CD27hi pre-plasmablast subset containing a higher frequency of ASC than the corresponding CD27lo subset, although IgG secretion rates among ASC in either group are identical18.
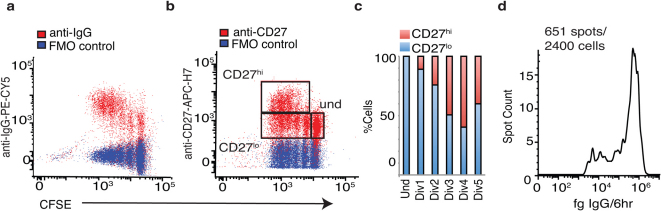
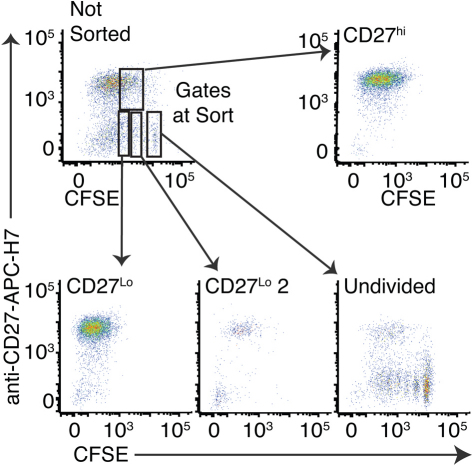
At 96 hours of stimulation with CpG plus a combination of cytokines (IL-2, IL-10, IL-15; CpG + CK), IgG class switched human mBc show increased intracellular IgG after two cell divisions (Fig. 1a), and heterogeneous expression of CD27 (Fig. 1b). The fraction of CD27hi cells increased in each generation up to division 4 (Fig. 1c). Quantitative ELISPOT (qELISPOT) assays (Fig.1d) show a typical bimodal distribution of single-cell IgG secretion rates, and an IgG secreting cell frequency of only 27%. To examine the functional activities of CpG-activated and dividing CD27lo cells, we sorted CD19+ Bc at 96 hours into CD27hi, CD27lo, and undivided cell populations (sort gates shown in Supplementary Fig. S1 online), and performed genome-wide transcriptome analysis. While CD19 levels decreased modestly on proliferating cells, >95% of cells were within the CD19+ sort gate. Relative CD27 gene levels were as expected for the sorted subsets (see Supplementary Fig. S2 online). Control analysis of CD3, CD4, CD8A and CD8B mRNA expression by gene array and analysis of CD3 by qRT-PCR showed no evidence of contamination by T cells (see Supplementary Fig. S2 online).
Figure 1. Human Memory B Cell CD27 Expression is Heterogeneous at 96 hr CpG+CK Stimulation.
Normal human peripheral blood Bc were depleted of IgM+ cells, stained with carboxyfluoroscein succinimidyl ester (CFSE), which decreases in concentration with each cell division, and placed in culture with CpG ODN + IL2, IL10, IL15, and BAFF. (a) At 96 hrs of stimulation, flow cytometric analysis showed intracellular IgG content increased in divisions 2–5 and (b) only a subset of cells in divisions 2–5 have increased in CD27 expression as compared with undivided cells. (c) The percentage of CD27hi cells per generation increases in divisions 2 through 4, with an apparent decrease in division 5+ that may be due to the spread of the data. Representative data from 1 subject, n = 10 subjects in 5 separate experiments. (d) Histogram of semi-quantitative ELISPOT data at this timepoint shows biphasic IgG secretion rate of the early recall response, but only 27% of cells secreting IgG. Cells were FACS sorted into CD27lo, CD27hi, and undivided cells subsets as in (b) for gene array analysis. Representative data from 1 subject, n = 10 subjects in 5 separate experiments.
We first analyzed gene array results for significant differences between the CD27hi, CD27lo, and undivided cell populations from the same subject by using a paired SAM analysis. 6,954 probe sets were found to be significantly differentially expressed between CD27lo and undivided at false discovery rate (FDR) level 0.01. We selected 4,615 differentially expressed genes based on the following criterion: a gene is declared differentially expressed if at least one associated probe set was significantly differentially expressed. When a more stringent significance level was used (FDR = 0.008), 3,093 probe sets (2,238 genes) were found significant (Table 1). This represents a large difference between these two cell populations. When comparing CD27hi to CD27lo cells at FDR = 0.01 level, only 7 probe sets were found to be significantly different. The lack of statistical power is due to two reasons: a) some overlap of the cell populations as sorted b) the sample size of this study (six individuals) is relatively small and we are using a very rigorous significant level for testing gene differentiation. When FDR = 0.03 was used for this comparison, we were able to find 2,771 significant probe sets (2,033 genes). Expression levels of several biologically relevant genes were confirmed by subsequent quantitative RT-PCR experiments (p-values in Supplementary Table S2). Row-normalized heatmaps (Fig. 2, Fig. 3, Fig. 4 and Fig. 5) show genes with at least one probe set that was significantly different between CD27lo and CD27hi (†), genes in the comparison of CD27lo to undivided cells that had at least one probe that was within the set of significant probe sets at FDR = 0.01 level (*) or FDR = 0.008 (**).
Table 1. Statistical Analysis of Microarray (SAM). SAM analysis was performed on matched B cell subpopulations isolated from 6 human subjects. Genes appearing in these sets with are noted in the following heatmaps by the symbols indicating their corresponding FDR.
| Comparison | FDR | Number of Differentially Expressed Probes | Number Differentially Expressed Genes | Symbol |
|---|---|---|---|---|
| Undivided: CD27Hi | 0.008 | 8,316 | 5,299 | |
| Undivided: CD27Hi | 0.01 | 11,174 | 6,607 | |
| Undivided: CD27Lo | 0.008 | 3,093 | 2,238 | (**) |
| Undivided: CD27Lo | 0.01 | 7,500 | 4,434 | (*) |
| CD27Lo: CD27Hi | 0.008 | 4 | 4 | |
| CD27Lo: CD27Hi | 0.01 | 7 | 5 | |
| CD27Lo: CD27Hi | 0.03 | 2,746 | 1,815 | (†) |
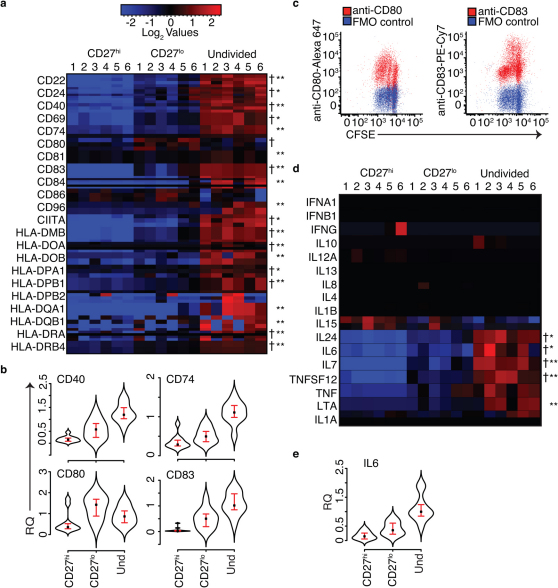
Figure 2. CD27lo cells do not predominantly express antigen presentation/co-stimulation molecules or cytokines.
(a) Row-normalized heatmaps of genome-wide transcriptional analysis using Affymetrix U133 2.0 Plus gene arrays (n = 6 subjects). All probes are shown for each gene plotted. With the exception of CD80, most genes for antigen presentation/co-stimulation were expressed at higher levels in undivided cells. A dagger (†) indicates genes with at least one probe that was found by SAM analysis to be significantly different between CD27lo and CD27hi at FDR = 0.03 (n = 2746 probes). An asterisk is (*) indicates genes in the comparison of undivided to CD27lo cells that had at least one probe significantly different at FDR = 0.01 (n = 7500 probes) and two asterisks (**) are next to genes that were still significant (CD27lo/undivided comparison) at FDR = 0.008 (n = 3093 probes). (b) Quantitative RT-PCR confirmed relative RNA levels of CD40, CD74, CD80, and CD83 (n = 10 subjects, including the same 6 subjects analyzed by gene array) (paired T-test p values in Supplementary Table 2). The point in each violinplot is the median value and the brackets indicate the interquartile range with the addition of the probability density of the data at different values. (c) Flow cytometric analyses of normal human Bc stimulated for 96 hr with CpG+CK confirmed relative protein levels per generation consistent with RNA levels for CD80 and CD83 and a higher percentage of CD27lo cells in divisions 1 and 2 (n = 3 subjects). The undivided subpopulation expresses higher levels of these genes. (d) Gene array analysis shows messages for IL-6, IL-7, IL-24, TNFSF12 (April), Lymphotoxin A (LTA), produced in larger amounts by undivided cells than the other cell subsets. While TNF and IL1A were significantly different between groups at an FDR = 0.03, they were not significant at lower FDRs (n = 6 subjects). (e) Relative IL-6 RNA levels were confirmed by qRT-PCR. (n = 10, paired T-test p values in Supplementary Table 2). The point in each violinplot is the median value and the brackets indicate the interquartile range with addition of the probability density of the data at different values. Except for IL-15, the undivided cells produce higher amounts of these cytokine messages. IL-15 message was significantly higher in CD27hi cells than in CD27lo.
Figure 3. CD27lo cells produce activation-induced cytidine deaminase (AICDA, AID).
AICDA and EXO1, which have been implicated in somatic hypermutation and class-switch recombination, have more abundant mRNA levels in CD27lo cells. This suggests that the cells may be undergoing receptor editing. IL2RA was also expressed at higher levels by CD27lo cells. CCL22, CD80, and BCL2L11 were also expressed at higher levels in CD27lo cells than in CD27hi or in undivided cells (n = 6 subjects, SAM analysis q values in Supplementary Table 1). (†) at least one probe significantly different between CD27lo and CD27hi at FDR = 0.03, SAM analysis, (*)CD27lo/Undivided, at least one probe significant, FDR = 0.01, (**)CD27lo/undivided at least one probe significant at FDR = 0.008. (b) AICDA, IL2RA, and CCL22 mRNA levels were confirmed by qRT-PCR. CCL22 levels between CD27hi and undivided cells were not significantly different (n = 10 subjects, paired T-test p values in Supplementary Table 2). The point in each violinplot is the median value and the brackets indicate the interquartile range. AID protein was expressed in higher levels in divisions 1 and 2, where CD27lo cells predominate. Representative data from 1 subject shown, n = 4 subjects in 2 separate experiments. (c) Gene array analysis showed that while AICDA, IL2RA, and EXO1 were more abundant in CD27lo cells, many other germinal center markers were at higher levels in undivided cells. Naïve cell markers CD5 and ABCB1 were not expressed in amounts that would indicate naïve cell contamination. (d) Expression levels of 7 GC-associated genes were confirmed by qRT-PCR.
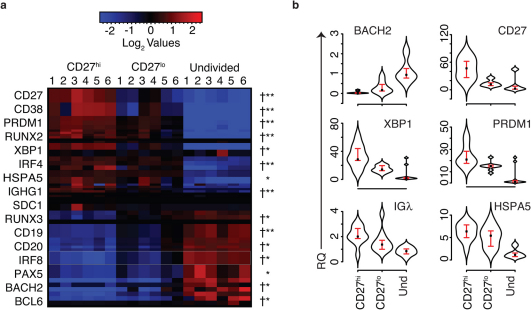
Figure 4. B cell development molecules are expressed at intermediate levels by CD27lo cells.
(a) Intermediate levels of many B cell development molecules in CD27lo cells suggest a transient phenotype consistent with developing pre-plasmablasts engaged in receptor editing before differentiation (n = 6 subjects, SAM analysis q values in Supplementary Table 1). (†) at least one probe significantly different between CD27lo and CD27hi at FDR = 0.03, SAM analysis, (*)CD27lo/Undivided, at least one probe significant, FDR = 0.01, (**)CD27lo/undivided at least one probe significant at FDR = 0.008. (b) Quantitative RT-PCR confirmed relative RNA levels of BACH2, CD27, PRDM1 (BLIMP-1), XBP-1, IgL, and HSPA5 (BiP) (n = 10 subjects, paired T-test p values in Supplementary Table 2). The point in each violinplot is the median value and the brackets indicate the interquartile range.
Figure 5. The Fate of CD27lo cells.
We sorted normal human mBc at 60 hrs of stimulation with CpG+CK and placed isolated CD27hi, CD27lo, and undivided cells back into culture with fresh CpG+cytokines. Flow cytometric analysis 48 hrs later showed that CD27hi cells remained CD27hi, while CD27lo cells became CD27hi. This supports the hypothesis that CD27lo cells are a transient population which differentiate into CD27hi cells. N = 4 subjects in 3 separate experiments.
Do CpG induced CD27lo Bc function as antigen presenting, co-stimulating or cytokine secreting cells?
As co-stimulation molecules have been shown to be expressed in mBc culture systems19,20 we hypothesized that CpG-stimulated CD27lo Bc would have transcriptome patterns rich in antigen presentation and co-stimulation molecules. We therefore examined the relative gene expression levels of: CD22, CD24, CD40, CD69, CD74, CD80, CD81, CD83, CD84, CD86, CD96, Class II Transcriptional Activator (CIITA), and HLA antigens.
Figure 2a shows expression levels for selected genes in each subset (CD27hi , CD27lo and undivided). Undivided Bc had higher expression of transcripts for proteins involved in T cell-B cell adhesion, co-stimulation and antigen presentation, including: CD40, CD83, CD74, CD96, and CIITA. RNA levels for HLA antigens including, HLA-DP HLA-DQ, and HLA-DR were at higher levels in undivided cells. Consistent with increased antigen presenting activity, transcripts for proteins involved in MHC class II assembly, HLA-DO and HLA-DM were also more highly expressed in the undivided cells than the CD27lo and the CD27hi cells. In contrast, the co-stimulatory protein CD80 had higher expression levels in CD27lo cells than in the other cell subsets. CD86 transcript levels were slightly higher in CD27lo cells, however were not significantly different from CD27hi and Undivided cells. We confirmed the gene array results for CD40, CD74, CD80 and CD83 with quantitative RT-PCR (Fig. 2b). Supplementary Table 2 contains p-values from unadjusted paired t-test for comparisons of qRT-PCR data from all 3 cell subsets. The few samples with zero values for gene expression were compared using an unpaired t-test.
We also examined variation in cell surface protein levels with cell division by flow cytometry. CD80 protein levels were higher in earlier cell divisions, which is consistent with higher numbers of CD27lo Bc in these divisions. In contrast, CD83 protein expression levels dropped after division 2 (Fig. 2c), which correlated with higher levels of CD27hi cells in later generations. While these data do not preclude antigen presentation/co-stimulation by CD27lo cells, they better support an alternative hypothesis that undivided Bc might better serve this function.
We next examined cytokine transcript expression between the three cell types (Fig. 2d). A number of murine models have demonstrated phenotypes of regulatory Bc (Breg), characterized by patterns of cytokine secretion (reviewed in3). Human and murine Breg produce several cytokines including: IFN-γ, IL-4, IL-6, IL-10, LT-α, TNF-α and IL123 as well as IL-1α, IL-1β, IL-814, and IL-721. After CpG stimulation, we found very few cytokine transcripts produced in CD27lo cells. IFNA1, IFNB1, IFNG, IL-1β, IL-4, IL-8, IL10, IL-12α and IL13, were not differentially expressed. Transcripts for IL-6, IL-7, and IL-24 were differentially expressed in the undivided cells, with minimal expression in CD27hi and CD27lo proliferating Bc subsets (Fig. 2d). Relative changes in IL-6 transcript levels were confirmed by qRT-PCR (Fig. 2e). Also of interest, low but statistically significant levels of IL-15 mRNA were produced by CD27hi cells (Fig. 2d). This finding is consistent with IL-15 production reported by others in plasma cells22. IL-15 expression suggests the possibility of paracrine stimulation, with CD27hi cells secreting IL-15 to support proliferation of CD27lo cells.
Overall, it was the CpG-stimulated but undivided mBc, not the CD27lo cells, that produced higher transcript levels for proteins with key roles in antigen presentation, co-stimulation and cytokine production.
AID is upregulated in CD27lo populations
As the non-antibody secreting CD27lo cells resulting from CpG stimulation of IgG class switched mBc did not express antigen presenting or cytokine transcripts, we looked for other transcriptome patterns differentially expressed in CD27lo cells to identify other potential functional roles for these cells.
CD27lo cells expressed significantly higher levels of activation-induced cytidine deaminase (gene symbol AICDA, protein abbreviated AID) mRNA than CD27hi or undivided CpG stimulated mBc (Fig. 3a). This was also confirmed by quantitative RT-PCR (Fig. 3b). Flow cytometric data showed increased intracellular AID protein after divisions 1 and 2, where CD27lo cells predominate, and a reduced amount beyond division 3 (Fig. 3c). Upregulation of AICDA, a regulator of Ig gene rearrangements23, suggested an alternative hypothesis that these CpG stimulated, IgG+ mBc were capable of receptor editing before committing to a plasma cell phenotype. AID activity can be induced by TLR activation and can also be an indicator of receptor editing which typically occurs in the germinal center24,25. The CD27lo subpopulation also had higher levels of EXO1, a double-stranded DNA exonuclease required for somatic hypermutation23 (Fig. 3a), as well as the IL-2 receptor alpha (IL-2Rα/CD25), a marker of Bc activation in the germinal center26. However, CD27lo Bc did not differentially express other genes found in germinal center Bc such as BCL6, CD19, CD40, CD44, LTA, CD20, PAX5, SPIB, STAT6, TNF, LRMP, TCL1A, PTPRC (CD45), WEE1, and BCL3 (Fig. 3d). Quantitative rt- PCR confirmed expression levels of BCL6, CD19, CD40, PAX5, PTPRC, STAT6, and TCL1A (Fig. 3e). Although one possible interpretation of increased AICDA in CD27lo cells is contamination by naïve Bc, we did not see differential expression of CD5 or ABCB1 by CD27lo cells (Fig. 3d).
Other transcriptome elements significantly upregulated in CD27lo B cells
Genes of interest found to be significantly upregulated in CD27lo cells in comparison with CD27hi cells included not only AICDA and CD80 (Fig. 3a), but also chemokine ligand 22 (CCL22), BCL2L11 (Bim) a regulator of apoptosis in Bc, and suppressor of cytokines 2 (SOCS2), a cytokine-induced modulator of Jak/Stat pathways that down-regulates cytokine signaling27, IL-5-induced CISH28, and HRAS, which had been shown to mediate ERK activation in response to B cell receptor activation29. This suggested a mechanism for cross-talk between BCR ligation and cytokine receptor signaling of the adaptive immune response, and the antigen independent activation of the TLR9 triggered Bc response. As shown in Figure 2b, qRT-PCR for relative RNA levels of CD80 showed statistically significant differences between each subset. While the differences in CCL22 expression between CD27hi and CD27lo as assessed by qRT-PCR were significant, the differences between CD27hi and undivided cells were not (Fig. 3b). Also, the IL2Rα gene was differentially expressed between CD27hi and CD27lo cells by qRT-PCR but not between CD27lo and undivided cells.
Several other genes were identified by SAM analysis as significantly higher in expression in CD27lo cells, although at low overall expression levels (Fig. 3a). These included ITGAL (CD11a), which along with ITGB2 (CD18) can form LFA-1, a germinal center adhesion complex, but corresponding expression levels of ITGB2 were not seen. Interestingly, RCAN1, which inhibits calcineurin, a signal transduction modulator between TLR, cytokines, and NF-κB30, was also differentially expressed in CD27lo cells. Like SOCS2, this could provide feedback inhibition of cytokine-induced signaling in activated Bc after CpG stimulation.
CD27lo cells express intermediate levels of B cell differentiation transcripts
If CpG-stimulated CD27lo cells were a transient population, undergoing receptor editing before further differentiation, we hypothesized that they should have levels of Bc differentiation markers that are higher than undivided cells and lower than CD27hi cells. Indeed, we found differential expression of many proteins associated with Bc and plasma cell differentiation (Fig. 4a). These included CD27 and CD38, markers associated with antibody secreting plasmablasts in vivo and in vitro17, and immunoglobulin genes. Similarly, expression of HSPA5 (BiP) mRNA, a protein involved in the secretion of immunoglobulin and other proteins31 was also higher in CD27hi cells. Decreases in expression levels between CD27lo and CD27hi cells were seen for CD19, a pan-B cell marker which markedly down-regulated in plasma cell differentiation, and CD20 (MS4A1; reviewed in11). Consistent with previous observations9, large increases in the mature plasma cell marker syndecan-1 (SDC1 or CD138) transcripts were not seen in this pre-plasmablast population.
Overall, the data suggested a progression of transcription patterns towards plasma cell differentiation starting with undivided cells, progressing through CD27lo and then CD27hi cells with the most pronounced plasma cell transcriptome pattern. CD27hi cells exhibited a transcription pattern associated with plasma cell differentiation. This included increased PRDM1 (Blimp), a transcription factor that drives plasma cells differentiation and represses the mature Bc program; and the transcription factor XBP1 which is induced by Blimp and is associated with antibody secretion32. Consistent with this pattern, expression of BACH2, a transcriptional repressor of PRDM133 was decreased. IRF4, an inhibitor of Blimp and a repressor of BCL634 increased in expression level in the CD27hi population as compared with CD27lo. PAX5, which induces BACH211 was differentially expressed in undivided cells, as were BCL6 and IRF8, which is consistent with a mBc phenotype and repression of the plasma cell phenotype11. RUNX2, was produced at higher levels in CD27hi cells than in undivided cells, while the inverse was true of RUNX3, a mBc transcript. For all these transcripts, intermediate levels were expressed in CD27lo cells, consistent with CD27lo cells having a transient phenotype between that of undivided cells and CD27hi cells. Quantitative RT-PCR confirmed differential expression of CD27, Igλ, HSPA5, BACH2, XBP1, and PRDM1 genes (Fig. 4b).
To further explore the outcome of CD27lo cell division we sorted cells at 60hrs of CpG+CK stimulation (Fig. 5) and placed them back into culture for 48 hrs. As expected, CD27hi cells remained CD27hi and continued to proliferate, consistent with a pre-plasmablast or plasmablast phenotype. CD27lo cells did not remain CD27lo, but increased in CD27 expression as they proliferated. This is consistent with a cell population that had completed transition to a plasmablast phenotype and supports the conclusion that the CD27lo population is transient.
Network Analysis
We next performed transcriptome network analysis through the use of Ingenuity Pathways Analysis, looking for possible connecting pathways between the input stimuli of our in vitro stimulation protocol (IL-2, IL-10, IL-15, BAFF, and CpG) and the observed phenotypes of CD27lo and CD27hi cells (Fig. 6). There are at least two pathways active in Bc not involving the Bc receptor that can result in the CD27lo phenotype. First, TLR9 can activate NF-κB35, which has been shown to be a marker of receptor editing in Bc36. Gene targets of NF-κB that we found to be upregulated in CD27lo cells include IL2RA, BCL2L11, CD80, and AICDA. This cluster of upregulated NF-κB targets suggests that NF-κB may be active in the CD27lo subpopulation. In a second activation pathway, IL-2 and IL-15 through action of the common IL-2 receptor γ-chain, can activate various Jak/STAT proteins37,38. IL-10 can also activate STAT339 which can increase production of IRF4, a key regulator of Bc differentiation.
Figure 6. Model of possible network connecting stimulation conditions and induced proteins based on available findings.
The same stimulation conditions can result in either CD27lo or CD27hi phenotypes and progression from one to the other. CpG, through TLR9, can activate ΝΦ−κΒ, a marker of receptor editing in Bc. Gene targets of NF-κB include several genes shown to be upregulated in CD27lo cells (in gold), including AICDA. BAFF (TNFSF13) also has been shown to increase AICDA production. In a separate pathway, Jak proteins, including Jak3 can be activated through action of the common gamma chain cytokines, IL2 or IL-15, resulting in the phosphorylation of STAT proteins such as STAT3, STAT5, and STAT6. IL-10 can also activate STAT3. STAT3, by itself or with STAT6 can increase production of IRF4, a key regulator of Bc differentiation. Induction of IRF4 at low levels can stimulate the production of AICDA, a product of NF-κB and at higher levels, IRF-4 induces production of PRDM-1, a key plasmablast regulator. Thus, as IRF-4 levels increase cells may progress from CD27lo phenotype to a CD27hi phenotype. NF-κB can also activate transcription of SPIB which negatively regulates PRDM1 (BLIMP1). PRDM1 then represses BCL6 and AICDA. PRDM1, STAT6, and IRF4 can also enhance production of XBP1 which is necessary for immunoglobulin production. HSPA5 (BiP), a mediator of immunoglobulin folding is also a target of XBP1 and is upregulated in CD27hi cells.
Graded production of IRF4 can trigger different B cell responses34. Induced at low levels, of IRF4 can trigger production of AICDA, which we found upregulated in CD27lo cells. Induced at higher levels, IRF4 induces production of PRDM-1, a key plasmablast development gene, which was upregulated in CD27hi cells. This provides a means by which the same stimulation conditions can result in either cell phenotype and a mechanism with which CD27lo cells might progress to a CD27hi phenotype as IRF4 levels increase over time. IRF4 also increases the activation of XBP1, a critical regulator of immunoglobulin production upregulated in CD27hi cells. HSPA5 (BiP), a mediator of immunoglobulin folding in the ER is also a target of XBP1 and was found to be upregulated in CD27hi cells.
NF-κB can also activate transcription of SPIB, which may also provide another point of interaction of the CD27lo network with PRDM1 (BLIMP1), a key regulator of plasmablast (CD27hi) differentiation11. PRDM1 expression, in turn, represses BCL634,40 and AICDA34. So while there are separate pathways to the two phenotypes, these pathways appear to have points of reciprocal regulation.
Extensive differences in transcriptome profiles between CpG-stimulated memory B cell subsets
Supplementary Table S1 lists 2,747 probes with statistically significant differences between CD27lo and CD27hi cell subpopulations (SAM analysis, FDR = 0.03). In addition to the gene sets examined for our hypotheses, other genes of interest were differentially expressed between the cell subsets. These are clustered here into gene sets that are relatively upregulated or downregulated as cells transit from undivided to CD27lo to CD27hi phenotypes.
Figure 7A shows a heatmap of selected genes. Increased in expression with cell division were messages including: SLAMF7, a CD2 family member that induces Bc proliferation41; TNFRSF17 (BCMA or CD269) a receptor for the plasma cell survival factor BAFF; IL6 receptor and IL6 signal transducer (IL6ST), which together transmit IL-6 signals to stimulate Bc division; low-density-lipoprotein receptor (LDLR) which can be a marker of Bc activation; and IFN-α receptor 2 (IFNAR2); as well as CD59, a potent complement inhibitor and possible signaling molecule42. Relative RNA levels of CD59, IL6R, and TNFRSF17 (BCMA) were confirmed by qRT-PCR (Fig. 7b).
Figure 7. Other genes of interest differentially expressed between the cell subsets.
(a) Heatmap of selected genes of interest. Genes that increase in expression with cell division include: SLAMF7, TNFRSF17 (BCMA) IL6 receptor and IL6 signal transducer (IL6ST), low-density-lipoprotein receptor (LDLR) a marker of Bc activation; and IFN-α receptor 2 (IFNAR2); as well as CD59, an inhibitor of complement and possible signaling molecule. Genes of interest downregulated with cell division include FCRL family members, FAM129C, CCR6, CCR7 and CXCR5; FOXP1, IL-4 receptor and SLAMF6. CD27lo cells showed intermediate expression of FCRL4, and did not co-express other molecules associated with FCRL4+ cells including RANKL (TNFSF11), and CCNB2. (b) Relative expression levels of select genes were confirmed by qRT-PCR. The point in each violin plot is the median value and the brackets indicate the interquartile range. (n = 10 subjects, paired T-test p values in Supplementary Table 2). We explored the utility of CD59 as a surrogate marker of IgG expression. (c) Relative gene expression levels of CD59 were confirmed by quantitative RT-PCR. (d) CD59 and CD27 are both expressed at higher levels in the same population of cells as assessed by flow cytometric analysis. (e) CD59 expression increased with cell division, but no more than 50% of CD59hi cells in any single generation stained for intracellular IgG. Representative data from 1 subject shown, n = 4 subjects in 2 separate experiments.
A number of genes of interest were downregulated with cell division (Fig. 7a) including FCRL family members FCRLA, FCRL1, FCRL3, and FCRL4. While CD27lo cells showed intermediate expression of FCRL4, they did not co-express other molecules associated with FCRL4+ cells including RANKL (TNFSF11), CCNB2 (Fig. 7a), or RUNX2 (Fig. 4a), so this is a unique population. Also downregulated was FAM129C, a Bc specific transcript, chemokine receptors CCR6, CCR7 and CXCR5; FOXP1, an essential early Bc transcriptional regulator43; IL-4 receptor and SLAMF6, a member of the CD2 family expressed on resting Bc44. Relative levels of FCRL3, FAM129C, CCR6, CCR7, IL4R, and CXCR5 were confirmed by qRT-PCR (Fig. 7b). These changes are consistent with resting Bc differentiating into plasmablasts and point to new molecules that could be explored as markers of antibody production.
Novel markers of Bc subsets that secrete antibody are of interest in that isolation of Ig-secreting cells based on surface expression of Ig or other known cell surface markers is difficult. Even CD27hi cell subsets generally have only 50% of cells producing antibody18. Given the increase in CD59 transcripts in the CD27hi subset gene array, we confirmed expression with qRT-PCR (Fig. 7c) and examined the utility of CD59 as a marker for IgG-producing cells. We stimulated normal human mBc for 96 hrs with CpG and cytokines and found that at the protein level, CD27hi cells were CD59hi (Fig. 7d). Also, CD59 protein expression increased with cell division, but no more than 50% of CD59hi cells in any generation stained for intracellular IgG (Fig. 7e). CD59 appeared similar to, but not better than, CD27 as a marker for discriminating these populations.
Discussion
An increasingly sophisticated understanding of the human CD27+ class switched Bc recall response is emerging from studies identifying subpopulations of activated human mBc. These responses are known to be heterogeneous4,11, with antibody secreting cells, cytokine secreting cells45, antigen presenting and co-stimulatory20 cells, and a mBc pool46 all emerging within the same memory recall response. Understanding the division of labor between the various activated mBc subsets, particularly when altered by adjuvant during vaccination, may allow us to bias the response to favor one set of functions. It is for this reason that we focused on TLR-9 activated CD27+ mBc, as CpG ODN adjuvants are being actively developed to augment vaccine responses47,48.
Previous transcriptome analyses of Bc subsets have examined differential gene expression in mouse naïve and memory subsets from spleen, or human Bc from tonsil49,50, identifying different numbers of genes or probes (50-450 human genes, ∼3,000 murine genes, ∼5,500 murine probes) differentially expressed between Bc subsets. Our results demonstrated ∼2,000 differentially expressed genes between the pairs of the activated memory Bc subsets at a FDR = 0.03. There are several key differences in study methodology that account for this difference with the human studies. First, a number of these papers used a mathematical pattern recognition method which did not specify a false discovery or level of statistical significance, so the quantity of differentially expressed genes are not directly comparable. Also, these studies did not examine the differences between dividing, antibody-secreting, and non-dividing subpopulations of mBc. In contrast, we report transcriptome analysis on subpopulations of stimulated mBc based on generation and the expression of CD27. In addition, many of these reports used an older gene array with 12,000 probe sets, compared to our use of second generation gene arrays with ∼54,000 probe sets. Indeed, many B cell and cell division specific genes lacked corresponding probes in the older arrays. Finally, our results suggest a novel functional division between the proliferating and undivided populations of CpG activated, class switched mBc, as well as several potential feedback mechanisms that may regulate the partitioning of CpG activated, class switched mBc into antibody secreting, receptor editing, and antigen-presenting phenotypes.
Several groups have described patterns of cytokine secretion by activated murine and human Bc in vivo and in vitro3,51. Stimulation by synthetic CpG-B class ODN, such as the CpG2006 used in our experiments, has been reported to induce up-regulation of activation markers, IL-12, and IL-6 by Bc in bulk culture52. We found CpG-induced IL-6, IL-7, IL-15, and IL-24 transcripts at the highest levels in the activated but undivided Bc population. IL-24 belongs to the IL-10 family of cytokines, and has been reported to inhibit plasma cell differentiation of human germinal center Bc53. It is notable that IL-6 and IL-7 were the other predominant cytokine genes highly expressed in the undivided sub-population. IL-7 expression and signaling is associated with recombination activating genes in germinal center Bc54, and IL-6 acts as an autocrine growth and differentiation factor. However, in our experimental system, CpG-stimulation of class-switched mBc does not appear to induce strong, polarizing cytokine secretion associated with classical Breg functions of Th1 versus Th2 immune response deviation3.
Although derived from an in vitro system, our finding of contemporaneous transcription of IL-6, IL-7, and IL-24 genes do suggest a possible mechanism for activated but undivided mBc to regulate CD27lo sub-populations within a germinal center or T cell independent lymphoid follicle. Active gene expression and secretion of these cytokines by undivided Bc could regulate Bc subsets expressing high levels of the IL-6R, IL-7R, and IL20/22R complexes, leading to suppression of plasma cell differentiation while supporting proliferation and receptor editing in a subset of the CpG activated Bc. Consistent with this hypothesis is our finding that CD27lo cells are a proliferating but non-antibody secreting population, with a gene expression pattern suggesting receptor editing and affinity maturation potential. The availability of such adaptive mechanisms after antigen-independent Bc activation also suggests a linkage between innate and adaptive Bc responses, suggesting further mechanisms for the actions of vaccine adjuvants, and targets for future in vivo studies.
Of note, we also found that the undivided cells were not inactive, but appear to have an antigen presentation/co-stimulation phenotype, as suggested by high transcript levels of class II HLA antigens and CD83. We show that these transcripts were generally downregulated by CpG+CK, with intermediate levels in the proliferating CD27lo population. The exceptions are CD80 and CD86 whose transcripts are increased in CD27lo cells. In addition to their role in T cell co-stimulation, these proteins are upregulated on CpG-activated mBc and, when engaged can increase the antibody production55. This suggests that CD80 and CD86 may play a role in activating antibody production within the CD27lo population. This might provide a pathway for CD4 potentiation of antibody secretion during the transition from T cell-independent CpG activation to a more T cell-dependent adaptive immune response.
Also of interest, we found that the CD27lo Bc subpopulation expresses AICDA and precedes CD27hi cells in Bc development. Transcriptome profiling by others found AICDA expression in bulk cultures of stimulated mBc56. Our study points to the CD27lo subpopulation as being the primary producers of AICDA in such systems. CpG is known to stimulate proliferation in CD27- mBc population found in healthy human subjects and enriched in SLE patients57. This Bc population was also reported to be FCRL4-, while the proliferating CD27lo population that we describe here had a higher FCRL4 expression compared to expanding CD27hi cells. Like FCRL4+ Bc identified in other studies2, the CD27lo cells also expressed higher amounts of AICDA, SOX5, and ITGAX, than CD27hi cells, while other markers of FCRL4+ cells (RUNX2, CCNB2, and TNFSF11) were not co-expressed. Thus, while there are similarities between the CD27lo subpopulation in CpG+CK-stimulated mBc and the populations described by others, the CD27lo transitional phenotype described here is unique.
Finally, our work suggests another potential point of interaction between co-cultured CpG-stimulated mBc subsets that could stimulate further studies. IL-6 receptors and IL6ST were upregulated in CD27hi subpopulation, indicating a mechanism through which the activated but undivided Bc subpopulation, which produces IL-6 transcripts, might support plasmablast development. Others have proposed a Bc IL-6 signaling autocrine loop58, although it is beyond the scope of this study to confirm it in this system.
The gene regulatory network model generated from our data implies a dependency of CD27lo subpopulation development and AICDA expression upon NF-κB activation. Although receptor editing in early Bc has been associated with NF-κB activation36, it is difficult to assess alterations of NF-κB signaling in mBc, as early Bc development can be severely compromised by NF-κB disruption59. There is a recent case report of reduced NF-κB signaling in EBV-transformed Bc from two patients with mBc deficiencies60. While our analyses suggest that NF-κB may play a role in development of the CD27lo subpopulation, due to the redundancy of cellular signaling, other pathways may yet be found that provide that role as well.
Methods
Human Subjects Protection.
This study was approved by the Research Subjects Review Board at the University of Rochester Medical Center. Informed consent was obtained from all participants. Research data were coded such that subjects could not be identified, directly or through linked identifiers, in compliance with the Department of Health and Human Services Regulations for the Protection of Human Subjects (45 CFR 46.101(b)(4)).
Myeloma Cell Culture
Myeloma cell lines were maintained in log-phase growth as previously described61 and used for controls in flow cytometry: MPR-1130 (established in our laboratory), MC/CAR and Ramos cell lines (ATCC, Manassas, VA).
B Cell Isolation and CpG activation
Human peripheral blood mononuclear cells were isolated by Ficoll gradient centrifugation as previously described9. Negative magnetic immunoaffinity bead separation (Miltenyi Biotec, Auburn, CA) was used to isolate total Bc. Anti-IgM-PE antibody (BD Biosciences, San Diego, CA) and anti-PE beads (Miltenyi Biotec, Auburn, CA) were used to deplete IgM expressing naïve and mBc. Pacific Blue Succinimidyl Ester (PBSE) (Carlsbad, CA, USA). Flow cytometric analysis was performed on all isolates, showing >90% purity of the isolates.
Freshly isolated CD27+ IgG enriched human peripheral blood Bc were cultured in the presence of CpG 2006 (10 ng/ml, Oligos, etc., Wilsonville, OR), plus recombinant human cytokines IL-2 (20 IU/ml), IL-10 (50 ng/ml), IL-15 (10 ng/ml) (all from BD Biosciences, San Diego, CA), and recombinant human BAFF (75 ng/ml, Chemicon, Temecula, CA) PC-L medium (IMDM medium, lacromin (50µg/ml, Seracare, Milford, MA), insulin (5µg/ml, Sigma-Aldrich, St. Louis, MO), penicillin/streptomycin (1x, Invitrogen, Carlsbad, CA), gentamicin (15µg/ml, Invitrogen), heat-inactivated fetal bovine serum (10% v/v, Invitrogen), normocin (0.1% v/v, Invivogen, San Diego, CA) in round-bottomed 96-well plates (BD Biosciences, San Diego, CA). All cells were cultured at 37°C, 5% CO2.
Flow Cytometric Analysis
Data on Bc surface and intracellular markers and CFSE labeling was collected as previously described9 on an LSR II cytometer with FACS Diva data acquisition software. Antibodies used for cytometry included anti-CD27-APC-H7, anti-IgG-PE-CY5, CD83-PECy7 (BD Bioscience, San Diego, CA) anti-AID (Cell Signalling), antiCD59-APC (abCAM), anti-CD80-Alexa 647 (Serotec), anti-CD19-PE-Cy7, anti-murine IgG-Alexa A680 (Invitrogen), Data was gated, analyzed and displayed using Flowjo software (Treestar, Ashland, OR).
Fluorescence-Activated Cell Sorting
CFSE-stained cells were harvested, washed, and counted. The cells were stained with Live/Dead Violet (Invitrogen, Carlsbad, CA), washed and stained with anti-CD27-APC-H7 antibody (BD Bioscience, San Diego, CA) on ice. FACS cell sorts were performed on live cells. All cell sorting was performed at the University of Rochester Medical Center Flow Cytometry Core Facility.
Transcriptional Analysis
Human CD27+ IgG enriched mBc labeled with PBSE (Invitrogen) and stimulated with the CpG+CK or CD40L+IL-4 for 80 hours round-bottomed 96-well plates. The cells were harvested, washed, counted, stained with anti-CD27-APC-H7 (BD biosciences), and Live/Dead Green (LifeTechnologies, Carlsbad, CA). The cells from each donor were sorted into three samples – Undivided, Dividing CD27 low, and Dividing CD27 high. The cells were suspended in RLT buffer (RNeasy Kit for RNA Isolation, Qiagen, Hilden, Germany), homogenized with a QIAshredder column, and snap frozen in liquid nitrogen. A core facility, the Functional Genomics Center at the University of Rochester, carried out sample carried out cDNA generation using the WR-Ovation Pico System (NuGEN Technologies, San Carlos, CA). Data from the hybridized Affymetrix U133 Plus 2.0 chips was normalized using the GCRMA method. The open-source statistical package R-Bioconductor, Excel (Microsoft, Redmond,WA), Partek Genomic Suite (St. Louis, MO) and Ingenuity Pathways Analysis (Ingenuity, Redwood City, CA) software programs were used for analysis and comparisons. Custom software developed in Mathematica (Wolfram, Champaign, IL) was used to generate heatmaps. A copy of the software is available for download at https://cbim.urmc.rochester.edu/software.
Quantitative RT-PCR
Quantitative RT-PCR was performed by the Functional Genomics Center at the University of Rochester, a core facility. Ten sample sets were analyzed by qRT-PCR, the same 6 sets of RNA analyzed by gene array, and an additional 4 set of cells from different subjects, stimulated and sorted with the same conditions. Immediately after collection, sorted cells were lysed in RLT buffer and passed through a Qiashredder column (both from Qiagen, Germantown, MD) and snap frozen in liquid nitrogen. Samples were stored at −70°C until RNA extraction. RNA was extracted using the RNeasy Micro Kit with on-column DNAse as recommended by the manufacturer (Qiagen). Quantitative RT-PCR was performed on three experimental replicates per sample in TaqMan® Array Fast Plates with TaqMan® Universal PCR Master Mix, No AmpErase® UNG on the StepOnePlus™ Real-Time PCR System (Life Technologies, Carlsbad, CA) with 10 ng cDNA into each 10 ul reaction. RNA quality was confirmed by the presence of intact rRNA with an Agilent 2100 Bioanalyzer (Santa Clara, CA).
For analysis, relative quantity (RQ) values for each gene were calculated using ΔΔCT (Cq) method62. Cq values for each gene were normalized to total RNA content using 18S RNA, normalized to each other using the first sample in the submitted batch, an Undivided sample and then re-normalized to the mean of four control genes chosen from the gene array data, B2M, TERF21P, USP11, and RPL8.
Statistical Analysis
For gene array significance estimates, we applied a modified t-statistic (paired version) by Significance Analysis of Micorarrays (SAM) method63 using the Bioconductor package in R (www.bioconductor.org). A paired t-test was used to compare qRT-PCR datasets. For the genes that contained negative (zero) values, significance was confirmed using an independent t-test. Violin plots for each gene, showing mean and standard deviation of normalized RQ values for each cell population were constructed using the Using R package in R.
Antibody Coated Paramagnetic Bead Preparation
Antibody coated paramagnetic beads were prepared by a modification of our previously published protocol18. Paramagnetic microspheres 8 μm (Bangs Lab, Fishers, IN) 10, 12, 16 or 30 μm in diameter (Micromod DE, Germany) were coated with streptavidin (Sigma-Aldrich, St. Louis, MO) using a commercial kit (PolyLink Coupling Kit, Bangs Lab, Fishers, IN). Additional SA was attached to 10 or 16 micron beads using an 8-branch PEG polymer with amino linker groups (NOF America, White Plains, NY). Streptavidin-coated beads were incubated in photocleavable-biotin-IgG solution (PC-IgG) (Human IgG from Sigma, PC-biotinylation kit (Ambergen, Watertown, MA). All beads were washed overnight in phosphate-buffered saline (PBS) plus 2% w/v bovine serum albumin at room temp with slow mixing on a rotator. Bead concentration was determined by manual count using a hemocytometer (VWR, Westchester, PA).
Paired ELISA-ELISPOT assays
For both qELISPOT and ELISA assays, capture Ab-coated, control wells of 96 well assay plates were seeded with IgG bearing beads, each bead type in triplicate wells. The ELISPOT plates were placed on large rare-earth magnets to settle the magnetic beads rapidly to the well membrane for antibody release. The IgG was released from the beads with a 35 min 365 nm UV exposure from a Black-Ray UV lamp (UVP, Upland, CA), with a 10 min incubation in place before washing.
Matching ELISPOT and ELISA plates were plated with 9 replicate wells of beads for standard curve generation to relate total IgG released with spot density. Plates for cellular ELISPOT assay were pre-spotted with triplicate wells of each bead standard. Bc were collected, washed, counted with trypan blue for viability to calculate live cell numbers for plating. At each time point, ELISPOT plates were developed as previously published18.
ELISA plates were processed by incubation for 1 hour at 37°C with horseradish peroxidase-conjugated goat-anti-human IgG in PBS with 2% BSA, washed, and then incubated with bovine-anti-goat IgG-HRP in PBS + 2% BSA for 1 at room temperature. Color was developed using ABTS One Component Microwell Substrate (Southern Biotech, Birmingham, AL) and read at 450 nm on a Benchmark Plus microplate spectrophotometer (Bio-Rad, Hercules, CA).
ELISPOT and ELISA Reagents
Capture Ab, for ELISPOT and ELISA assays consisted of Mouse anti-Human IgG (H+L) (Jackson, West Grove, PA). For ELISPOT detection, we employed phosphatase-conjugated Goat anti-Human IgG (Jackson, West Grove, PA), for ELISA detection, peroxidase-conjugated Goat anti-Human IgG and peroxidase-conjugated bovine anti-goat IgG, standard curves made with human IgG F(ab)2 fragment (Jackson, West Grove, PA).
Network Analysis
A data set containing genes of interest and corresponding expression values was uploaded into Ingenuity Pathways Analysis (http://ingenuity.com/). Each identifier was mapped to its corresponding object in Ingenuity's Knowledge Base. Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in Ingenuity's Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity.
Author Contributions
A.H. and M.Z. designed the experiments, analyzed the data, and wrote the manuscript. A.H. and S.W. performed the experiments, C.T.B, H.Y., X.Q., H.W., and H.M. performed the statistical analysis of the data, S.W. supervised the gene array data collection and pre-analysis processing, C.T.B. assisted with gene regulatory network analysis, M.L. performed data pre-processing and analysis.
Supplementary Material
Supplementary Material
Acknowledgments
We would like to thank Tim Mosmann, David Topham, Tim Bushnell, and the members of the University of Rochester Center for Biodefense Immune Modeling for spirited discussions which greatly improved the manuscript. We would like to thank Michelle Zanche, Tina Pellegrin, Danielle Morsch and the URMC Flow Core for their expert technical assistance. This work was supported by NIH grants HHSN272201000055C (M.Z., O.H., M.L., H. Y., H.M., H.W., S.W.), N01-AI-50020 (A.H., M.L., H.W., H.M., M.Z.), N01-AI-50029 (M.Z., M.L.) and R01 AI069351 (M.Z., A.H.).
References
- Sanz I., Wei C., Lee F. E. & Anolik J. Phenotypic and functional heterogeneity of human memory B cells. Semin Immunol 20, 67–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt G. R. et al. Discriminating gene expression profiles of memory B cell subpopulations. J Exp Med 205, 1807–17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund F. E. & Randall T. D. Effector and regulatory B cells: modulators of CD4(+) T cell immunity. Nature reviews. Immunology 10, 236–47 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlinton D. B-cell memory: are subsets necessary? Nature reviews. Immunology 6, 785–90 (2006). [DOI] [PubMed] [Google Scholar]
- Bryant V. L. et al. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. Journal of immunology 179, 8180–90 (2007). [DOI] [PubMed] [Google Scholar]
- Jacob J. & Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath-associated foci and germinal centers. J Exp Med 176, 679–87 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatreau S. Novel regulatory functions for Toll-like receptor-activated B cells during intracellular bacterial infection. Immunol Rev 240, 52–71 (2011). [DOI] [PubMed] [Google Scholar]
- Agematsu K. et al. Plasma cell generation from B-lymphocytes via CD27/CD70 interaction. Leukemia & lymphoma 35, 219–25 (1999). [DOI] [PubMed] [Google Scholar]
- Huggins J. et al. CpG DNA activation and plasma-cell differentiation of CD27- naive human B cells. Blood 109, 1611–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J., Zhang J., Lane P. J., Chan E. Y. & MacLennan I. C. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol 21, 2951–62 (1991). [DOI] [PubMed] [Google Scholar]
- Oracki S. A., Walker J. A., Hibbs M. L., Corcoran L. M. & Tarlinton D. M. Plasma cell development and survival. Immunol Rev 237, 140–59 (2010). [DOI] [PubMed] [Google Scholar]
- Tross D. & Klinman D. M. Effect of CpG oligonucleotides on vaccine-induced B cell memory. Journal of immunology 181, 5785–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. D. et al. A Phase 1 study of the blood-stage malaria vaccine candidate AMA1-C1/Alhydrogel with CPG 7909, using two different formulations and dosing intervals. Vaccine 27, 4104–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S. & Gupta S. TLR1/2, TLR7, and TLR9 signals directly activate human peripheral blood naive and memory B cell subsets to produce cytokines, chemokines, and hematopoietic growth factors. J Clin Immunol 31, 89–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard O. S. et al. Improving the immunogenicity of pneumococcal conjugate vaccine in HIV-infected adults with a toll-like receptor 9 agonist adjuvant: a randomized, controlled trial. Clin Infect Dis 51, 42–50 (2010). [DOI] [PubMed] [Google Scholar]
- Johnson T. R., Rao S., Seder R. A., Chen M. & Graham B. S. TLR9 agonist, but not TLR7/8, functions as an adjuvant to diminish FI-RSV vaccine-enhanced disease, while either agonist used as therapy during primary RSV infection increases disease severity. Vaccine 27, 3045–52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery D. T. et al. Increased expression of CD27 on activated human memory B cells correlates with their commitment to the plasma cell lineage. Journal of immunology 174, 4034–42 (2005). [DOI] [PubMed] [Google Scholar]
- Henn A. D. et al. Modulation of single-cell IgG secretion frequency and rates in human memory B cells by CpG DNA, CD40L, IL-21, and cell division. Journal of immunology 183, 3177–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. C. et al. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med 204, 819–30 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good K. L., Avery D. T. & Tangye S. G. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. Journal of immunology 182, 890–901 (2009). [DOI] [PubMed] [Google Scholar]
- Roifman C. M., Zhang J., Chitayat D. & Sharfe N. A partial deficiency of interleukin-7R alpha is sufficient to abrogate T-cell development and cause severe combined immunodeficiency. Blood 96, 2803–7 (2000). [PubMed] [Google Scholar]
- Tarte K., Zhan F., De Vos J., Klein B. & Shaughnessy J. Jr. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood 102, 592–600 (2003). [DOI] [PubMed] [Google Scholar]
- Odegard V. H. & Schatz D. G. Targeting of somatic hypermutation. Nature reviews. Immunology 6, 573–83 (2006). [DOI] [PubMed] [Google Scholar]
- Han J. H. et al. Class switch recombination and somatic hypermutation in early mouse B cells are mediated by B cell and Toll-like receptors. Immunity 27, 64–75 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D. et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. Journal of immunology 179, 6808–19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeren F. A. et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nature Immunology 6, 303–13 (2005). [DOI] [PubMed] [Google Scholar]
- Tannahill G. M. et al. SOCS2 can enhance interleukin-2 (IL-2) and IL-3 signaling by accelerating SOCS3 degradation. Mol Cell Biol 25, 9115–26 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K. et al. Essential role of Stat5 for IL-5-dependent IgH switch recombination in mouse B cells. Journal of immunology 167, 5018–26 (2001). [DOI] [PubMed] [Google Scholar]
- Imamura Y. et al. BLNK binds active H-Ras to promote B cell receptor-mediated capping and ERK activation. J Biol Chem 284, 9804–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. J. et al. Calcineurin negatively regulates TLR-mediated activation pathways. Journal of immunology 179, 4598–607 (2007). [DOI] [PubMed] [Google Scholar]
- Dul J. L. & Argon Y. A single amino acid substitution in the variable region of the light chain specifically blocks immunoglobulin secretion. Proceedings of the National Academy of Sciences of the United States of America 87, 8135–9 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A. L. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 (2004). [DOI] [PubMed] [Google Scholar]
- Ochiai K. et al. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J Biol Chem 281, 38226–34 (2006). [DOI] [PubMed] [Google Scholar]
- Sciammas R. et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25, 225–36 (2006). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology 11, 373–84 (2010). [DOI] [PubMed] [Google Scholar]
- Cadera E. J. et al. NF-kappaB activity marks cells engaged in receptor editing. The Journal of experimental medicine 206, 1803–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Totero D. et al. The opposite effects of IL-15 and IL-21 on CLL B cells correlate with differential activation of the JAK/STAT and ERK1/2 pathways. Blood 111, 517–24 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al.. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proceedings of the National Academy of Sciences of the United States of America 93, 9148–53 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alas S. & Bonavida B. Rituximab inactivates signal transducer and activation of transcription 3 (STAT3) activity in B-non-Hodgkin's lymphoma through inhibition of the interleukin 10 autocrine/paracrine loop and results in down-regulation of Bcl-2 and sensitization to cytotoxic drugs. Cancer research 61, 5137–44 (2001). [PubMed] [Google Scholar]
- Tunyaplin C. et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. Journal of immunology 173, 1158–65 (2004). [DOI] [PubMed] [Google Scholar]
- Lee J. K., Mathew S. O., Vaidya S. V., Kumaresan P. R. & Mathew P. A. CS1 (CRACC, CD319) induces proliferation and autocrine cytokine expression on human B lymphocytes. Journal of immunology 179, 4672–8 (2007). [DOI] [PubMed] [Google Scholar]
- Menu E., Tsai B. C., Bothwell A. L., Sims P. J. & Bierer B. E. CD59 costimulation of T cell activation. CD58 dependence and requirement for glycosylation. Journal of immunology 153, 2444–56 (1994). [PubMed] [Google Scholar]
- Hu H. et al. Foxp1 is an essential transcriptional regulator of B cell development. Nature Immunology 7, 819–26 (2006). [DOI] [PubMed] [Google Scholar]
- Bottino C. et al. NTB-A [correction of GNTB-A], a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative disease. J Exp Med 194, 235–46 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagro A. et al. Type I cytokine profiles of human naive and memory B lymphocytes: a potential for memory cells to impact polarization. Immunology 118, 66–77 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye S. G. & Hodgkin P. D. Divide and conquer: the importance of cell division in regulating B-cell responses. Immunology 112, 509–20 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode C., Zhao G., Steinhagen F., Kinjo T. & Klinman D. M. CpG DNA as a vaccine adjuvant. Expert review of vaccines 10, 499–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coban C., Horii T., Akira S. & Ishii K. J. TLR9 and endogenous adjuvants of the whole blood-stage malaria vaccine. Expert review of vaccines 9, 775–84 (2010). [DOI] [PubMed] [Google Scholar]
- Klein U. et al. Transcriptional analysis of the B cell germinal center reaction. Proceedings of the National Academy of Sciences of the United States of America 100, 2639–44 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomayko M. M. et al. Systematic comparison of gene expression between murine memory and naive B cells demonstrates that memory B cells have unique signaling capabilities. Journal of immunology 181, 27–38 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R. et al. Regulatory B cells (B10 cells) have a suppressive role in murine lupus: CD19 and B10 cell deficiency exacerbates systemic autoimmunity. Journal of immunology 184, 4801–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg A. M. et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374, 546–9 (1995). [DOI] [PubMed] [Google Scholar]
- Wu P. W. et al. IL-22R, IL-10R2, and IL-22BP binding sites are topologically juxtaposed on adjacent and overlapping surfaces of IL-22. Journal of molecular biology 382, 1168–83 (2008). [DOI] [PubMed] [Google Scholar]
- Hikida M. et al. Expression of Recombination Activating Genes in Germinal Center B Cells: Involvement of Interleukin 7 (IL-7) and the IL-7 Receptor. The Journal of experimental medicine 188, 365–372 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau F. C., Dieter J., Luo Z., Priest S. O. & Baumgarth N. B7-1/2 (CD80/CD86) direct signaling to B cells enhances IgG secretion. Journal of immunology 183, 7661–71 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan M. et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood 114, 5173–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. et al. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. Journal of immunology 178, 6624–33 (2007). [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Hamano T., Fujimoto J. & Kakishita E. Regulation of interleukin-6 and interleukin-6R alpha (gp80) expression by murine immunoglobulin-secreting B-cell hybridomas. Immunology 93, 498–504 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derudder E. et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nature immunology 10, 647–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooster J. L. et al. Immune deficiency caused by impaired expression of nuclear factor-kappaB essential modifier (NEMO) because of a mutation in the 5′ untranslated region of the NEMO gene. The Journal of allergy and clinical immunology 126, 127–32 e7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zand M. S. et al. Apoptosis and complement-mediated lysis of myeloma cells by polyclonal rabbit antithymocyte globulin. Blood 107, 2895–903 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R. & Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America 98, 5116–21 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material