Abstract
Gene expression data from microarrays are being applied to predict preclinical and clinical endpoints, but the reliability of these predictions has not been established. In the MAQC-II project, 36 independent teams analyzed six microarray data sets to generate predictive models for classifying a sample with respect to one of 13 endpoints indicative of lung or liver toxicity in rodents, or of breast cancer, multiple myeloma or neuroblastoma in humans. In total, >30,000 models were built using many combinations of analytical methods. The teams generated predictive models without knowing the biological meaning of some of the endpoints and, to mimic clinical reality, tested the models on data that had not been used for training. We found that model performance depended largely on the endpoint and team proficiency and that different approaches generated models of similar performance. The conclusions and recommendations from MAQC-II should be useful for regulatory agencies, study committees and independent investigators that evaluate methods for global gene expression analysis.
As part of the United States Food and Drug Administration’s (FDA’s) Critical Path Initiative to medical product development (http://www.fda.gov/oc/initiatives/criticalpath/), the MAQC consortium began in February 2005 with the goal of addressing various microarray reliability concerns raised in publications1–9 pertaining to reproducibility of gene signatures. The first phase of this project (MAQC-I) extensively evaluated the technical performance of microarray platforms in identifying all differentially expressed genes that would potentially constitute biomarkers. The MAQC-I found high intra-platform reproducibility across test sites, as well as inter-platform concordance of differentially expressed gene lists10–15 and confirmed that microarray technology is able to reliably identify differentially expressed genes between sample classes or populations16,17. Importantly, the MAQC-I helped produce companion guidance regarding genomic data submission to the FDA (http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm079855.pdf ).
Although the MAQC-I focused on the technical aspects of gene expression measurements, robust technology platforms alone are not sufficient to fully realize the promise of this technology. An additional requirement is the development of accurate and reproducible multivariate gene expression–based prediction models, also referred to as classifiers. Such models take gene expression data from a patient as input and as output produce a prediction of a clinically relevant outcome for that patient. Therefore, the second phase of the project (MAQC-II) has focused on these predictive models18, studying both how they are developed and how they are evaluated. For any given microarray data set, many computational approaches can be followed to develop predictive models and to estimate the future performance of these models. Understanding the strengths and limitations of these various approaches is critical to the formulation of guidelines for safe and effective use of preclinical and clinical genomic data. Although previous studies have compared and bench-marked individual steps in the model development process19, no prior published work has, to our knowledge, extensively evaluated current community practices on the development and validation of microarray-based predictive models.
Microarray-based gene expression data and prediction models are increasingly being submitted by the regulated industry to the FDA to support medical product development and testing applications20. For example, gene expression microarray–based assays that have been approved by the FDA as diagnostic tests include the Agendia MammaPrint microarray to assess prognosis of distant metastasis in breast cancer patients21,22 and the Pathwork Tissue of Origin Test to assess the degree of similarity of the RNA expression pattern in a patient’s tumor to that in a database of tumor samples for which the origin of the tumor is known23. Gene expression data have also been the basis for the development of PCR-based diagnostic assays, including the xDx Allomap test for detection of rejection of heart transplants24.
The possible uses of gene expression data are vast and include diagnosis, early detection (screening), monitoring of disease progression, risk assessment, prognosis, complex medical product characterization and prediction of response to treatment (with regard to safety or efficacy) with a drug or device labeling intent. The ability to generate models in a reproducible fashion is an important consideration in predictive model development.
A lack of consistency in generating classifiers from publicly available data is problematic and may be due to any number of factors including insufficient annotation, incomplete clinical identifiers, coding errors and/or inappropriate use of methodology25,26. There are also examples in the literature of classifiers whose performance cannot be reproduced on independent data sets because of poor study design27, poor data quality and/or insufficient cross-validation of all model development steps28,29. Each of these factors may contribute to a certain level of skepticism about claims of performance levels achieved by microarray-based classifiers.
Previous evaluations of the reproducibility of microarray-based classifiers, with only very few exceptions30,31, have been limited to simulation studies or reanalysis of previously published results. Frequently, published benchmarking studies have split data sets at random, and used one part for training and the other for validation. This design assumes that the training and validation sets are produced by unbiased sampling of a large, homogeneous population of samples. However, specimens in clinical studies are usually accrued over years and there may be a shift in the participating patient population and also in the methods used to assign disease status owing to changing practice standards. There may also be batch effects owing to time variations in tissue analysis or due to distinct methods of sample collection and handling at different medical centers. As a result, samples derived from sequentially accrued patient populations, as was done in MAQC-II to mimic clinical reality, where the first cohort is used for developing predictive models and subsequent patients are included in validation, may differ from each other in many ways that could influence the prediction performance.
The MAQC-II project was designed to evaluate these sources of bias in study design by constructing training and validation sets at different times, swapping the test and training sets and also using data from diverse preclinical and clinical scenarios. The goals of MAQC-II were to survey approaches in genomic model development in an attempt to understand sources of variability in prediction performance and to assess the influences of endpoint signal strength in data. By providing the same data sets to many organizations for analysis, but not restricting their data analysis protocols, the project has made it possible to evaluate to what extent, if any, results depend on the team that performs the analysis. This contrasts with previous benchmarking studies that have typically been conducted by single laboratories. Enrolling a large number of organizations has also made it feasible to test many more approaches than would be practical for any single team. MAQC-II also strives to develop good modeling practice guidelines, drawing on a large international collaboration of experts and the lessons learned in the perhaps unprecedented effort of developing and evaluating >30,000 genomic classifiers to predict a variety of endpoints from diverse data sets.
MAQC-II is a collaborative research project that includes participants from the FDA, other government agencies, industry and academia. This paper describes the MAQC-II structure and experimental design and summarizes the main findings and key results of the consortium, whose members have learned a great deal during the process. The resulting guidelines are general and should not be construed as specific recommendations by the FDA for regulatory submissions.
RESULTS
Generating a unique compendium of >30,000 prediction models
The MAQC-II consortium was conceived with the primary goal of examining model development practices for generating binary classifiers in two types of data sets, preclinical and clinical (Supplementary Tables 1 and 2). To accomplish this, the project leader distributed six data sets containing 13 preclinical and clinical endpoints coded A through M (Table 1) to 36 voluntary participating data analysis teams representing academia, industry and government institutions (Supplementary Table 3). Endpoints were coded so as to hide the identities of two negative-control endpoints (endpoints I and M, for which class labels were randomly assigned and are not predictable by the microarray data) and two positive-control endpoints (endpoints H and L, representing the sex of patients, which is highly predictable by the microarray data). Endpoints A, B and C tested teams’ ability to predict the toxicity of chemical agents in rodent lung and liver models. The remaining endpoints were predicted from microarray data sets from human patients diagnosed with breast cancer (D and E), multiple myeloma (F and G) or neuroblastoma (J and K). For the multiple myeloma and neuroblastoma data sets, the endpoints represented event free survival (abbreviated EFS), meaning a lack of malignancy or disease recurrence, and overall survival (abbreviated OS) after 730 days (for multiple myeloma) or 900 days (for neuroblastoma) post treatment or diagnosis. For breast cancer, the endpoints represented estrogen receptor status, a common diagnostic marker of this cancer type (abbreviated ‘erpos’), and the success of treatment involving chemotherapy followed by surgical resection of a tumor (abbreviated ‘pCR’). The biological meaning of the control endpoints was known only to the project leader and not revealed to the project participants until all model development and external validation processes had been completed.
Table 1.
Microarray data sets used for model development and validation in the MAQC-II project
| Date set code |
Endpoint code |
Endpoint description |
Microarray platform |
Training seta |
Validation seta |
Comments and references | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of samples |
Positives (P) |
Negatives (N) |
P/N ratio |
Number of samples |
Positives (P) |
Negatives (N) |
P/N ratio |
|||||
| Hamner | A | Lung tumorigen vs. non-tumorigen (mouse) | Affymetrix Mouse 430 2.0 | 70 | 26 | 44 | 0.59 | 88 | 28 | 60 | 0.47 | The training set was first published in 2007 (ref. 50) and the validation set was generated for MAQC-II |
| Iconix | B | Non-genotoxic liver carcinogens vs. non-carcinogens (rat) | Amersham Uniset Rat 1 Bioarray | 216 | 73 | 143 | 0.51 | 201 | 57 | 144 | 0.40 | The data set was first published in 2007 (ref. 51). Raw microarray intensity data, instead of ratio data, were provided for MAQC-II data analysis |
| NIEHS | C | Liver toxicants vs. non-toxicants based on overall necrosis score (rat) | Affymetrix Rat 230 2.0 | 214 | 79 | 135 | 0.58 | 204 | 78 | 126 | 0.62 | Exploratory visualization of the data set was reported in 2008 (ref. 53). However, the phenotype classification problem was formulated specifically for MAQC-II. A large amount of additional microarray and phenotype data were provided to MAQC-II for cross-platform and cross-tissue comparisons |
| Breast cancer (BR) | D | Pre-operative treatment response (pCR, pathologic complete response) | Affymetrix Human U133A | 130 | 33 | 97 | 0.34 | 100 | 15 | 85 | 0.18 | The training set was first published in 2006 (ref. 56) and the validation set was specifically generated for MAQC-II. In addition, two distinct endpoints (D and E) were analyzed in MAQC-II |
| E | Estrogen receptor status (erpos) | 130 | 80 | 50 | 1.6 | 100 | 61 | 39 | 1.56 | |||
| Multiple myeloma (MM) | F | Overall survival milestone outcome (OS, 730-d cutoff) | Affymetrix Human U133Plus 2.0 | 340 | 51 | 289 | 0.18 | 214 | 27 | 187 | 0.14 | The data set was first published in 2006 (ref. 57) and 2007 (ref. 58). However, patient survival data were updated and the raw microarray data (CEL files) were provided specifically for MAQC-II data analysis. In addition, endpoints H and I were designed and analyzed specifically in MAQC-II |
| G | Event-free survival milestone outcome (EFS, 730-d cutoff) | 340 | 84 | 256 | 0.33 | 214 | 34 | 180 | 0.19 | |||
| H | Clinical parameter S1 (CPS1). The actual class label is the sex of the patient. Used as a “positive” control endpoint | 340 | 194 | 146 | 1.33 | 214 | 140 | 74 | 1.89 | |||
| I | Clinical parameter R1 (CPR1). The actual class label is randomly assigned. Used as a “negative” control endpoint | 340 | 200 | 140 | 1.43 | 214 | 122 | 92 | 1.33 | |||
| Neuro-blastoma (NB) | J | Overall survival milestone outcome (OS, 900-d cutoff) | Different versions of Agilent human microarrays | 238 | 22 | 216 | 0.10 | 177 | 39 | 138 | 0.28 | The training data set was first published in 2006 (ref. 63). The validation set (two-color Agilent platform) was generated specifically for MAQC-II. In addition, one-color Agilent platform data were also generated for most samples used in the training and validation sets specifically for MAQC-II to compare the prediction performance of two-color versus one-color platforms. Patient survival data were also updated. In addition, endpoints L and M were designed and analyzed specifically in MAQC-II |
| K | Event-free survival milestone outcome (EFS, 900-d cutoff) | 239 | 49 | 190 | 0.26 | 193 | 83 | 110 | 0.75 | |||
| L | Newly established parameter S (NEP_S). The actual class label is the sex of the patient. Used as a “positive” control endpoint | 246 | 145 | 101 | 1.44 | 231 | 133 | 98 | 1.36 | |||
| M | Newly established parameter R (NEP_R). The actual class label is randomly assigned. Used as a “negative” control endpoint | 246 | 145 | 101 | 1.44 | 253 | 143 | 110 | 1.30 | |||
The first three data sets (Hamner, Iconix and NIEHS) are from preclinical toxicogenomics studies, whereas the other three data sets are from clinical studies. Endpoints H and L are positive controls (sex of patient) and endpoints I and M are negative controls (randomly assigned class labels). The nature of H, I, L and M was unknown to MAQC-II participants except for the project leader until all calculations were completed.
Numbers shown are the actual number of samples used for model development or validation.
To evaluate the reproducibility of the models developed by a data analysis team for a given data set, we asked teams to submit models from two stages of analyses. In the first stage (hereafter referred to as the ‘original’ experiment), each team built prediction models for up to 13 different coded endpoints using six training data sets. Models were ‘frozen’ against further modification, submitted to the consortium and then tested on a blinded validation data set that was not available to the analysis teams during training. In the second stage (referred to as the ‘swap’ experiment), teams repeated the model building and validation process by training models on the original validation set and validating them using the original training set.
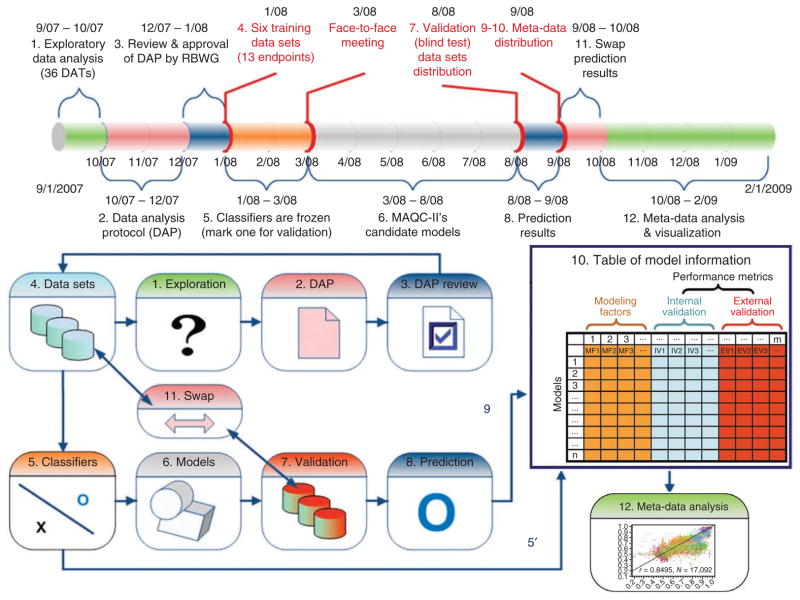
To simulate the potential decision-making process for evaluating a microarray-based classifier, we established a process for each group to receive training data with coded endpoints, propose a data analysis protocol (DAP) based on exploratory analysis, receive feedback on the protocol and then perform the analysis and validation (Fig. 1). Analysis protocols were reviewed internally by other MAQC-II participants (at least two reviewers per protocol) and by members of the MAQC-II Regulatory Biostatistics Working Group (RBWG), a team from the FDA and industry comprising biostatisticians and others with extensive model building expertise. Teams were encouraged to revise their protocols to incorporate feedback from reviewers, but each team was eventually considered responsible for its own analysis protocol and incorporating reviewers’ feedback was not mandatory (see Online Methods for more details).
Figure 1.
Experimental design and timeline of the MAQC-II project. Numbers (1–11) order the steps of analysis. Step 11 indicates when the original training and validation data sets were swapped to repeat steps 4–10. See main text for description of each step. Every effort was made to ensure the complete independence of the validation data sets from the training sets. Each model is characterized by several modeling factors and seven internal and external validation performance metrics (Supplementary Tables 1 and 2). The modeling factors include: (i) organization code; (ii) data set code; (iii) endpoint code; (iv) summary and normalization; (v) feature selection method; (vi) number of features used; (vii) classification algorithm; (viii) batch-effect removal method; (ix) type of internal validation; and (x) number of iterations of internal validation. The seven performance metrics for internal validation and external validation are: (i) MCC; (ii) accuracy; (iii) sensitivity; (iv) specificity; (v) AUC; (vi) mean of sensitivity and specificity; and (vii) r.m.s.e. s.d. of metrics are also provided for internal validation results.
We assembled two large tables from the original and swap experiments (Supplementary Tables 1 and 2, respectively) containing summary information about the algorithms and analytic steps, or ‘modeling factors’, used to construct each model and the ‘internal’ and ‘external’ performance of each model. Internal performance measures the ability of the model to classify the training samples, based on cross-validation exercises. External performance measures the ability of the model to classify the blinded independent validation data. We considered several performance metrics, including Matthews Correlation Coefficient (MCC), accuracy, sensitivity, specificity, area under the receiver operating characteristic curve (AUC) and root mean squared error (r.m.s.e.). These two tables contain data on >30,000 models. Here we report performance based on MCC because it is informative when the distribution of the two classes in a data set is highly skewed and because it is simple to calculate and was available for all models. MCC values range from +1 to −1, with +1 indicating perfect prediction (that is, all samples classified correctly and none incorrectly), 0 indicates random prediction and −1 indicating perfect inverse prediction.
The 36 analysis teams applied many different options under each modeling factor for developing models (Supplementary Table 4) including 17 summary and normalization methods, nine batch-effect removal methods, 33 feature selection methods (between 1 and >1,000 features), 24 classification algorithms and six internal validation methods. Such diversity suggests the community’s common practices are well represented. For each of the models nominated by a team as being the best model for a particular endpoint, we compiled the list of features used for both the original and swap experiments (see the MAQC Web site at http://edkb.fda.gov/MAQC/). These comprehensive tables represent a unique resource. The results that follow describe data mining efforts to determine the potential and limitations of current practices for developing and validating gene expression–based prediction models.
Performance depends on endpoint and can be estimated during training
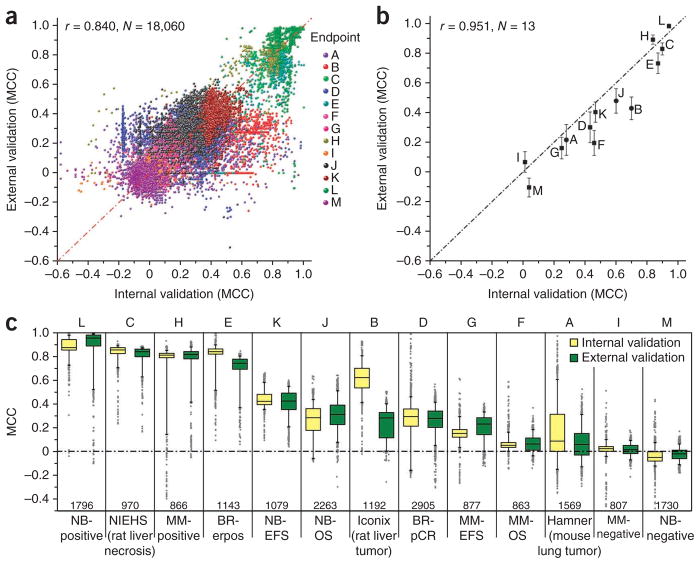
Unlike many previous efforts, the study design of MAQC-II provided the opportunity to assess the performance of many different modeling approaches on a clinically realistic blinded external validation data set. This is especially important in light of the intended clinical or preclinical uses of classifiers that are constructed using initial data sets and validated for regulatory approval and then are expected to accurately predict samples collected under diverse conditions perhaps months or years later. To assess the reliability of performance estimates derived during model training, we compared the performance on the internal training data set with performance on the external validation data set for of each of the 18,060 models in the original experiment (Fig. 2a). Models without complete metadata were not included in the analysis.
Figure 2.
Model performance on internal validation compared with external validation. (a) Performance of 18,060 models that were validated with blinded validation data. (b) Performance of 13 candidate models. r, Pearson correlation coefficient; N, number of models. Candidate models with binary and continuous prediction values are marked as circles and squares, respectively, and the standard error estimate was obtained using 500-times resampling with bagging of the prediction results from each model. (c) Distribution of MCC values of all models for each endpoint in internal (left, yellow) and external (right, green) validation performance. Endpoints H and L (sex of the patients) are included as positive controls and endpoints I and M (randomly assigned sample class labels) as negative controls. Boxes indicate the 25% and 75% percentiles, and whiskers indicate the 5% and 95% percentiles.
We selected 13 ‘candidate models’, representing the best model for each endpoint, before external validation was performed. We required that each analysis team nominate one model for each endpoint they analyzed and we then selected one candidate from these nominations for each endpoint. We observed a higher correlation between internal and external performance estimates in terms of MCC for the selected candidate models (r = 0.951, n = 13, Fig. 2b) than for the overall set of models (r = 0.840, n = 18,060, Fig. 2a), suggesting that extensive peer review of analysis protocols was able to avoid selecting models that could result in less reliable predictions in external validation. Yet, even for the hand-selected candidate models, there is noticeable bias in the performance estimated from internal validation. That is, the internal validation performance is higher than the external validation performance for most endpoints (Fig. 2b). However, for some endpoints and for some model building methods or teams, internal and external performance correlations were more modest as described in the following sections.
To evaluate whether some endpoints might be more predictable than others and to calibrate performance against the positive- and negative-control endpoints, we assessed all models generated for each endpoint (Fig. 2c). We observed a clear dependence of prediction performance on endpoint. For example, endpoints C (liver necrosis score of rats treated with hepatotoxicants), E (estrogen receptor status of breast cancer patients), and H and L (sex of the multiple myeloma and neuroblastoma patients, respectively) were the easiest to predict (mean MCC > 0.7). Toxicological endpoints A and B and disease progression endpoints D, F, G, J and K were more difficult to predict (mean MCC ~0.1–0.4). Negative-control endpoints I and M were totally unpredictable (mean MCC ~0), as expected. For 11 endpoints (excluding the negative controls), a large proportion of the submitted models predicted the endpoint significantly better than chance (MCC > 0) and for a given endpoint many models performed similarly well on both internal and external validation (see the distribution of MCC in Fig. 2c). On the other hand, not all the submitted models performed equally well for any given endpoint. Some models performed no better than chance, even for some of the easy-to-predict endpoints, suggesting that additional factors were responsible for differences in model performance.
Data analysis teams show different proficiency
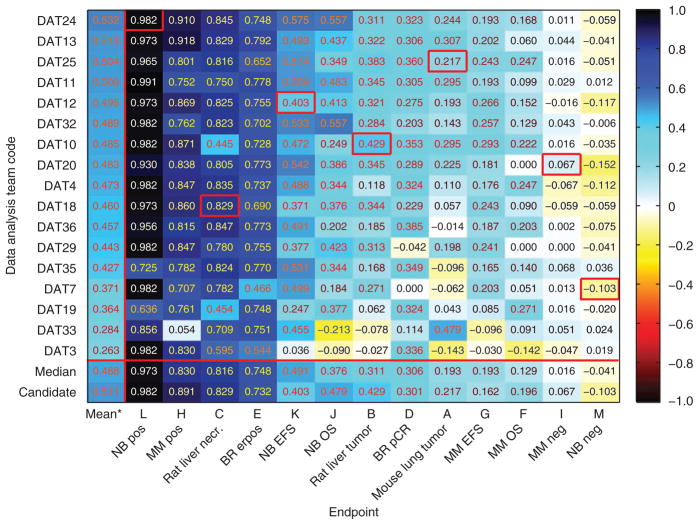
Next, we summarized the external validation performance of the models nominated by the 17 teams that analyzed all 13 endpoints (Fig. 3). Nominated models represent a team’s best assessment of its model-building effort. The mean external validation MCC per team over 11 endpoints, excluding negative controls I and M, varied from 0.532 for data analysis team (DAT)24 to 0.263 for DAT3, indicating appreciable differences in performance of the models developed by different teams for the same data. Similar trends were observed when AUC was used as the performance metric (Supplementary Table 5) or when the original training and validation sets were swapped (Supplementary Tables 6 and 7). Table 2 summarizes the modeling approaches that were used by two or more MAQC-II data analysis teams.
Figure 3.
Performance, measured using MCC, of the best models nominated by the 17 data analysis teams (DATs) that analyzed all 13 endpoints in the original training-validation experiment. The median MCC value for an endpoint, representative of the level of predicability of the endpoint, was calculated based on values from the 17 data analysis teams. The mean MCC value for a data analysis team, representative of the team’s proficiency in developing predictive models, was calculated based on values from the 11 non-random endpoints (excluding negative controls I and M). Red boxes highlight candidate models. Lack of a red box in an endpoint indicates that the candidate model was developed by a data analysis team that did not analyze all 13 endpoints.
Table 2.
Modeling factor options frequently adopted by MAQC-II data analysis teams
| Modeling factor | Option | Original analysis (training => validation)
|
||
|---|---|---|---|---|
| Number of teams | Number of endpoints | Number of models | ||
| Summary and normalization | Loess | 12 | 3 | 2,563 |
| RMA | 3 | 7 | 46 | |
| MAS5 | 11 | 7 | 4,947 | |
| Batch-effect removal | None | 10 | 11 | 2,281 |
| Mean shift | 3 | 11 | 7,279 | |
| Feature selection | SAM | 4 | 11 | 3,771 |
| FC+P | 8 | 11 | 4,711 | |
| T-Test | 5 | 11 | 400 | |
| RFE | 2 | 11 | 647 | |
| Number of features | 0~9 | 10 | 11 | 393 |
| 10~99 | 13 | 11 | 4,445 | |
| ≥1,000 | 3 | 11 | 474 | |
| 100~999 | 10 | 11 | 4,298 | |
| Classification algorithm | DA | 4 | 11 | 103 |
| Tree | 5 | 11 | 358 | |
| NB | 4 | 11 | 924 | |
| KNN | 8 | 11 | 6,904 | |
| SVM | 9 | 11 | 986 | |
Analytic options used by two or more of the 14 teams that submitted models for all endpoints in both the original and swap experiments. RMA, robust multichip analysis; SAM, significance analysis of microarrays; FC, fold change; RFE, recursive feature elimination; DA, discriminant analysis; Tree, decision tree; NB, naive Bayes; KNN, K-nearest neighbors; SVM, support vector machine.
Many factors may have played a role in the difference of external validation performance between teams. For instance, teams used different modeling factors, criteria for selecting the nominated models, and software packages and code. Moreover, some teams may have been more proficient at microarray data modeling and better at guarding against clerical errors. We noticed substantial variations in performance among the many K-nearest neighbor algorithm (KNN)-based models developed by four analysis teams (Supplementary Fig. 1). Follow-up investigations identified a few possible causes leading to the discrepancies in performance32. For example, DAT20 fixed the parameter ‘number of neighbors’ K = 3 in its data analysis protocol for all endpoints, whereas DAT18 varied K from 3 to 15 with a step size of 2. This investigation also revealed that even a detailed but standardized description of model building requested from all groups failed to capture many important tuning variables in the process. The subtle modeling differences not captured may have contributed to the differing performance levels achieved by the data analysis teams. The differences in performance for the models developed by various data analysis teams can also be observed from the changing patterns of internal and external validation performance across the 13 endpoints (Fig. 3, Supplementary Tables 5–7 and Supplementary Figs. 2–4). Our observations highlight the importance of good modeling practice in developing and validating microarray-based predictive models including reporting of computational details for results to be replicated26. In light of the MAQC-II experience, recording structured information about the steps and parameters of an analysis process seems highly desirable to facilitate peer review and reanalysis of results.
Swap and original analyses lead to consistent results
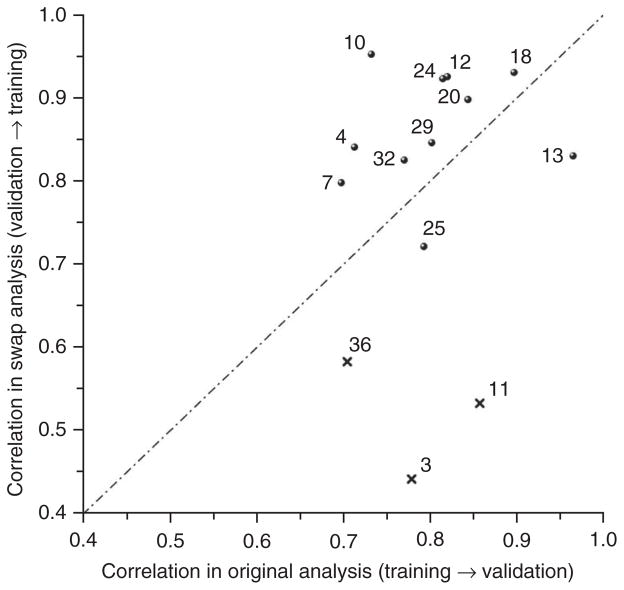
To evaluate the reproducibility of the models generated by each team, we correlated the performance of each team’s models on the original training data set to performance on the validation data set and repeated this calculation for the swap experiment (Fig. 4). The correlation varied from 0.698–0.966 on the original experiment and from 0.443–0.954 on the swap experiment. For all but three teams (DAT3, DAT10 and DAT11) the original and swap correlations were within ±0.2, and all but three others (DAT4, DAT13 and DAT36) were within ±0.1, suggesting that the model building process was relatively robust, at least with respect to generating models with similar performance. For some data analysis teams the internal validation performance drastically overestimated the performance of the same model in predicting the validation data. Examination of some of those models revealed several reasons, including bias in the feature selection and cross-validation process28, findings consistent with what was observed from a recent literature survey33.
Figure 4.
Correlation between internal and external validation is dependent on data analysis team. Pearson correlation coefficients between internal and external validation performance in terms of MCC are displayed for the 14 teams that submitted models for all 13 endpoints in both the original (x axis) and swap (y axis) analyses. The unusually low correlation in the swap analysis for DAT3, DAT11 and DAT36 is a result of their failure to accurately predict the positive endpoint H, likely due to operator errors (Supplementary Table 6).
Previously, reanalysis of a widely cited single study34 found that the results in the original publication were very fragile—that is, not reproducible if the training and validation sets were swapped35. Our observations, except for DAT3, DAT11 and DAT36 with correlation <0.6, mainly resulting from failure of accurately predicting the positive-control endpoint H in the swap analysis (likely owing to operator errors), do not substantiate such fragility in the currently examined data sets. It is important to emphasize that we repeated the entire model building and evaluation processes during the swap analysis and, therefore, stability applies to the model building process for each data analysis team and not to a particular model or approach. Supplementary Figure 5 provides a more detailed look at the correlation of internal and external validation for each data analysis team and each endpoint for both the original (Supplementary Fig. 5a) and swap (Supplementary Fig. 5d) analyses.
As expected, individual feature lists differed from analysis group to analysis group and between models developed from the original and the swapped data. However, when feature lists were mapped to biological processes, a greater degree of convergence and concordance was observed. This has been proposed previously but has never been demonstrated in a comprehensive manner over many data sets and thousands of models as was done in MAQC-II36.
The effect of modeling factors is modest
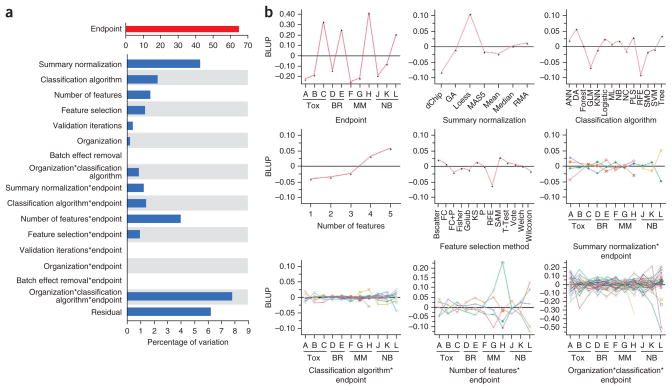
To rigorously identify potential sources of variance that explain the variability in external-validation performance (Fig. 2c), we applied random effect modeling (Fig. 5a). We observed that the endpoint itself is by far the dominant source of variability, explaining >65% of the variability in the external validation performance. All other factors explain <8% of the total variance, and the residual variance is ~6%. Among the factors tested, those involving interactions with endpoint have a relatively large effect, in particular the interaction between endpoint with organization and classification algorithm, highlighting variations in proficiency between analysis teams.
Figure 5.
Effect of modeling factors on estimates of model performance. (a) Random-effect models of external validation performance (MCC) were developed to estimate a distinct variance component for each modeling factor and several selected interactions. The estimated variance components were then divided by their total in order to compare the proportion of variability explained by each modeling factor. The endpoint code contributes the most to the variability in external validation performance. (b) The BLUP plots of the corresponding factors having proportion of variation larger than 1% in a. Endpoint abbreviations (Tox., preclinical toxicity; BR, breast cancer; MM, multiple myeloma; NB, neuroblastoma). Endpoints H and L are the sex of the patient. Summary normalization abbreviations (GA, genetic algorithm; RMA, robust multichip analysis). Classification algorithm abbreviations (ANN, artificial neural network; DA, discriminant analysis; Forest, random forest; GLM, generalized linear model; KNN, K-nearest neighbors; Logistic, logistic regression; ML, maximum likelihood; NB, Naïve Bayes; NC, nearest centroid; PLS, partial least squares; RFE, recursive feature elimination; SMO, sequential minimal optimization; SVM, support vector machine; Tree, decision tree). Feature selection method abbreviations (Bscatter, between-class scatter; FC, fold change; KS, Kolmogorov-Smirnov algorithm; SAM, significance analysis of microarrays).
To further investigate the impact of individual levels within each modeling factor, we estimated the empirical best linear unbiased predictors (BLUPs)37. Figure 5b shows the plots of BLUPs of the corresponding factors in Figure 5a with proportion of variation >1%. The BLUPs reveal the effect of each level of the factor to the corresponding MCC value. The BLUPs of the main endpoint effect show that rat liver necrosis, breast cancer estrogen receptor status and the sex of the patient (endpoints C, E, H and L) are relatively easier to be predicted with ~0.2–0.4 advantage contributed on the corresponding MCC values. The rest of the endpoints are relatively harder to be predicted with about −0.1 to −0.2 disadvantage contributed to the corresponding MCC values. The main factors of normalization, classification algorithm, the number of selected features and the feature selection method have an impact of −0.1 to 0.1 on the corresponding MCC values. Loess normalization was applied to the endpoints (J, K and L) for the neuroblastoma data set with the two-color Agilent platform and has 0.1 advantage to MCC values. Among the Microarray Analysis Suite version 5 (MAS5), Robust Multichip Analysis (RMA) and dChip normalization methods that were applied to all endpoints (A, C, D, E, F, G and H) for Affymetrix data, the dChip method has a lower BLUP than the others. Because normalization methods are partially confounded with endpoints, it may not be suitable to compare methods between different confounded groups. Among classification methods, discriminant analysis has the largest positive impact of 0.056 on the MCC values. Regarding the number of selected features, larger bin number has better impact on the average across endpoints. The bin number is assigned by applying the ceiling function to the log base 10 of the number of selected features. All the feature selection methods have a slight impact of −0.025 to 0.025 on MCC values except for recursive feature elimination (RFE) that has an impact of −0.006. In the plots of the four selected interactions, the estimated BLUPs vary across endpoints. The large variation across endpoints implies the impact of the corresponding modeling factor on different endpoints can be very different. Among the four interaction plots (see Supplementary Fig. 6 for a clear labeling of each interaction term), the corresponding BLUPs of the three-way interaction of organization, classification algorithm and endpoint show the highest variation. This may be due to different tuning parameters applied to individual algorithms for different organizations, as was the case for KNN32.
We also analyzed the relative importance of modeling factors on external-validation prediction performance using a decision tree model38. The analysis results revealed observations (Supplementary Fig. 7) largely consistent with those above. First, the endpoint code was the most influential modeling factor. Second, feature selection method, normalization and summarization method, classification method and organization code also contributed to prediction performance, but their contribution was relatively small.
Feature list stability is correlated with endpoint predictability
Prediction performance is the most important criterion for evaluating the performance of a predictive model and its modeling process. However, the robustness and mechanistic relevance of the model and the corresponding gene signature is also important (Supplementary Fig. 8). That is, given comparable prediction performance between two modeling processes, the one yielding a more robust and reproducible gene signature across similar data sets (e.g., by swapping the training and validation sets), which is therefore less susceptible to sporadic fluctuations in the data, or the one that provides new insights to the underlying biology is preferable. Reproducibility or stability of feature sets is best studied by running the same model selection protocol on two distinct collections of samples, a scenario only possible, in this case, after the blind validation data were distributed to the data analysis teams that were asked to perform their analysis after swapping their original training and test sets. Supplementary Figures 9 and 10 show that, although the feature space is extremely large for microarray data, different teams and protocols were able to consistently select the best-performing features. Analysis of the lists of features indicated that for endpoints relatively easy to predict, various data analysis teams arrived at models that used more common features and the overlap of the lists from the original and swap analyses is greater than those for more difficult endpoints (Supplementary Figs. 9–11). Therefore, the level of stability of feature lists can be associated to the level of difficulty of the prediction problem (Supplementary Fig. 11), although multiple models with different feature lists and comparable performance can be found from the same data set39. Functional analysis of the most frequently selected genes by all data analysis protocols shows that many of these genes represent biological processes that are highly relevant to the clinical outcome that is being predicted36. The sex-based endpoints have the best overlap, whereas more difficult survival endpoints (in which disease processes are confounded by many other factors) have only marginally better overlap with biological processes relevant to the disease than that expected by random chance.
Summary of MAQC-II observations and recommendations
The MAQC-II data analysis teams comprised a diverse group, some of whom were experienced microarray analysts whereas others were graduate students with little experience. In aggregate, the group’s composition likely mimicked the broad scientific community engaged in building and publishing models derived from microarray data. The more than 30,000 models developed by 36 data analysis teams for 13 endpoints from six diverse clinical and preclinical data sets are a rich source from which to highlight several important observations.
First, model prediction performance was largely endpoint (biology) dependent (Figs. 2c and 3). The incorporation of multiple data sets and endpoints (including positive and negative controls) in the MAQC-II study design made this observation possible. Some endpoints are highly predictive based on the nature of the data, which makes it possible to build good models, provided that sound modeling procedures are used. Other endpoints are inherently difficult to predict regardless of the model development protocol.
Second, there are clear differences in proficiency between data analysis teams (organizations) and such differences are correlated with the level of experience of the team. For example, the top-performing teams shown in Figure 3 were mainly industrial participants with many years of experience in microarray data analysis, whereas bottom-performing teams were mainly less-experienced graduate students or researchers. Based on results from the positive and negative endpoints, we noticed that simple errors were sometimes made, suggesting rushed efforts due to lack of time or unnoticed implementation flaws. This observation strongly suggests that mechanisms are needed to ensure the reliability of results presented to the regulatory agencies, journal editors and the research community. By examining the practices of teams whose models did not perform well, future studies might be able to identify pitfalls to be avoided. Likewise, practices adopted by top-performing teams can provide the basis for developing good modeling practices.
Third, the internal validation performance from well-implemented, unbiased cross-validation shows a high degree of concordance with the external validation performance in a strict blinding process (Fig. 2). This observation was not possible from previously published studies owing to the small number of available endpoints tested in them.
Fourth, many models with similar performance can be developed from a given data set (Fig. 2). Similar prediction performance is attainable when using different modeling algorithms and parameters, and simple data analysis methods often perform as well as more complicated approaches32,40. Although it is not essential to include the same features in these models to achieve comparable prediction performance, endpoints that were easier to predict generally yielded models with more common features, when analyzed by different teams (Supplementary Fig. 11).
Finally, applying good modeling practices appeared to be more important than the actual choice of a particular algorithm over the others within the same step in the modeling process. This can be seen in the diverse choices of the modeling factors used by teams that produced models that performed well in the blinded validation (Table 2) where modeling factors did not universally contribute to variations in model performance among good performing teams (Fig. 5).
Summarized below are the model building steps recommended to the MAQC-II data analysis teams. These may be applicable to model building practitioners in the general scientific community.
Step one (design). There is no exclusive set of steps and procedures, in the form of a checklist, to be followed by any practitioner for all problems. However, normal good practice on the study design and the ratio of sample size to classifier complexity should be followed. The frequently used options for normalization, feature selection and classification are good starting points (Table 2).
Step two (pilot study or internal validation). This can be accomplished by bootstrap or cross-validation such as the ten repeats of a fivefold cross-validation procedure adopted by most MAQC-II teams. The samples from the pilot study are not replaced for the pivotal study; rather they are augmented to achieve ‘appropriate’ target size.
Step three (pivotal study or external validation). Many investigators assume that the most conservative approach to a pivotal study is to simply obtain a test set completely independent of the training set(s). However, it is good to keep in mind the exchange34,35 regarding the fragility of results when the training and validation sets are swapped. Results from further resampling (including simple swapping as in MAQC-II) across the training and validation sets can provide important information about the reliability of the models and the modeling procedures, but the complete separation of the training and validation sets should be maintained41.
Finally, a perennial issue concerns reuse of the independent validation set after modifications to an originally designed and validated data analysis algorithm or protocol. Such a process turns the validation set into part of the design or training set42. Ground rules must be developed for avoiding this approach and penalizing it when it occurs; and practitioners should guard against using it before such ground rules are well established.
DISCUSSION
MAQC-II conducted a broad observational study of the current community landscape of gene-expression profile–based predictive model development. Microarray gene expression profiling is among the most commonly used analytical tools in biomedical research. Analysis of the high-dimensional data generated by these experiments involves multiple steps and several critical decision points that can profoundly influence the soundness of the results43. An important requirement of a sound internal validation is that it must include feature selection and parameter optimization within each iteration to avoid overly optimistic estimations of prediction performance28,29,44. To what extent this information has been disseminated and followed by the scientific community in current microarray analysis remains unknown33. Concerns have been raised that results published by one group of investigators often cannot be confirmed by others even if the same data set is used26. An inability to confirm results may stem from any of several reasons: (i) insufficient information is provided about the methodology that describes which analysis has actually been done; (ii) data preprocessing (normalization, gene filtering and feature selection) is too complicated and insufficiently documented to be reproduced; or (iii) incorrect or biased complex analytical methods26 are performed. A distinct but related concern is that genomic data may yield prediction models that, even if reproducible on the discovery data set, cannot be extrapolated well in independent validation. The MAQC-II project provided a unique opportunity to address some of these concerns.
Notably, we did not place restrictions on the model building methods used by the data analysis teams. Accordingly, they adopted numerous different modeling approaches (Table 2 and Supplementary Table 4). For example, feature selection methods varied widely, from statistical significance tests, to machine learning algorithms, to those more reliant on differences in expression amplitude, to those employing knowledge of putative biological mechanisms associated with the endpoint. Prediction algorithms also varied widely. To make internal validation performance results comparable across teams for different models, we recommended that a model’s internal performance was estimated using a ten times repeated fivefold cross-validation, but this recommendation was not strictly followed by all teams, which also allows us to survey internal validation approaches. The diversity of analysis protocols used by the teams is likely to closely resemble that of current research going forward, and in this context mimics reality. In terms of the space of modeling factors explored, MAQC-II is a survey of current practices rather than a randomized, controlled experiment; therefore, care should be taken in interpreting the results. For example, some teams did not analyze all endpoints, causing missing data (models) that may be confounded with other modeling factors.
Overall, the procedure followed to nominate MAQC-II candidate models was quite effective in selecting models that performed reasonably well during validation using independent data sets, although generally the selected models did not do as well in validation as in training. The drop in performance associated with the validation highlights the importance of not relying solely on internal validation performance, and points to the need to subject every classifier to at least one external validation. The selection of the 13 candidate models from many nominated models was achieved through a peer-review collaborative effort of many experts and could be described as slow, tedious and sometimes subjective (e.g., a data analysis team could only contribute one of the 13 candidate models). Even though they were still subject to over-optimism, the internal and external performance estimates of the candidate models were more concordant than those of the overall set of models. Thus the review was productive in identifying characteristics of reliable models.
An important lesson learned through MAQC-II is that it is almost impossible to retrospectively retrieve and document decisions that were made at every step during the feature selection and model development stage. This lack of complete description of the model building process is likely to be a common reason for the inability of different data analysis teams to fully reproduce each other’s results32. Therefore, although meticulously documenting the classifier building procedure can be cumbersome, we recommend that all genomic publications include supplementary materials describing the model building and evaluation process in an electronic format. MAQC-II is making available six data sets with 13 endpoints that can be used in the future as a benchmark to verify that software used to implement new approaches performs as expected. Subjecting new software to benchmarks against these data sets could reassure potential users that the software is mature enough to be used for the development of predictive models in new data sets. It would seem advantageous to develop alternative ways to help determine whether specific implementations of modeling approaches and performance evaluation procedures are sound, and to identify procedures to capture this information in public databases.
The findings of the MAQC-II project suggest that when the same data sets are provided to a large number of data analysis teams, many groups can generate similar results even when different model building approaches are followed. This is concordant with studies29,33 that found that given good quality data and an adequate number of informative features, most classification methods, if properly used, will yield similar predictive performance. This also confirms reports6,7,39 on small data sets by individual groups that have suggested that several different feature selection methods and prediction algorithms can yield many models that are distinct, but have statistically similar performance. Taken together, these results provide perspective on the large number of publications in the bioinformatics literature that have examined the various steps of the multivariate prediction model building process and identified elements that are critical for achieving reliable results.
An important and previously underappreciated observation from MAQC-II is that different clinical endpoints represent very different levels of classification difficulty. For some endpoints the currently available data are sufficient to generate robust models, whereas for other endpoints currently available data do not seem to be sufficient to yield highly predictive models. An analysis done as part of the MAQC-II project and that focused on the breast cancer data demonstrates these points in more detail40. It is also important to point out that for some clinically meaningful endpoints studied in the MAQC-II project, gene expression data did not seem to significantly outperform models based on clinical covariates alone, highlighting the challenges in predicting the outcome of patients in a heterogeneous population and the potential need to combine gene expression data with clinical covariates (unpublished data).
The accuracy of the clinical sample annotation information may also play a role in the difficulty to obtain accurate prediction results on validation samples. For example, some samples were misclassified by almost all models (Supplementary Fig. 12). It is true even for some samples within the positive control endpoints H and L, as shown in Supplementary Table 8. Clinical information of neuroblastoma patients for whom the positive control endpoint L was uniformly misclassified were rechecked and the sex of three out of eight cases (NB412, NB504 and NB522) was found to be incorrectly annotated.
The companion MAQC-II papers published elsewhere give more in-depth analyses of specific issues such as the clinical benefits of genomic classifiers (unpublished data), the impact of different modeling factors on prediction performance45, the objective assessment of microarray cross-platform prediction46, cross-tissue prediction47, one-color versus two-color prediction comparison48, functional analysis of gene signatures36 and recommendation of a simple yet robust data analysis protocol based on the KNN32. For example, we systematically compared the classification performance resulting from one- and two-color gene-expression profiles of 478 neuroblastoma samples and found that analyses based on either platform yielded similar classification performance48. This newly generated one-color data set has been used to evaluate the applicability of the KNN-based simple data analysis protocol to future data sets32. In addition, the MAQC-II Genome-Wide Association Working Group assessed the variabilities in genotype calling due to experimental or algorithmic factors49.
In summary, MAQC-II has demonstrated that current methods commonly used to develop and assess multivariate gene-expression based predictors of clinical outcome were used appropriately by most of the analysis teams in this consortium. However, differences in proficiency emerged and this underscores the importance of proper implementation of otherwise robust analytical methods. Observations based on analysis of the MAQC-II data sets may be applicable to other diseases. The MAQC-II data sets are publicly available and are expected to be used by the scientific community as benchmarks to ensure proper modeling practices. The experience with the MAQC-II clinical data sets also reinforces the notion that clinical classification problems represent several different degrees of prediction difficulty that are likely to be associated with whether mRNA abundances measured in a specific data set are informative for the specific prediction problem. We anticipate that including other types of biological data at the DNA, microRNA, protein or metabolite levels will enhance our capability to more accurately predict the clinically relevant endpoints. The good modeling practice guidelines established by MAQC-II and lessons learned from this unprecedented collaboration provide a solid foundation from which other high-dimensional biological data could be more reliably used for the purpose of predictive and personalized medicine.
METHODS
Methods and any associated references are available here.
Supplementary Material
Acknowledgments
The MAQC-II project was funded in part by the FDA’s Office of Critical Path Programs (to L.S.). Participants from the National Institutes of Health (NIH) were supported by the Intramural Research Program of NIH, Bethesda, Maryland or the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (NIEHS), Research Triangle Park, North Carolina. J.F. was supported by the Division of Intramural Research of the NIEHS under contract HHSN273200700046U. Participants from the Johns Hopkins University were supported by grants from the NIH (1R01GM083084-01 and 1R01RR021967-01A2 to R.A.I. and T32GM074906 to M.M.). Participants from the Weill Medical College of Cornell University were partially supported by the Biomedical Informatics Core of the Institutional Clinical and Translational Science Award RFA-RM-07-002. F.C. acknowledges resources from The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine and from the David A. Cofrin Center for Biomedical Information at Weill Cornell. The data set from The Hamner Institutes for Health Sciences was supported by a grant from the American Chemistry Council’s Long Range Research Initiative. The breast cancer data set was generated with support of grants from NIH (R-01 to L.P.), The Breast Cancer Research Foundation (to L.P. and W.F.S.) and the Faculty Incentive Funds of the University of Texas MD Anderson Cancer Center (to W.F.S.). The data set from the University of Arkansas for Medical Sciences was supported by National Cancer Institute (NCI) PO1 grant CA55819-01A1, NCI R33 Grant CA97513-01, Donna D. and Donald M. Lambert Lebow Fund to Cure Myeloma and Nancy and Steven Grand Foundation. We are grateful to the individuals whose gene expression data were used in this study. All MAQC-II participants freely donated their time and reagents for the completion and analyses of the MAQC-II project. The MAQC-II consortium also thanks R. O’Neill for his encouragement and coordination among FDA Centers on the formation of the RBWG. The MAQC-II consortium gratefully dedicates this work in memory of R.F. Wagner who enthusiastically worked on the MAQC-II project and inspired many of us until he unexpectedly passed away in June 2008.
Leming Shi1, Gregory Campbell2, Wendell D Jones3, Fabien Campagne4, Zhining Wen1, Stephen J Walker5, Zhenqiang Su6, Tzu-Ming Chu7, Federico M Goodsaid8, Lajos Pusztai9, John D Shaughnessy Jr10, André Oberthuer11, Russell S Thomas12, Richard S Paules13, Mark Fielden14, Bart Barlogie10, Weijie Chen2, Pan Du15, Matthias Fischer11, Cesare Furlanello16, Brandon D Gallas2, Xijin Ge17, Dalila B Megherbi18, W Fraser Symmans19, May D Wang20, John Zhang21, Hans Bitter22, Benedikt Brors23, Pierre R Bushel13, Max Bylesjo24, Minjun Chen1, Jie Cheng25, Jing Cheng26, Jeff Chou13, Timothy S Davison27, Mauro Delorenzi28, Youping Deng29, Viswanath Devanarayan30, David J Dix31, Joaquin Dopazo32, Kevin C Dorff33, Fathi Elloumi31, Jianqing Fan34, Shicai Fan35, Xiaohui Fan36, Hong Fang6, Nina Gonzaludo37, Kenneth R Hess38, Huixiao Hong1, Jun Huan39, Rafael A Irizarry40, Richard Judson31, Dilafruz Juraeva23, Samir Lababidi41, Christophe G Lambert42, Li Li7, Yanen Li43, Zhen Li31, Simon M Lin15, Guozhen Liu44, Edward K Lobenhofer45, Jun Luo21, Wen Luo46, Matthew N McCall40, Yuri Nikolsky47, Gene A Pennello2, Roger G Perkins1, Reena Philip2, Vlad Popovici28, Nathan D Price48, Feng Qian6, Andreas Scherer49, Tieliu Shi50, Weiwei Shi47, Jaeyun Sung48, Danielle Thierry-Mieg51, Jean Thierry-Mieg51, Venkata Thodima52, Johan Trygg24, Lakshmi Vishnuvajjala2, Sue Jane Wang8, Jianping Wu53, Yichao Wu54, Qian Xie55, Waleed A Yousef56, Liang Zhang53, Xuegong Zhang35, Sheng Zhong57, Yiming Zhou10, Sheng Zhu53, Dhivya Arasappan6, Wenjun Bao7, Anne Bergstrom Lucas58, Frank Berthold11, Richard J Brennan47, Andreas Buness59, Jennifer G Catalano41, Chang Chang50, Rong Chen60, Yiyu Cheng36, Jian Cui50, Wendy Czika7, Francesca Demichelis61, Xutao Deng62, Damir Dosymbekov63, Roland Eils23, Yang Feng34, Jennifer Fostel13, Stephanie Fulmer-Smentek58, James C Fuscoe1, Laurent Gatto64, Weigong Ge1, Darlene R Goldstein65, Li Guo66, Donald N Halbert67, Jing Han41, Stephen C Harris1, Christos Hatzis68, Damir Herman69, Jianping Huang36, Roderick V Jensen70, Rui Jiang35, Charles D Johnson71, Giuseppe Jurman16, Yvonne Kahlert11, Sadik A Khuder72, Matthias Kohl73, Jianying Li74, Li Li75, Menglong Li76, Quan-Zhen Li77, Shao Li36, Zhiguang Li1, Jie Liu1, Ying Liu35, Zhichao Liu1, Lu Meng35, Manuel Madera18, Francisco Martinez-Murillo2, Ignacio Medina78, Joseph Meehan6, Kelci Miclaus7, Richard A Moffitt20, David Montaner78, Piali Mukherjee33, George J Mulligan79, Padraic Neville7, Tatiana Nikolskaya47, Baitang Ning1, Grier P Page80, Joel Parker3, R Mitchell Parry20, Xuejun Peng81, Ron L Peterson82, John H Phan20, Brian Quanz39, Yi Ren83, Samantha Riccadonna16, Alan H Roter84, Frank W Samuelson2, Martin M Schumacher85, Joseph D Shambaugh86, Qiang Shi1, Richard Shippy87, Shengzhu Si88, Aaron Smalter39, Christos Sotiriou89, Mat Soukup8, Frank Staedtler85, Guido Steiner90, Todd H Stokes20, Qinglan Sun53, Pei-Yi Tan7, Rong Tang2, Zivana Tezak2, Brett Thorn1, Marina Tsyganova63, Yaron Turpaz91, Silvia C Vega92, Roberto Visintainer16, Juergen von Frese93, Charles Wang62, Eric Wang21, Junwei Wang50, Wei Wang94, Frank Westermann23, James C Willey95, Matthew Woods21, Shujian Wu96, Nianqing Xiao97, Joshua Xu6, Lei Xu1, Lun Yang1, Xiao Zeng44, Jialu Zhang8, Li Zhang8, Min Zhang1, Chen Zhao50, Raj K Puri41, Uwe Scherf2, Weida Tong1 & Russell D Wolfinger7
Footnotes
National Center for Toxicological Research, US Food and Drug Administration, Jefferson, Arkansas, USA
Center for Devices and Radiological Health, US Food and Drug Administration, Silver Spring, Maryland, USA
Expression Analysis Inc., Durham, North Carolina, USA
Department of Physiology and Biophysics and HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Medical College of Cornell University, New York, New York, USA
Wake Forest Institute for Regenerative Medicine, Wake Forest University, Winston-Salem, North Carolina, USA
Z-Tech, an ICF International Company at NCTR/FDA, Jefferson, Arkansas, USA
SAS Institute Inc., Cary, North Carolina, USA
Center for Drug Evaluation and Research, US Food and Drug Administration, Silver Spring, Maryland, USA
Breast Medical Oncology Department, University of Texas (UT) M.D. Anderson Cancer Center, Houston, Texas, USA
Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, Little Rock, Arkansas, USA
Department of Pediatric Oncology and Hematology and Center for Molecular Medicine (CMMC), University of Cologne, Cologne, Germany
The Hamner Institutes for Health Sciences, Research Triangle Park, North Carolina, USA
National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, North Carolina, USA
Roche Palo Alto LLC, South San Francisco, California, USA
Biomedical Informatics Center, Northwestern University, Chicago, Illinois, USA
Fondazione Bruno Kessler, Povo-Trento, Italy
Department of Mathematics & Statistics, South Dakota State University, Brookings, South Dakota, USA
CMINDS Research Center, Department of Electrical and Computer Engineering, University of Massachusetts Lowell, Lowell, Massachusetts, USA
Department of Pathology, UT M.D. Anderson Cancer Center, Houston, Texas, USA
Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, Georgia, USA
Systems Analytics Inc., Waltham, Massachusetts, USA
Hoffmann-LaRoche, Nutley, New Jersey, USA
Department of Theoretical Bioinformatics, German Cancer Research Center (DKFZ), Heidelberg, Germany
Computational Life Science Cluster (CLiC), Chemical Biology Center (KBC), Umeå University, Umeå, Sweden
GlaxoSmithKline, Collegeville, Pennsylvania, USA
Medical Systems Biology Research Center, School of Medicine, Tsinghua University, Beijing, China
Almac Diagnostics Ltd., Craigavon, UK
Swiss Institute of Bioinformatics, Lausanne, Switzerland
Department of Biological Sciences, University of Southern Mississippi, Hattiesburg, Mississippi, USA
Global Pharmaceutical R&D, Abbott Laboratories, Souderton, Pennsylvania, USA
National Center for Computational Toxicology, US Environmental Protection Agency, Research Triangle Park, North Carolina, USA
Department of Bioinformatics and Genomics, Centro de Investigación Príncipe Felipe (CIPF), Valencia, Spain
HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Medical College of Cornell University, New York, New York, USA
Department of Operation Research and Financial Engineering, Princeton University, Princeton, New Jersey, USA
MOE Key Laboratory of Bioinformatics and Bioinformatics Division, TNLIST/Department of Automation, Tsinghua University, Beijing, China
Institute of Pharmaceutical Informatics, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, Zhejiang, China
Roche Palo Alto LLC, Palo Alto, California, USA
Department of Biostatistics, UT M.D. Anderson Cancer Center, Houston, Texas, USA
Department of Electrical Engineering & Computer Science, University of Kansas, Lawrence, Kansas, USA
Department of Biostatistics, Johns Hopkins University, Baltimore, Maryland, USA
Center for Biologics Evaluation and Research, US Food and Drug Administration, Bethesda, Maryland, USA
Golden Helix Inc., Bozeman, Montana, USA
Department of Computer Science, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA
SABiosciences Corp., a Qiagen Company, Frederick, Maryland, USA
Cogenics, a Division of Clinical Data Inc., Morrisville, North Carolina, USA
Ligand Pharmaceuticals Inc., La Jolla, California, USA
GeneGo Inc., Encinitas, California, USA
Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA
Spheromics, Kontiolahti, Finland
The Center for Bioinformatics and The Institute of Biomedical Sciences, School of Life Science, East China Normal University, Shanghai, China
National Center for Biotechnology Information, National Institutes of Health, Bethesda, Maryland, USA
Rockefeller Research Laboratories, Memorial Sloan-Kettering Cancer Center, New York, New York, USA
CapitalBio Corporation, Beijing, China
Department of Statistics, North Carolina State University, Raleigh, North Carolina, USA
SRA International (EMMES), Rockville, Maryland, USA
Helwan University, Helwan, Egypt
Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA
Agilent Technologies Inc., Santa Clara, California, USA
F. Hoffmann-La Roche Ltd., Basel, Switzerland
Stanford Center for Biomedical Informatics Research, Stanford University, Stanford, California, USA
Department of Pathology and Laboratory Medicine and HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Medical College of Cornell University, New York, New York, USA
Cedars-Sinai Medical Center, UCLA David Geffen School of Medicine, Los Angeles, California, USA
Vavilov Institute for General Genetics, Russian Academy of Sciences, Moscow, Russia
DNAVision SA, Gosselies, Belgium
École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
State Key Laboratory of Multi-phase Complex Systems, Institute of Process Engineering, Chinese Academy of Sciences, Beijing, China
Abbott Laboratories, Abbott Park, Illinois, USA
Nuvera Biosciences Inc., Woburn, Massachusetts, USA
Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences, Little Rock, Arkansas, USA
VirginiaTech, Blacksburg, Virgina, USA
BioMath Solutions, LLC, Austin, Texas, USA
Bioinformatic Program, University of Toledo, Toledo, Ohio, USA
Department of Mathematics, University of Bayreuth, Bayreuth, Germany
Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, North Carolina, USA
Pediatric Department, Stanford University, Stanford, California, USA
College of Chemistry, Sichuan University, Chengdu, Sichuan, China
University of Texas Southwestern Medical Center (UTSW), Dallas, Texas, USA
Centro de Investigación Príncipe Felipe (CIPF), Valencia, Spain
Millennium Pharmaceuticals Inc., Cambridge, Massachusetts, USA
RTI International, Atlanta, Georgia, USA
Takeda Global R & D Center, Inc., Deerfield, Illinois, USA
Novartis Institutes of Biomedical Research, Cambridge, Massachusetts, USA
W.M. Keck Center for Collaborative Neuroscience, Rutgers, The State University of New Jersey, Piscataway, New Jersey, USA
Entelos Inc., Foster City, California, USA
Biomarker Development, Novartis Institutes of BioMedical Research, Novartis Pharma AG, Basel, Switzerland
Genedata Inc., Lexington, Massachusetts, USA
Affymetrix Inc., Santa Clara, California, USA
Department of Chemistry and Chemical Engineering, Hefei Teachers College, Hefei, Anhui, China
Institut Jules Bordet, Brussels, Belgium
Biostatistics, F. Hoffmann-La Roche Ltd., Basel, Switzerland
Lilly Singapore Centre for Drug Discovery, Immunos, Singapore
Microsoft Corporation, US Health Solutions Group, Redmond, Washington, USA
Data Analysis Solutions DA-SOL GmbH, Greifenberg, Germany
Cornell University, Ithaca, New York, USA
Division of Pulmonary and Critical Care Medicine, Department of Medicine, University of Toledo Health Sciences Campus, Toledo, Ohio, USA
Bristol-Myers Squibb, Pennington, New Jersey, USA
OpGen Inc., Gaithersburg, Maryland, USA
Accession codes. All MAQC-II data sets are available through GEO (series accession number: GSE16716), the MAQC Web site (http://www.fda.gov/nctr/science/centers/toxicoinformatics/maqc/), ArrayTrack (http://www.fda.gov/nctr/science/centers/toxicoinformatics/ArrayTrack/) or CEBS (http://cebs.niehs.nih.gov/) accession number: 009-00002-0010-000-3.
Supplementary information is available on the Nature Biotechnology website.
Publisher's Disclaimer: DISCLAIMER
This work includes contributions from, and was reviewed by, individuals at the FDA, the Environmental Protection Agency (EPA) and the NIH. This work has been approved for publication by these agencies, but it does not necessarily reflect official agency policy. Certain commercial materials and equipment are identified in order to adequately specify experimental procedures. In no case does such identification imply recommendation or endorsement by the FDA, the EPA or the NIH, nor does it imply that the items identified are necessarily the best available for the purpose.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Marshall E. Getting the noise out of gene arrays. Science. 2004;306:630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 2.Frantz S. An array of problems. Nat Rev Drug Discov. 2005;4:362–363. doi: 10.1038/nrd1746. [DOI] [PubMed] [Google Scholar]
- 3.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 4.Ntzani EE, Ioannidis JP. Predictive ability of DNA microarrays for cancer outcomes and correlates: an empirical assessment. Lancet. 2003;362:1439–1444. doi: 10.1016/S0140-6736(03)14686-7. [DOI] [PubMed] [Google Scholar]
- 5.Ioannidis JP. Microarrays and molecular research: noise discovery? Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- 6.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 7.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi L, et al. QA/QC: challenges and pitfalls facing the microarray community and regulatory agencies. Expert Rev Mol Diagn. 2004;4:761–777. doi: 10.1586/14737159.4.6.761. [DOI] [PubMed] [Google Scholar]
- 9.Shi L, et al. Cross-platform comparability of microarray technology: intra-platform consistency and appropriate data analysis procedures are essential. BMC Bioinformatics. 2005;6 (Suppl 2):S12. doi: 10.1186/1471-2105-6-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24:1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 12.Canales RD, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 13.Patterson TA, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 14.Shippy R, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong W, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat Biotechnol. 2006;24:1132–1139. doi: 10.1038/nbt1237. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 17.Strauss E. Arrays of hope. Cell. 2006;127:657–659. doi: 10.1016/j.cell.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Perkins RG, Fang H, Tong W. Reproducible and reliable microarray results through quality control: good laboratory proficiency and appropriate data analysis practices are essential. Curr Opin Biotechnol. 2008;19:10–18. doi: 10.1016/j.copbio.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97:77–87. [Google Scholar]
- 20.Goodsaid FM, et al. Voluntary exploratory data submissions to the US FDA and the EMA: experience and impact. Nat Rev Drug Discov. 2010;9:435–445. doi: 10.1038/nrd3116. [DOI] [PubMed] [Google Scholar]
- 21.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 22.Buyse M, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 23.Dumur CI, et al. Interlaboratory performance of a microarray-based gene expression test to determine tissue of origin in poorly differentiated and undifferentiated cancers. J Mol Diagn. 2008;10:67–77. doi: 10.2353/jmoldx.2008.070099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng MC, et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 25.Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nat Med. 2007;13:1276–1277. doi: 10.1038/nm1107-1276b. author reply 1277–1278. [DOI] [PubMed] [Google Scholar]
- 26.Ioannidis JPA, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 27.Baggerly KA, Edmonson SR, Morris JS, Coombes KR. High-resolution serum proteomic patterns for ovarian cancer detection. Endocr Relat Cancer. 2004;11:583–584. doi: 10.1677/erc.1.00868. author reply 585–587. [DOI] [PubMed] [Google Scholar]
- 28.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon R. Using DNA microarrays for diagnostic and prognostic prediction. Expert Rev Mol Diagn. 2003;3:587–595. doi: 10.1586/14737159.3.5.587. [DOI] [PubMed] [Google Scholar]
- 30.Dobbin KK, et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005;11:565–572. [PubMed] [Google Scholar]
- 31.Shedden K, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parry RM, et al. K-nearest neighbors (KNN) models for microarray gene-expression analysis and reliable clinical outcome prediction. Pharmacogenomics J. 2010;10:292–309. doi: 10.1038/tpj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 34.Dave SS, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 35.Tibshirani R. Immune signatures in follicular lymphoma. N Engl J Med. 2005;352:1496–1497. doi: 10.1056/NEJM200504073521422. author reply 1496–1497. [DOI] [PubMed] [Google Scholar]
- 36.Shi W, et al. Functional analysis of multiple genomic signatures demonstrates that classification algorithms choose phenotype-related genes. Pharmacogenomics J. 2010;10:310–323. doi: 10.1038/tpj.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson GK. That BLUP is a good thing: the estimation of random effects. Stat Sci. 1991;6:15–32. [Google Scholar]
- 38.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Statist. 2006;15:651–674. [Google Scholar]
- 39.Boutros PC, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci USA. 2009;106:2824–2828. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popovici V, et al. Effect of training sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12:R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yousef WA, Wagner RF, Loew MH. Assessing classifiers from two independent data sets using ROC analysis: a nonparametric approach. IEEE Trans Pattern Anal Mach Intell. 2006;28:1809–1817. doi: 10.1109/TPAMI.2006.218. [DOI] [PubMed] [Google Scholar]
- 42.Gur D, Wagner RF, Chan HP. On the repeated use of databases for testing incremental improvement of computer-aided detection schemes. Acad Radiol. 2004;11:103–105. doi: 10.1016/s1076-6332(03)00511-7. [DOI] [PubMed] [Google Scholar]
- 43.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 44.Wood IA, Visscher PM, Mengersen KL. Classification based upon gene expression data: bias and precision of error rates. Bioinformatics. 2007;23:1363–1370. doi: 10.1093/bioinformatics/btm117. [DOI] [PubMed] [Google Scholar]
- 45.Luo J, et al. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J. 2010;10:278–291. doi: 10.1038/tpj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan X, et al. Consistency of predictive signature genes and classifiers generated using different microarray platforms. Pharmacogenomics J. 2010;10:247–257. doi: 10.1038/tpj.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J, et al. Genomic indicators in the blood predict drug-induced liver injury. Pharmacogenomics J. 2010;10:267–277. doi: 10.1038/tpj.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberthuer A, et al. Comparison of performance of one-color and two-color geneexpression analyses in predicting clinical endpoints of neuroblastoma patients. Pharmacogenomics J. 2010;10:258–266. doi: 10.1038/tpj.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong H, et al. Assessing sources of inconsistencies in genotypes and their effects on genome-wide association studies with HapMap samples. Pharmacogenomics J. 2010;10:364–374. doi: 10.1038/tpj.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.