Abstract
We have developed a novel in vitro mutagenesis technique that allows us to introduce mutations at the level of double-stranded DNA and then transcribe the mutant DNA directly. The technique is useful for those wishing to produce recombinant RNA, particularly if the desired recombinant is the result of an insertion or deletion. It is also useful for the preparation of 3'-truncated RNAs with a defined end. The technique is not dependent on the presence of a convenient restriction site within the target gene, and does not involve construction of a clone or amplification of the mutant DNA within a bacterial host. It is intended as a simple and rapid method for the preparation of roughly 100-200 pmol of mutant RNA, which would be sufficient for obtaining sequence information and assessing the functional consequences of the mutation.
Full text
PDF





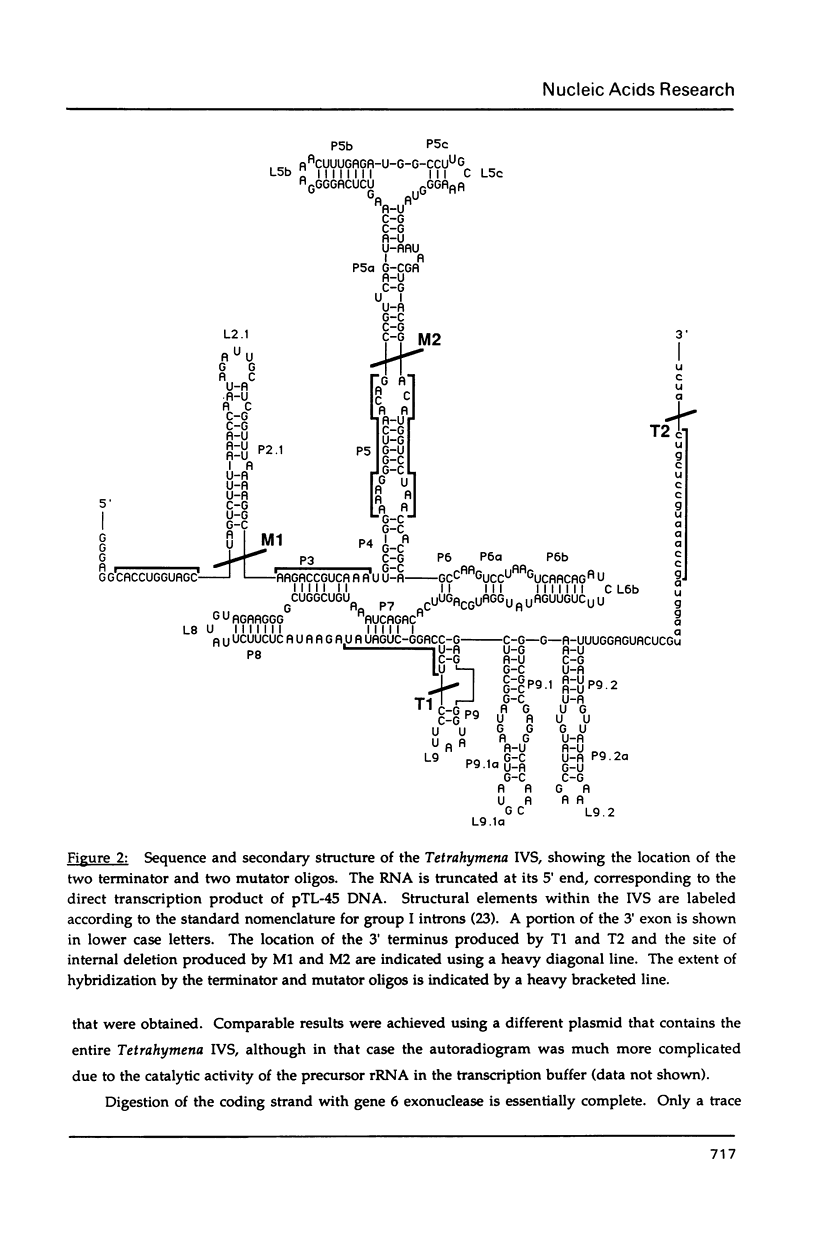





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger S. L., Wallace D. M., Puskas R. S., Eschenfeldt W. H. Reverse transcriptase and its associated ribonuclease H: interplay of two enzyme activities controls the yield of single-stranded complementary deoxyribonucleic acid. Biochemistry. 1983 May 10;22(10):2365–2372. doi: 10.1021/bi00279a010. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler E. T., Chamberlin M. J. Bacteriophage SP6-specific RNA polymerase. I. Isolation and characterization of the enzyme. J Biol Chem. 1982 May 25;257(10):5772–5778. [PubMed] [Google Scholar]
- Chisaka O., Iwai S., Ohtsuka E., Matsubara K. Cloning a defined region of DNA using a limited action of DNA polymerase: application to dissection of hepatitis B virus surface antigen gene. Gene. 1986;45(1):19–25. doi: 10.1016/0378-1119(86)90127-7. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: I. Optimum conditions and minimum ologodeoxyribonucleotide length. Gene. 1979 Dec;8(1):81–97. doi: 10.1016/0378-1119(79)90009-x. [DOI] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Kerr C., Sadowski P. D. Gene 6 exonuclease of bacteriophage T7. I. Purification and properties of the enzyme. J Biol Chem. 1972 Jan 10;247(1):305–310. [PubMed] [Google Scholar]
- Krupp G., Söll D. Simplified in vitro synthesis of mutated RNA molecules. An oligonucleotide promoter determines the initiation site of T7RNA polymerase on ss M13 phage DNA. FEBS Lett. 1987 Feb 23;212(2):271–275. doi: 10.1016/0014-5793(87)81359-5. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal N. G. DNA synthesis on a double-stranded DNA template by the T4 bacteriophage DNA polymerase and the T4 gene 32 DNA unwinding protein. J Biol Chem. 1974 Sep 10;249(17):5668–5676. [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S. J., Horn G. T., Erlich H. A. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. Science. 1986 Sep 5;233(4768):1076–1078. doi: 10.1126/science.3461561. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]




