Abstract
Background
Recent studies suggest reduction of radical-propagating fatty acid hydroperoxides to inert hydroxides by interaction with apolipoprotein-D (apoD) Met93 may represent an antioxidant function for apoD. The nature and structural consequences of this selective interaction are unknown.
Methodology/Principal Findings
Herein we used molecular dynamics (MD) analysis to address these issues. Long-timescale simulations of apoD suggest lipid molecules are bound flexibly, with the molecules free to explore multiple conformations in a binding site at the entrance to the classical lipocalin ligand-binding pocket. Models of 5s- 12s- and 15s-hydroperoxyeicosatetraenoic acids were created and the lipids found to wrap around Met93 thus providing a plausible mechanism by which eicosatetraenoic acids bearing hydroperoxides on different carbon atoms can interact with Met93 to yield Met93 sulfoxide (Met93SO). Simulations of glycosylated apoD indicated that a second solvent exposed Met at position 49 was shielded by a triantennerary N-glycan attached to Asn45 thereby precluding lipid interactions. MD simulations of apoD showed B-factors of the loop containing Met93SO were higher in the oxidized protein, indicating increased flexibility that is predicted to destabilize the protein and promote self-association.
Conclusions/Significance
These studies provide novel insights into the mechanisms that may contribute to the antioxidant function of apoD and the structural consequences that result if Met93SO is not redox-cycled back to its native state.
Introduction
Apolipoprotein-D (apoD) is a 29 kDa glycoprotein member of the lipocalin family that comprises an eight-stranded antiparallel β-barrel flanked by an α-helix [1]. The β-barrel encloses a hydrophobic ligand pocket that binds arachidonic acid and progesterone with high affinity [1], [2]. The eicosanoids 12-hydroxyeicosatetraenoic acid (12-HETE) and 5,15- dihydroxyeicosatetraenoic acid have also been shown to bind to apoD albeit with lower affinity [3]–[5]. In addition to the classical lipocalin pocket [6], apoD may also interact with lipids via a cluster of exposed hydrophobic residues residing in three of its extended loops [1]. These exposed residues generate a hydrophobic surface region close to the open end of the binding pocket that facilitates apoD association with high-density lipoprotein (HDL) particles and is also thought to permit insertion of apoD into cellular lipid membranes [1].
Although apoD is well known to bind lipids and thereby thought to play a role in lipid transport in the brain and in plasma [7], it has also more recently been associated with lipid antioxidant protection. This has been clearly demonstrated using apoD null and apoD over-expressing mice [8] and in an ageing Drosophila model [9]. In these studies apoD overexpression protected against insults that promote cerebral lipid peroxidation whereas deletion of the APOD gene increased susceptibility to oxidative stress. This antioxidant function might also explain why apoD levels are increased in the human brain in association with ageing and Alzheimer's disease [10]–[12] as it could serve as a protective response to combat the increased levels of lipid peroxidation that are know to occur under these conditions [13], [14].
We have recently shown that apoD catalyzes the reduction of potential free-radical generating lipid hydroperoxides (L-OOHs) to relatively inert lipid hydroxides (L-OHs) via a selective interaction with apoD Met93 [15]. As a consequence of this reaction Met93 is selectively converted to MetSO [15]. We have proposed that this reduction of L-OOH to L-OH competes with transition metal-catalyzed Fenton-type reactions that could otherwise generate chain-propagating radicals from L-OOHs [15]. Although this pathway may contribute to the apoD antioxidant mechanism, it is presently unclear how different lipid hydroperoxides may interact selectively with apoD Met93 when high affinity lipid binding within the apoD ligand pocket is known to be selective [2]. In addition, details of the structural changes that may occur in apoD as a result of Met93SO generation remain to be defined.
The aim of the present study was therefore to use MD simulations and molecular modeling approaches to investigate the interactions of arachidonic acid and its lipoxygenase-derived HpETE products with apoD and to assess the potential structural consequences of apoD Met93 oxidation.
Materials and Methods
ApoD molecular dynamics simulations and modelling
In order to understand the structural basis for the behavior of apoD, modeling and MD simulations were performed. All MD trajectories were calculated using NAMD 2.8 [16]. For all simulations of apoD, models were embedded in a water box with overlapping water molecules removed and sodium ions added to ensure that the systems had no net charge. Temperature control at 310 K was maintained with Langevin dynamics (damping constant: 5 ps−1) applied to non-hydrogen atoms. Periodic boundary conditions were used with the Nosé-Hoover Langevin piston method (piston period 100 fs, decay rate 50 fs) to maintain a constant pressure of 1.013 Bar. The Particle-mesh Ewald algorithm was used to account for long-range electrostatic effects (grid resolution<1 Å). All other non-bonded interactions were calculated using a switching function to smooth interactions to zero between 10 and 12 Å. The integration timesteps were 1, 2, and 4 fs for bonded, nonbonded, and long-range electrostatic interactions respectively. Every system was initially equilibrated for 100 ps, after which the MD run was extended as described below. Coordinates were saved every 1 ps for analysis.
To determine the effects of Met93 oxidation on stability of apoD, 50 ns simulations of apoD and ApoD-Met93SO without bound ligands were calculated. The starting configuration for these calculations was the published structure of ligand-free apoD (PDB: 2HZR), with missing residues modeled, selenoMet residues changed to Met and any mutated residues reverted to wild-type. To assess the ability of the simulations to reproduce native protein-ligand conformations, a 100 ns simulation of the complex of apoD with progesterone was performed using the crystal structure of this complex (PDB: 2HZQ) for the starting configuration. To understand the behavior of lipid molecules bound to apoD, four 200 ns runs were produced with arachidonic acid modeled in the binding site in different extended conformations. Low energy conformations were used to model arachidonate metabolites 5s-, 12s- and 15s-HpETE bound to apoD. Finally, a 50 ns simulation of the apoD-progesterone complex with experimentally determined glycosylation was performed. In all cases the CHARMM protein force field [17] with backbone 2D dihedral energy correction (CMAP) [18] was used. Parameters for MetSO were adapted from parameters for DMSO [19]. The CHARMM general force-field [20] and carbohydrate derivative forcefield [21] was used for progesterone and glycosylations respectively. Trajectory data were analyzed using VMD [22]. Protein-ligand interaction energies were estimated using the generalized Born implicit solvent model [23]. Energies of isolated protein and ligand entities were calculated and subtracted from the energy of the combined system. For modeling HpETE compounds, parameters were taken from the CHARMM General Force Field (CGenFF) [20] or were derived from ab inito calculations on the model compound 2-hydroperoxypropane using Gaussian09 [24], performed at the MP2/6-31G* level of theory.
ApoD analysis by circular dichroism (CD)
Recombinant forms of human apoD (NM_001647.3) containing a C-terminal linker, FLAG-tag and poly His tail (SGGGGSDYKDDDDKHHHHHH) were synthesized in HEK293 cells (American Type Culture Collection (ATCC), Catalogue No. CRL-1573, Manassas, VA, USA) and purified using a Ni-HiTrap column as described previously [15]. To generate apoD containing Met93SO, wild type apoD (0.5 mg/ml) was incubated in PBS containing 15s-HpETE (0.05 mg/ml) for 4 h. The protein fraction was precipitated with 9 volumes of ice-cold ethanol for 1 h at −20° and the pellet re-suspended in 20 µl of PBS in preparation for CD analysis. Far UV CD spectra for apoD and its mutants (apoD, M49-A, M93-A, M157-A) were acquired using Jasco J-810 CD spectropolarimeter at 22°C. Protein samples were prepared to final concentration of 0.1 mg/ml in PBS and spectra recorded using 0.1 cm quartz cuvette. Measurements were taken from 195–260 nm using a scan speed of 100 nm/min, 1 nm bandwidth, 1 s response time and data pitch of 0.1 nm. Each spectrum is an average of 6 scans and corrected by subtraction of the PBS spectrum acquired under similar conditions.
Results
ApoD structure and position of Met residues
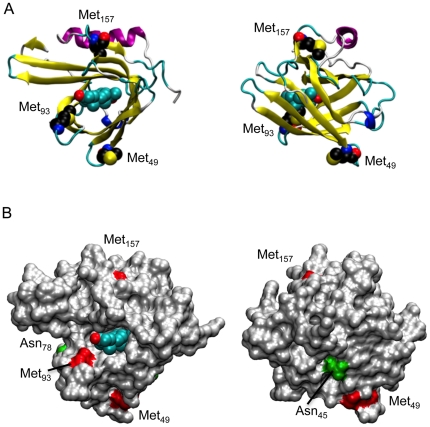
We first examined apoD amino acid sequence across a variety of mammalian species. ApoD contains three Met residues that are highly conserved (Figure 1). In humans, apoD Met residues are Met49, Met93 and Met157. Met93 is located in a hydrophobic region at the opening of the ligand-binding pocket and is clearly exposed to the surface (Figure 2). Met49 is also exposed to the surface, residing within a β-strand stretch four amino acids C-terminal to one of the two N-linked glycosylation sites (Asn45). Met157 appears to be largely buried beneath a surface-accessible α-helix (Figure 2).
Figure 1. ApoD Clustal alignment.
ApoD sequences were aligned using the UniProt Consortium Clustal alignment tool for the listed mammalian species. The positions of the three human Met residues are annotated (using the human residue numbering without the 20 amino acid signal peptide). Conserved Met residues are highlighted in yellow with substituted amino acids highlighted in red.
Figure 2. ApoD structure.
(A) Orthogonal views of apoD in cartoon form with progesterone bound. The ligand and methionine residues are represented as van der Waals spheres. (B) Same as (A) but with protein represented as a solvent-accessible surface. Methionine residues (red patches) and glycosylated asparagine residues (green patches) are indicated.
MD simulation of apoD interactions with progesterone and arachidonic acid
In a recent study we reported that apoD exhibits a lipid antioxidant activity that we propose is due to the direct interaction of the apoD Met93 side chain with lipid hydroperoxides such as 5s-, 12s- and 15s-HpETEs [15]. Previous work indicated the high affinity binding of lipids within the apoD ligand binding pocket is selective and it was therefore unclear how different lipid hydroperoxides may interact with Met93.
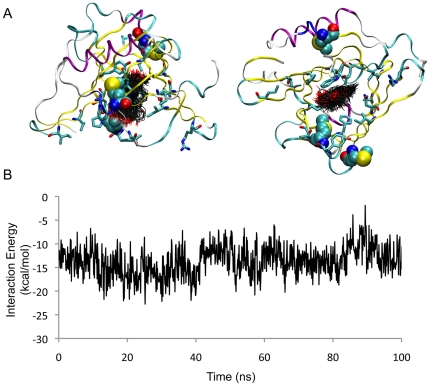
We first assessed the interaction of progesterone with apoD as crystallography studies indicate this lipid forms a stable complex within the ligand-binding pocket [1]. In a 100 ns simulation of progesterone bound to apoD, the ligand remained close to the crystallographic conformation, with binding energies fluctuating between −1 and −23 kcal/mol (Figure 3). The RMSD of ligand atoms with respect to the crystallographically observed conformation was between 2 and 3.5 Å. A small degree of lateral sliding of the ligand in the pocket was observed in our MD simulation. All crystallographically observed contacts between progesterone and apoD were maintained over 100 ns.
Figure 3. MD simulation of apoD with progesterone.
(A) Orthogonal views of apoD in ribbon form with ligand-binding residues shown (cyan carbon atoms). The conformation of progesterone, sampled at 1 ns intervals, is shown (black carbon atoms). Methionine residues are represented as van der Waals spheres. (B) Interaction energy is plotted as a function of time.
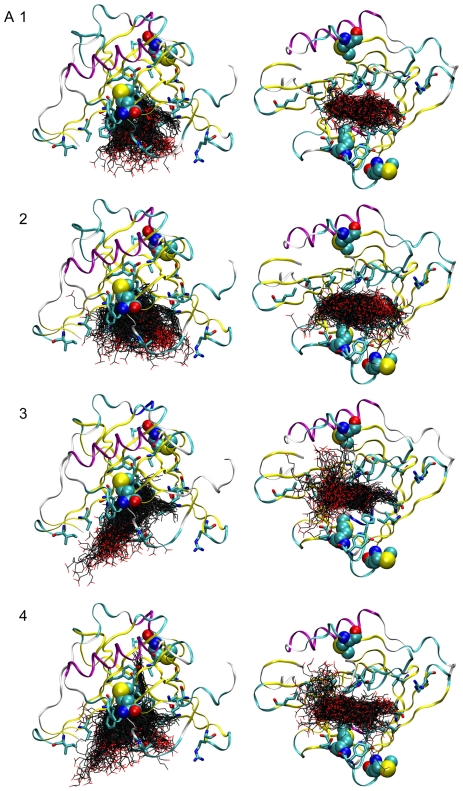
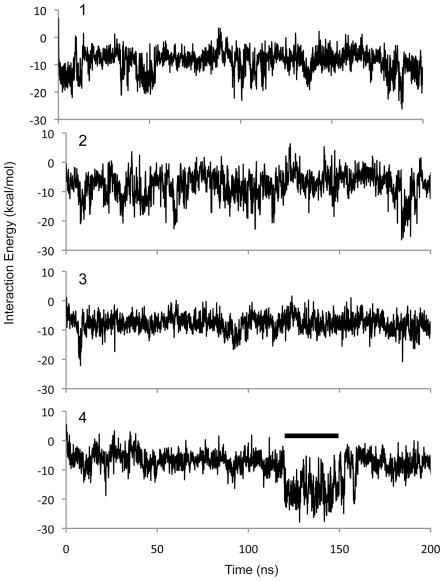
We next examined the interaction of arachidonic acid with apoD. Four simulations of apoD with bound arachidonic acid, totaling 800 ns, indicated that the principal binding site is at the opening of the ligand binding pocket and that this hydrophobic surface appears to behave as a “greasy slide”. In none of the four simulations, did arachidonate converge on a particular bound conformation (Figure 4). Instead, arachidonate continuously explores conformational space. In the majority of conformations sampled, arachidonate is extended length-ways across the binding site or is partly curled up in the binding site. In simulation 3, arachidonate became partly unbound but re-entered the binding site. In simulation 4, the tail of arachidonic acid became transiently buried in the binding site in a manner reminiscent of progesterone. Also in simulation 4, arachidonate adopted a conformation with low energy of interaction lasting approximately 30 ns (Figure 5), in which the carboxylic acid group of the fatty acid engaged in a salt bridge interaction with Arg62, with the remainder of the molecule lying in the binding site in a crescent-shaped conformation wrapped around Met93. This population of low-energy arachidonate conformations was used to model binding of 5s-, 12s- and 15s-HpETE to apoD.
Figure 4. MD simulation of apoD with arachidonate.
Orthogonal views of apoD in ribbon form with ligand-binding residues shown (cyan carbon atoms). The conformation of arachidonate, sampled at 1 ns intervals, is shown for simulations 1 to 4 (black carbon atoms). Methionine residues are represented as van der Waals spheres.
Figure 5. Interaction energy for MD simulation of apoD with arachidonate.
Interaction energy of the four runs shown in Figure 4 are plotted as a function of time. The conformations in Figure 4 simulation 4 used for modeling of HpETE are indicated by a bar.
Modeling of HpETE interaction with apoD
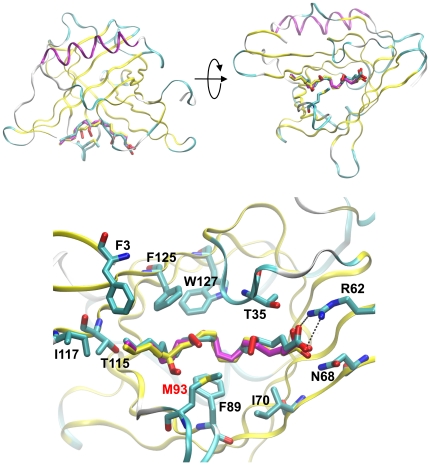
The absence of appropriate parameters for lipid hydroperoxides in the CHARMM force-field currently precludes simulation of this class of molecule with apoD. Nevertheless, in order to understand how L-OOHs might bind to apoD, the peroxidized arachidonate products 5s-, 12s-, and 15s-HpETE were modeled based on the most stable low-energy conformation observed for arachidonate. The models are shown overlaid in Figure 6. These models suggest that the L-OOH molecules wrap around Met93 thus providing a plausible mechanism by which eicosatetraenoic acids bearing peroxides on different carbon atoms can interact with Met93 and give rise to Met93SO.
Figure 6. Models of apoD interaction with 5-, 12- and 15-HpETE.
ApoD with 5- (cyan carbon atoms) 12- (magenta carbon atoms) and 15-HpETE (yellow carbon atoms) modeled in the binding site. Met93 is shown. The expanded view illustrates the close proximity of the Met93 side chain to the HpETE peroxide moiety. The salt bridges between Arg62 and the fatty acid are also indicted in the expanded view as dashed lines.
MD simulations of glycosylated apoD – Met49 is shielded
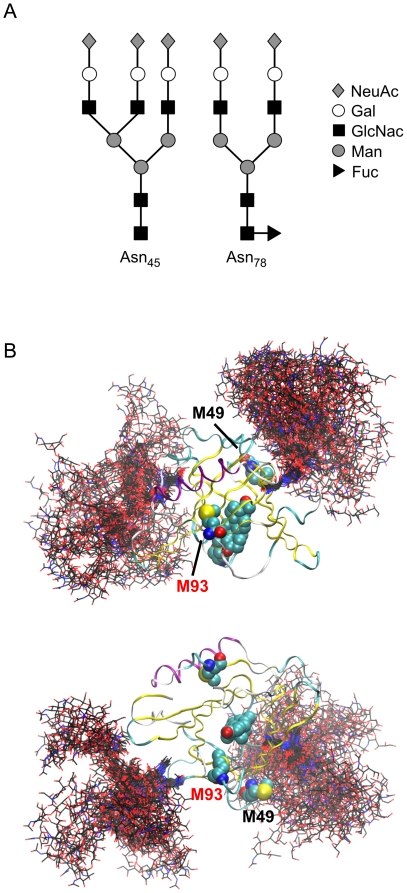
Data derived from our previous studies [15] and the simulations above clearly indicate that HpETEs can interact with Met93. The side-chain of Met157 is buried and a lack of interaction between this residue and L-OOHs is therefore expected. However, Met49 is relatively exposed in the crystal structure (Figure 2A) and was observed to interact with solvent in the simulations described above. It was therefore unclear why the interaction of HpETEs with apoD do not generate Met49SO. Given the location of N-glycan structures at Asn45 and Asn78 [25], we hypothesized that the N-glycan moiety at Asn45 might shield Met49 and prevent L-OOH mediated Met oxidation. We therefore ran an extended MD simulation of glycosylated apoD. The N-glycan structures present at Asn45 and Asn78 have been previously characterized and found to be mostly represented by trisialo triantennary and fucosylated disialo biantennary oligosaccharides, respectively [25], as graphically represented in Figure 7A. The conformations of the N-glycan chains are represented in Figure 7B. Clearly, access to Met49 is partially blocked by the trisialo triantennary N-glycan at Asn45 whereas neither of the N-glycans obstruct access to the apoD ligand binding pocket (Figure 7B). This provides a plausible explanation for the previously reported lack of reactivity between the apoD Met49 side chain and various HpETEs [15].
Figure 7. MD simulation of glycosylated apoD.
(A) Schematic representation of glycosylation patten found at Asn45 and Asn78. Symbols are: sialic acid (diamonds), mannose (filled circles), galactose (open circles), N-acetylglucosamine (squares) and fucose (triangle). (B) Orthogonal views of apoD in ribbon form with progesterone and methionine residues are represented as van der Waals spheres. The conformation of the glycosylations, sampled at 1 ns intervals, are shown (black carbon atoms).
CD analysis indicates MetSO formation does not induce major structural changes
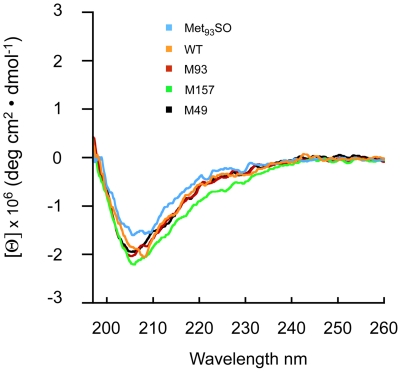
The above simulations give a plausible explanation for the selective oxidation of Met93 by L-OOHs. Based on this information and on our previous observations that the formation of Met93SO promotes apoD self-association and aggregation [15], we went on to investigate the possible structural changes that may be induced by apoD Met93SO formation. The CD spectrum of recombinant wild type apoD and all three Met to Ala mutants shows a negative minima in ellipticity at 208 nm (Figure 8) and overall is in very close agreement with previous studies [26]. Furthermore, the CD spectrum of HpETE-treated apoD (previously shown to convert apoD Met93 to Met93SO, [15]) was very similar to both the wild type apoD and the Met to Ala mutants (Figure 8). This indicates that neither mutation of apoD Met residues to Ala nor conversion of Met93 to Met93SO induce major changes to apoD secondary structure and, in particular, that the β-barrel structure remains in tact.
Figure 8. CD spectra of recombinant apoD.
Far-UV CD spectra of apoD and apoD-Met93SO and the Met to Ala mutants indicated. Spectra were obtained in PBS, pH 7.4 at 22°C.
MD simulaltion of apoD and apoD-Met93SO shows increased mobility in response to Met93 oxidation
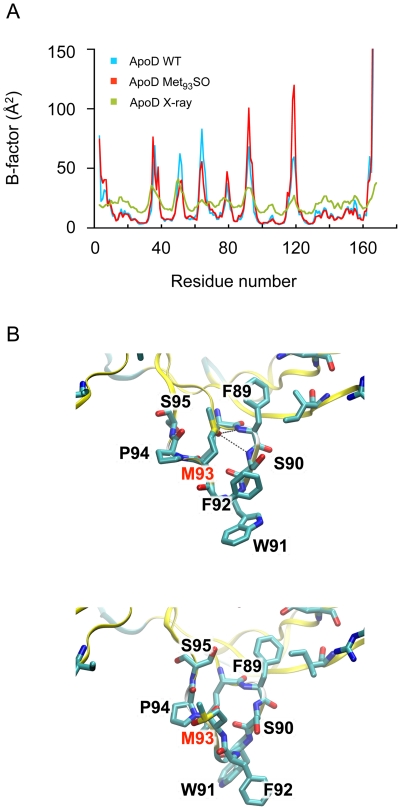
Because no major changes in apoD secondary structure were detected by CD spectroscopy, we next used MD simulations to probe for structural changes that may be induced by apoD Met93SO formation. ApoD and apoD-Met93SO were simulated for 50 ns. The proteins were stable over that timeframe. In both simulations the RMSD of Cα atoms initially increased and stabilized at around 2–2.5 Å (data not shown). Calculated B-factors from the simulations matched the pattern of B-factors in the X-ray structure (PDB: 2HZQ) (Figure 9A). The B-factors of the loop containing Met93SO were higher in the simulation of the oxidized protein, indicating increased flexibility. In the apoD simulation, Met93 remained associated with the side-chains of Phe89 and Phe92 via hydrophobic interactions. In the apoD-Met93SO simulation, the side-chain displayed more flexibility. Initially, the side-chain sulfoxide oxygen atom accepted hydrogen bonds from the main-chain amino groups of Phe89 and Ser90. After 7 ns, the Met93SO side-chain flipped out of the pocket and transiently associated with the side-chains of Phe89 and Phe92 in a similar fashion to native Met93. For the last 30 ns of the simulation, the side-chain of the oxidized residue was observed to be surrounded by water molecules and not directly interacting with other residues. A representative snapshot of this flipped-out conformation is shown in Figure 9B. The increase in the mobility of the Met93SO side chain that we have identified increases the thermal motion of the loop in which it is incorporated and this is predicted to destabilize apoD structure.
Figure 9. MD simulation of apoD and apoD-Met93SO.
(A) B-factors calculated from the MD simulations of apoD, apoD-Met93SO and crystallographic B-factors are shown. (B) Snapshots from the apoD-Met93SO simulation. Initially the sulfoxide oxygen of Met93SO remains in the initially modelled conformation, similar to Met93 in the ligand-free crystal structure. Simulation of apoD-Met93SO suggested that the sulfoxide oxygen of Met93SO could accept hydrogen bonds from the amide-nitrogen groups of Phe89 and Ser90. (upper structure). After 7 ns, the side-chain flips out of the pocket and is surrounded by water molecules. A representative snapshot of the populations of side-chain conformations is shown (lower structure).
Discussion
The results of the simulation of apoD with progesterone give us confidence that the CHARMM force-field is sufficiently accurate to investigate the interactions of this protein with representative ligands. The simulations of apoD with arachidonate indicate that this ligand has considerable flexibility. This is consistent with the structural plasticity of the fatty acid and the broad, open nature of the apoD binding site, which is ∼15 Å deep and 10 Å×15 Å wide at the entrance. It is noteworthy that the low-energy conformations of arachidonate used for the modeling of L-OOHs is similar to the mode of binding proposed by Eichinger and co-workers [1]. Although the simulations of apoD with arachidonate totaled 800 ns, it may be argued that this timeframe is insufficient for the system to converge on a global minimum, and that the optimum binding pose found in nature was not sampled in our simulations. However, even if such a state exists, fatty acids would be expected to explore a large number of conformations (such as those presented here) prior to converging on a hypothetical low-energy state. Consistent with our experimental results [15], our modeling indicate that the oxygenated fatty acid products of 5-, 12- and 15-lipoxygenase bind at the entrance of the ligand binding pocket of apoD in configurations that would permit direct interaction between the lipid hydroperoxide moiety and the crucial Met93 side chain. This interaction occurs as these highly flexible compounds can slide within the binding-site with many conformations of similar energy available.
Our data suggest that the solvent-exposed Met93, located in the hydrophobic region at the opening of the apoD ligand-binding pocket may also interact with lipid membrane surfaces from which phospholipid-bound fatty acid hydroperoxides protrude due to the addition of the polar peroxide group [27]. The side-chain of Met157 is buried and therefore not expected to be solvent exposed to lipid hydroperoxides. However, Met49 is solvent exposed and therefore has potential to interact with L-OOHs. The simulation of apoD with physiologically relevant glycosylation suggests that the ∼5 kDa N-linked oligosaccharide at Asn45 could have a shielding effect on Met49. From our simulation data, this oligosaccharide is also expected to sterically preclude apoD from approaching a membrane in an orientation favouring interaction with Met49. Glycosylation may therefore be one of the mechanisms by which aberrant oxidation of apoD Met49 is prevented.
It is noteworthy that apoD Met93 is highly conserved in mammals (Figure 1) and we speculate that this may afford an evolutionary advantage in terms of providing a lipid antioxidant function in the brain and possibly a protective function for the apoD ligand-binding pocket itself. Relevant to this latter point, the high propensity for redox-active Met residues to be located in close proximity to protein ligand binding sites/functional domains has previously been suggested to confer localized “guardian” antioxidant protection [28].
The mechanism underlying the increase in apoD aggregation that is associated with Met-SO formation [15] is not entirely clear. Previous research indicates that even though the introduction of an oxygen atom in the Met side chain would be expected to decrease the hydrophobicity of the protein (consistent with the decrease in reversed phase HPLC RT we have reported previously [15]), this modification can induce structural changes in the protein that increase the exposure of hydrophobic residues [29], [30]. This prediction is consistent with our simulation of apoD-Met93SO that suggested the sulfoxide oxygen could initially accept hydrogen bonds from the amide-nitrogen groups of Phe89 and Ser90 and then flip out of the entrance to the binding pocket to transiently associate with the side-chains of Phe89 and Phe92 and then adopt a conformation surrounded by water molecules without interacting with other residues. Based on previous studies [31], this may promote local unfolding of the protein structure and increase the propensity for apoD to self-associate and aggregate.
Previous studies have reported that the destabilization of protein structure as a consequence of MetSO formation can have profound functional consequences. The oxidation of p53 Met340 residue located within the hydrophobic core of the p53 tetramer significantly reduces p53 stability and is thought to represent one inactivation mechanism for p53 transcriptional function [32]. Interestingly, MD simulations assessing the oxidation of Met213 within Helix-3 of the cellular prion protein (PrPC) indicated a destabilization of the PrPC-like fold that favors more flexible states that are prone to conformation transition into the pathogenic infectious prion (PrPSc) form [33]. Furthermore, mutations of the PrPC that are associated with familial prion disorders increase the solvent exposure of Met213 and thus increase the probability for MetSO formation [34]. More directly related to the current study, previous research has shown that plasma apolipoprotein A-I (apoA-I) also has the capacity to promote the conversion of L-OOHs to L-OHs via a 2-electron redox reaction that concomitantly converts apoA-I Met residues to their respective MetSO moieties [35], [36]. Recent studies have shown that oxidation of apoA-I Met residues caused a partial unfolding of the protein, decreased its thermal stability and promoted its fibrillization [37]. It therefore appears that protein destabilization induced by selective Met oxidation may be common to several pathogenic processes.
In summary, our MD simulations provide a plausible mechanism that helps to explain how different forms of HpETEs can selectively interact with apoD Met93 and also reveal a potential basis for the destabilization of and self-association of apoD structure that results from Met93SO formation.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Australian National Health and Medical Research Council (ID# 1003886), the Australian Research Council Future Fellowships (BG: ID# FT0991986, AJO: ID# FT0990287, HE: ID# FT110100586) and by an award under the Merit Allocation Scheme on the NCI National Facility at the Australian National University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Eichinger A, Nasreen A, Kim HJ, Skerra A. Structural insight into the dual ligand specificity and mode of high density lipoprotein association of apolipoprotein D. J Biol Chem. 2007;282:31068–31075. doi: 10.1074/jbc.M703552200. [DOI] [PubMed] [Google Scholar]
- 2.Vogt M, Skerra A. Bacterially produced apolipoprotein D binds progesterone and arachidonic acid, but not bilirubin or E-3M2H. J Mol Recognit. 2001;14:79–86. doi: 10.1002/1099-1352(200101/02)14:1<79::AID-JMR521>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Lea OA. Binding properties of progesterone-binding Cyst protein, PBCP. Steroids. 1988;52:337–338. doi: 10.1016/0039-128x(88)90135-3. [DOI] [PubMed] [Google Scholar]
- 4.Dilley WG, Haagensen DE, Cox CE, Wells SA., Jr Immunologic and steroid binding properties of the GCDFP-24 protein isolated from human breast gross cystic disease fluid. Breast Cancer Res Treat. 1990;16:253–260. doi: 10.1007/BF01806333. [DOI] [PubMed] [Google Scholar]
- 5.Morais Cabral JH, Atkins GL, Sanchez LM, Lopez-Boado YS, Lopez-Otin C, et al. Arachidonic acid binds to apolipoprotein D: implications for the protein's function. FEBS Lett. 1995;366:53–56. doi: 10.1016/0014-5793(95)00484-q. [DOI] [PubMed] [Google Scholar]
- 6.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rassart E, Bedirian A, Do Carmo S, Guinard O, Sirois J, et al. Apolipoprotein D. Biochim Biophys Acta. 2000;1482:185–198. doi: 10.1016/s0167-4838(00)00162-x. [DOI] [PubMed] [Google Scholar]
- 8.Ganfornina MD, Do Carmo S, Lora JM, Torres-Schumann S, Vogel M, et al. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell. 2008;7:506–515. doi: 10.1111/j.1474-9726.2008.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muffat J, Walker DW, Benzer S. Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proc Natl Acad Sci U S A. 2008;105:7088–7093. doi: 10.1073/pnas.0800896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu T, Pan Y, Kao SY, Li C, Kohane I, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 11.Kim WS, Wong J, Weickert CS, Webster MJ, Bahn S, et al. Apolipoprotein-D expression is increased during development and maturation of the human prefrontal cortex. J Neurochem. 2009;109:1053–1066. doi: 10.1111/j.1471-4159.2009.06031.x. [DOI] [PubMed] [Google Scholar]
- 12.Terrisse L, Poirier J, Bertrand P, Merched A, Visvikis S, et al. Increased levels of apolipoprotein D in cerebrospinal fluid and hippocampus of Alzheimer's patients. J Neurochem. 1998;71:1643–1650. doi: 10.1046/j.1471-4159.1998.71041643.x. [DOI] [PubMed] [Google Scholar]
- 13.Butterfield DA, Bader Lange ML, Sultana R. Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer's disease. Biochim Biophys Acta. 2010;1801:924–929. doi: 10.1016/j.bbalip.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratico D. The neurobiology of isoprostanes and Alzheimer's disease. Biochim Biophys Acta. 2010;1801:930–933. doi: 10.1016/j.bbalip.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Bhatia S, Knoch B, Wong J, Kim WS, Else PL, et al. Selective reduction of hydroperoxyeicosatetraenoic acids to their hydroxy derivatives by apolipoprotein-D: Implications for lipid antioxidant activity and Alzheimer's disease. Biochem J. 2012;442:713–721. doi: 10.1042/BJ20111166. [DOI] [PubMed] [Google Scholar]
- 16.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 18.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of Computational Chemistry. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 19.Strader ML, Feller SE. A flexible all-atom model of dimethyl sulfoxide for molecular dynamics simulations. Journal of Physical Chemistry A. 2002;106:1074–1080. [Google Scholar]
- 20.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guvench O, Mallajosyula SS, Raman EP, Hatcher E, Vanommeslaeghe K, et al. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate-Protein Modeling. Journal of Chemical Theory and Computation. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 23.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 24.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al. Gaussian 09. Wallingford, CT: Gaussian, Inc; 2009. [Google Scholar]
- 25.Schindler PA, Settineri CA, Collet X, Fielding CJ, Burlingame AL. Site-specific detection and structural characterization of the glycosylation of human plasma proteins lecithin:cholesterol acyltransferase and apolipoprotein D using HPLC/electrospray mass spectrometry and sequential glycosidase digestion. Protein Sci. 1995;4:791–803. doi: 10.1002/pro.5560040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasreen A, Vogt M, Kim HJ, Eichinger A, Skerra A. Solubility engineering and crystallization of human apolipoprotein D. Protein Sci. 2006;15:190–199. doi: 10.1110/ps.051775606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg ME, Li XM, Gugiu BG, Gu X, Qin J, et al. The lipid whisker model of the structure of oxidized cell membranes. J Biol Chem. 2008;283:2385–2396. doi: 10.1074/jbc.M707348200. [DOI] [PubMed] [Google Scholar]
- 28.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 30.Chao CC, Ma YS, Stadtman ER. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc Natl Acad Sci U S A. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Nomura T, Kamada R, Ito I, Chuman Y, Shimohigashi Y, et al. Oxidation of methionine residue at hydrophobic core destabilizes p53 tetrameric structure. Biopolymers. 2009;91:78–84. doi: 10.1002/bip.21084. [DOI] [PubMed] [Google Scholar]
- 33.Colombo G, Meli M, Morra G, Gabizon R, Gasset M. Methionine sulfoxides on prion protein Helix-3 switch on the alpha-fold destabilization required for conversion. PLoS One. 2009;4:e4296. doi: 10.1371/journal.pone.0004296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meli M, Gasset M, Colombo G. Dynamic diagnosis of familial prion diseases supports the beta2-alpha2 loop as a universal interference target. PLoS One. 2011;6:e19093. doi: 10.1371/journal.pone.0019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garner B, Waldeck AR, Witting PK, Rye KA, Stocker R. Oxidation of high density lipoproteins II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J Biol Chem. 1998;273:6088–6095. doi: 10.1074/jbc.273.11.6088. [DOI] [PubMed] [Google Scholar]
- 36.Garner B, Witting PK, Waldeck AR, Christison JK, Raftery M, et al. Oxidation of high density lipoproteins I. Formation of methionine sulfoxide in apolipoproteins AI and AII is an early event that accompanies lipid peroxidation and can be enhanced by alpha-tocopherol. J Biol Chem. 1998;273:6080–6087. doi: 10.1074/jbc.273.11.6080. [DOI] [PubMed] [Google Scholar]
- 37.Wong YQ, Binger KJ, Howlett GJ, Griffin MD. Methionine oxidation induces amyloid fibril formation by full-length apolipoprotein A-I. Proc Natl Acad Sci U S A. 2010;107:1977–1982. doi: 10.1073/pnas.0910136107. [DOI] [PMC free article] [PubMed] [Google Scholar]