Abstract
In diploid eukaryotes, repair of double-stranded DNA breaks by homologous recombination often leads to loss of heterozygosity (LOH). Most previous studies of mitotic recombination in Saccharomyces cerevisiae have focused on a single chromosome or a single region of one chromosome at which LOH events can be selected. In this study, we used two techniques (single-nucleotide polymorphism microarrays and high-throughput DNA sequencing) to examine genome-wide LOH in a diploid yeast strain at a resolution averaging 1 kb. We examined both selected LOH events on chromosome V and unselected events throughout the genome in untreated cells and in cells treated with either γ-radiation or ultraviolet (UV) radiation. Our analysis shows the following: (1) spontaneous and damage-induced mitotic gene conversion tracts are more than three times larger than meiotic conversion tracts, and conversion tracts associated with crossovers are usually longer and more complex than those unassociated with crossovers; (2) most of the crossovers and conversions reflect the repair of two sister chromatids broken at the same position; and (3) both UV and γ-radiation efficiently induce LOH at doses of radiation that cause no significant loss of viability. Using high-throughput DNA sequencing, we also detected new mutations induced by γ-rays and UV. To our knowledge, our study represents the first high-resolution genome-wide analysis of DNA damage-induced LOH events performed in any eukaryote.
ALL organisms experience DNA damage from both exogenous and endogenous sources. Endogenous DNA damage includes spontaneous deamination of nucleotides, depurination/depyrimidination, oxidative damage, and double-stranded DNA breaks (DSBs) (Friedberg et al. 2006). DSBs are likely to be particularly deleterious, since unrepaired DSBs can lead to chromosome rearrangements or chromosome loss. Although the sources of endogenous DSBs have not been completely determined, some DSBs appear to reflect nuclease processing of secondary DNA structures (such as DNA “hairpins”) or head-on collisions between the replication and transcription machineries (Aguilera 2002). Below, in addition to examining spontaneous recombination events that presumably reflect the repair of endogenous DNA damage, we also analyze recombination events induced by two exogenous sources: γ-rays and ultraviolet (UV) radiation.
Both γ-rays and UV cause a variety of different types of DNA damage. γ-rays cause DSBs, single-stranded DNA nicks, and base damage (Ward 1990; Friedberg et al. 2006). UV results in pyrimidine dimers (Setlow 1966; Franklin et al. 1985), DNA–DNA or DNA–protein crosslinks (Peak and Peak 1986), and single-stranded DNA nicks resulting from the dimer excision (Breen and Murphy 1995).
In yeast, as in most eukaryotes, there are two recombination pathways that are used to repair DSBs: nonhomologous end-joining (NHEJ) and homologous recombination (HR). In NHEJ events, as the name implies, broken DNA molecules are rejoined by a mechanism that requires little or no homology (Daley et al. 2005). This mechanism is most active in haploid yeast cells during G1 of the cell cycle (Shrivastav et al. 2008). In diploid cells, the dominant pathway is HR. HR uses an intact homologous DNA molecule, either the sister chromatid or the homologous chromosome, as a template for repair of the broken chromosome.
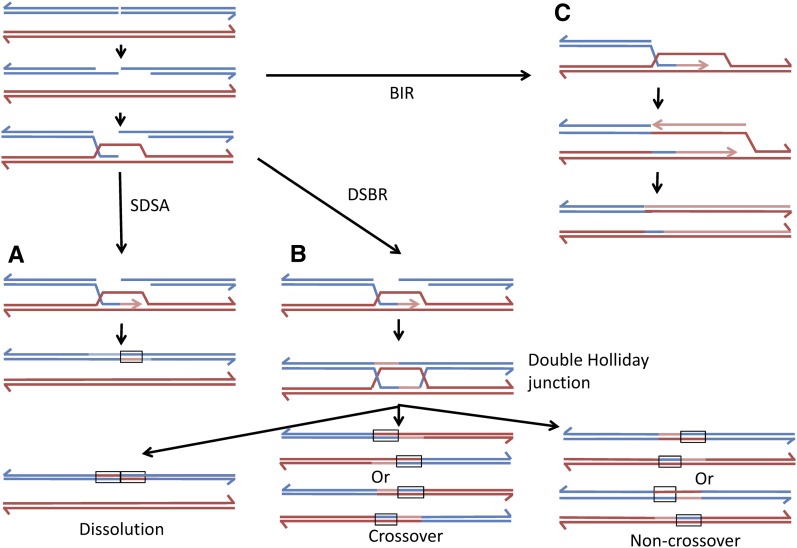
DSBs can be repaired by several different HR pathways (Heyer et al. 2010). The repair of a DSB by gene conversion unassociated with a crossover is shown in Figure 1A. This process involves the nonreciprocal transfer of sequences from the intact donor molecule to the broken chromosome in several steps: (1) invasion of one broken end into the intact template molecule, followed by DNA synthesis primed by the invading 3′ strand; (2) removal of the invading end and reannealing of this end back to the other broken end, forming a heteroduplex with mismatches; and (3) repair of the mismatches. This mechanism [synthesis-dependent strand-annealing (SDSA)] was first suggested to explain some features of meiotic recombination in yeast (Allers and Lichten 2001). In the second pathway (Figure 1B), gene conversion may be associated with crossovers. In this pathway, a double Holliday junction (dHJ) is formed that can be resolved to yield a crossover or noncrossover. In this pathway, heteroduplexes flank the original position of the DSB. Although the heteroduplex regions have the same size in Figure 1B, in both meiosis (Merker et al. 2003; Jessop et al. 2005) and mitosis (Mitchel et al. 2010; Tang et al. 2011), the conversion tracts flanking the DSB are often of different lengths. The dHJ can also be dissolved without nucleolytic cleavage of DNA strands to yield noncrossover products with heteroduplexes located in cis on one of the two interacting chromosomes (Heyer et al. 2010). In the third pathway (Figure 1C), one part of the broken DNA molecule is lost and a complete chromosome is then reconstructed by break-induced replication (BIR). In this mechanism, one of the broken ends invades the intact template molecule, and a replication fork is set up that duplicates the template from the site of invasion to the telomere.
Figure 1.
Pathways of DSB repair by homologous recombination. In this figure, we show accepted models of DSB repair by homologous recombination. DNA strands from two different homologs are shown in red and blue; light red and blue lines indicate newly synthesized DNA. Regions of the duplex that have strands of different colors represent heteroduplexes. These pathways are described in detail in the text. (A) Synthesis-dependent strand annealing (SDSA) pathway. Following processing of the DSB, the 3′ end of the left end of the broken DNA molecule invades the other duplex. Following DNA synthesis, the invading strand is displaced and hybridizes to the right end of the broken chromosome. This pathway results in conversion events unassociated with crossovers. (B) Double-strand break repair (DSBR) pathway. In this pathway, a double Holliday junction (dHJ) is formed. Resolution of these junctions by resolvase cleavage can result in two different crossover products (middle) and two different noncrossover products (bottom right). These products have two regions of heteroduplex located in trans. Alternatively, the dHJ can be dissolved by the action of topoisomerases/helicases, resulting in a noncrossover product with heteroduplexes located in cis. (C) Break-induced replication (BIR) pathway. One of the broken ends invades the homologous chromosome and duplicates sequences from the point of invasion to the telomere. The net result of BIR events is an apparent long terminal gene conversion event.
If HR involves an interaction between two homologs that can be distinguished by single-nucleotide polymorphisms (SNPs), conversions without crossovers will produce a small patch of loss of heterozygosity (LOH) within a chromosome that is otherwise heterozygous, whereas both crossovers and BIR result in LOH that extends from the site at which the event initiates to the end of the chromosome. Repair of a DSB by HR in which the homologous chromosome is used as a template will result in LOH, but repair events involving the sister chromatid will not. Although most sister-chromatid recombination events are genetically silent, unequal sister-chromatid exchanges can be detected in yeast by a variety of different systems (Petes and Hill 1988). Using one of these systems, Kadyk and Hartwell (1992) showed that, in diploid cells, sister chromatids are the preferred substrate for the repair of DSBs generated by X-rays. Despite this preference, it is clear that ionizing radiation and UV strongly stimulate HR events (both mitotic crossovers and gene conversions) between homologous chromosomes (Nakai and Mortimer 1969; Fabre 1978).
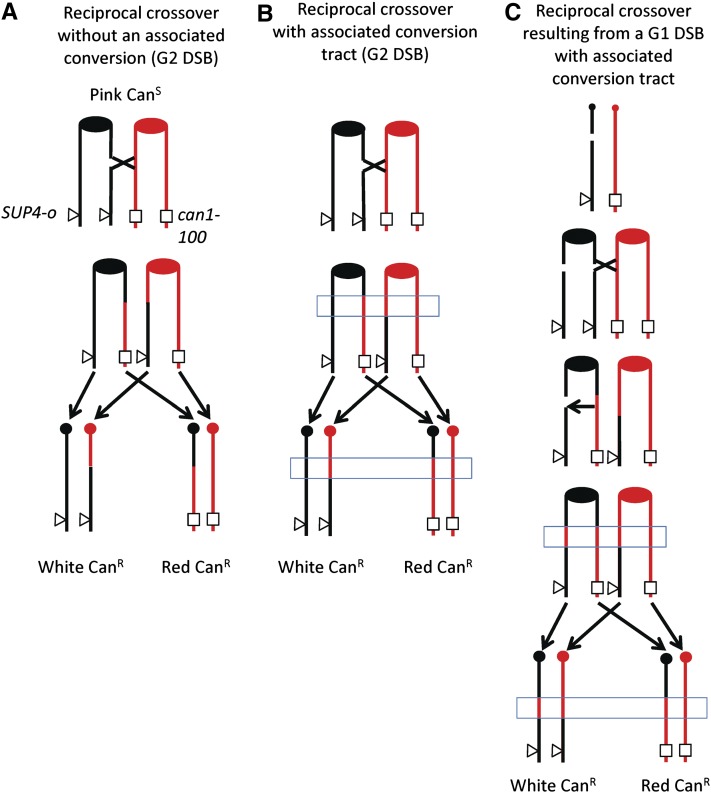
One problem with studying spontaneous mitotic recombination is that most analytic systems do not allow the selection of both daughter cells that contain the recombinant chromosomes. Several years ago, we developed a method of selecting reciprocal crossovers on chromosome V that surmounts this difficulty (Barbera and Petes 2006; Lee et al. 2009). One copy of chromosome V has the can1-100 allele (an ochre mutation) and, in the other copy, the CAN1 gene is replaced by SUP4-o, a gene encoding an ochre-suppressing transfer RNA (tRNA) (Figure 2A). In the absence of the suppressor, strains with the can1-100 allele are resistant to canavanine, but because of the suppressor, the diploid used in our experiments is canavanine-sensitive. In addition, the diploid is homozygous for the ade2-1 mutation (an ochre mutation). Strains with this mutation, in the absence of the SUP4-o gene, form red colonies, but form pink colonies if one copy of the SUP4-o gene is present. Thus, the diploid strain is canavanine-sensitive and forms pink colonies. If a crossover occurs between the centromere of chromosome V and the can1-100/SUP4-o markers (a distance of ∼120 kb), a canavanine-resistant red/white colony is formed (Figure 2A).
Figure 2.
Genetic system used to select mitotic crossovers and associated conversions on the left arm of chromosome V. The starting diploid strain PG311 has the ochre-suppressible can1-100 on one copy of chromosome V (shown in red) and the SUP4-o gene (encoding an ochre suppressor tRNA gene) at an allelic position on the other homolog (shown in black). The strain is homozygous for the ochre-suppressible ade2-1 allele. Strains with an unsuppressed ade2-1 mutation form red colonies. The starting diploid strain is canavanine-sensitive and forms pink colonies. (A) Reciprocal crossover without an associated gene conversion initiated by a single DSB in G2. This type of event produces a canavanine-resistant red/white sectored colony (Barbera and Petes 2006). The transition from heterozygous markers to LOH is identical in the two sectors. (B) Reciprocal crossover with an associated conversion event initiated by a single DSB in G2. If a DSB forms on one of the black chromatids, a conversion associated with the crossover may occur. This event will also result in a canavanine-resistant red/white sectored colony in which the transitions between heterozygous markers and LOH are different in the two sectors. The region of conversion is indicated by the blue rectangle. (C) Reciprocal crossover and conversion resulting from a DSB formed in G1. A black chromosome with a DSB is replicated to form two sister chromatids that are broken at the same place. One chromatid is repaired to yield a reciprocal crossover and an associated conversion; the second is repaired to yield a conversion without a crossover. The resulting red and black sectors will have a 4:0 conversion event, a region in which both sectors are homozygous for SNPs derived from the red chromatid (included within the blue rectangle).
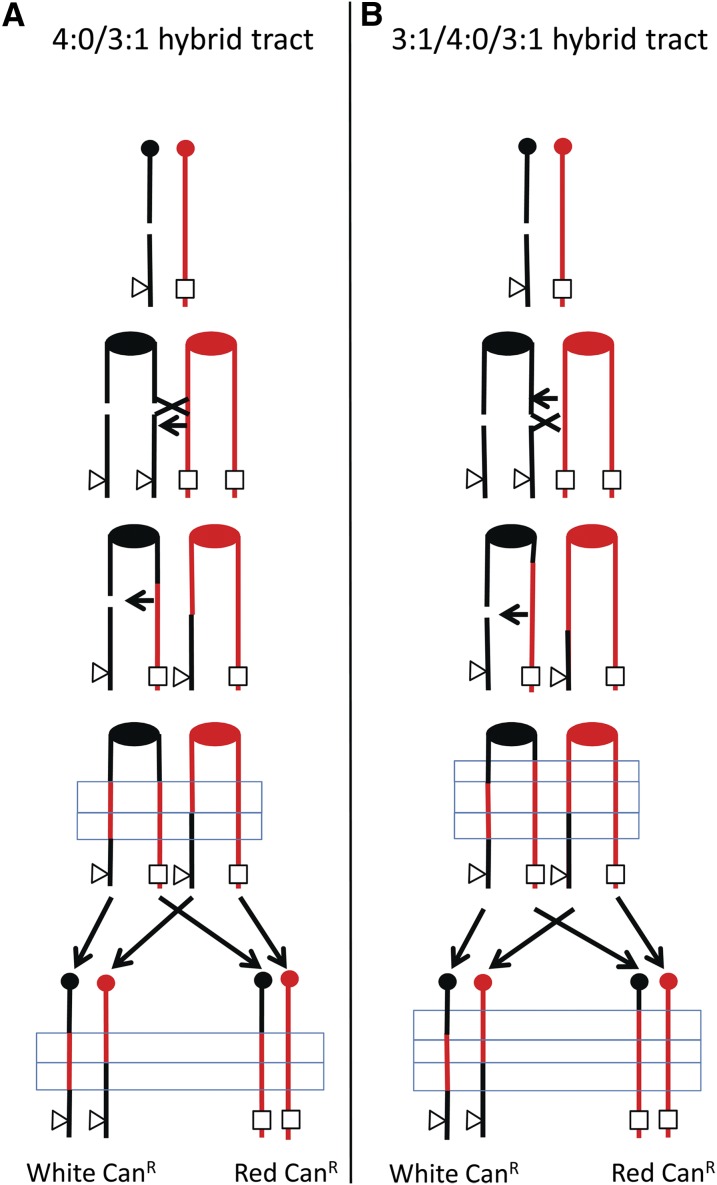
Although this method was first used in diploid strains lacking polymorphisms, subsequent studies were done in which a diploid was constructed using two haploid strains that had ∼0.5% sequence divergence (Lee et al. 2009; Lee and Petes 2010), resulting in ∼55,000 SNPs distributed throughout the genome. Crossovers and associated gene conversions on chromosome V were mapped to a resolution of ∼4 kb by using PCR, restriction digests, and gel electrophoresis to look for LOH. Although a few of the crossovers had no associated conversion, most of the crossovers were associated with an adjacent conversion event (boxed regions in Figure 2). In the 3:1 class of events (Figure 2B), in the boxed region, three of the four chromosomes had one of the two forms of the SNP, and one of the chromosomes had the other form (one sector being homozygous for a SNP with the other sector being heterozygous). In addition, ∼40% of the crossovers were associated with a conversion event in which the same SNP was homozygous in both sectors (Figure 2C); we term these events “4:0 conversions.” The observation of the 4:0 events argues that about half of mitotic crossovers result from the repair of two sister chromatids that are broken at approximately the same positions. One simple mechanism for generating this intermediate is to have the DSB occur in G1 and the broken chromosome replicate to form two broken chromatids (Lee et al. 2009). This proposed mechanism was confirmed by analysis of the types of conversion events stimulated by γ-rays in synchronized G1 and G2 cells (Lee and Petes 2010). In addition to 3:1 and 4:0 conversion events, 3:1/4:0 hybrid tracts are also observed (Lee et al. 2009; Lee and Petes 2010). Such tracts can also be explained as a consequence of the repair of two broken sister chromatids (Figure 3).
Figure 3.
Production of hybrid conversion tracts by repair of two broken sister chromatids. The black chromosome is broken in G1 and replicated to yield two broken sister chromatids. (A) Production of a 3:1/4:0 hybrid tract. If the two DSBs are repaired to yield conversion tracts that have the same centromere-proximal boundary, but different centromere-distal boundaries, a 3:1/4:0 hybrid will be generated (shown in the blue rectangle). (B) Production of a 3:1/4:0/3:1 hybrid tract. If one conversion event is extended beyond the other at both the centromere-proximal and centromere-distal boundaries, a 3:1/4:0/3:1 tract will be formed.
In this study, we use SNP microarrays and high-throughput DNA sequencing (HTS) to map selected events on chromosome V as well as unselected events throughout the genome at a resolution of ∼1 kb. To our knowledge, these studies are the first to measure the numbers and types of LOH events throughout the genome induced by doses of ionizing radiation (100 Gy) and UV (10–15 J/m2) that have no significant effect on cell viability. We also determined the number of mutations induced in the genomes by these treatments.
Materials and Methods
Strains and genetic methods
All experiments were conducted with the diploid strain PG311 (Lee et al. 2009). The relevant genotype of PG311 is MATα::NATR/MATa URA3/ura3-1 ade2-1/ade2-1 TRP1/trp1-1 HIS3/his3-11,15 GAL2/gal2SUP4-o/can1-100 V9229/V9229::HYG V261553/V261553::LEU2. This diploid was generated by crossing the haploid strains PSL2 and PSL5, which are isogenic with strains W303a and YJM789, respectively, except for alterations introduced by transformation (Lee et al. 2009). From this point on, we will refer to the haploid parents of PG311 as W303a and YJM789. In general, PG311 has the SNPs predicted from the haploid parents. The disruption of MATα in PG311 allows synchronization of this diploid by α-factor. Although diploids that lack MATα do not sporulate under normal conditions, such strains can be sporulated on plates containing 5 mM nicotinamide (J. Rine, personal communication). For experiments in which we analyzed meiotic products of PG311, the stains were pregrown on YPD plates with 5 mM of nicotinamide and then transferred to sporulation plates containing 5 mM nicotinamide. Plates were incubated at 25° for 2–4 days before tetrad dissection.
Standard media were used (Guthrie and Fink 1991) unless noted. To detect spontaneous crossovers, we first isolated single colonies of PG311 grown on rich growth medium (YPD) at 30° for 2 days. Individual colonies were suspended in 400 μl of dH2O, and 100 μl of this mixture was plated on canavanine-containing plates (SD-arg + 120 μg/ml canavanine). The plates were incubated 4 days at room temperature, followed by incubation for 16 hr at 4°; the 4° incubation allows better visualization of the red sectors. We purified cells from the red and white sectors for subsequent analysis.
In the experiments in which cells were irradiated, we synchronized cells in G1 using α-factor (Lee and Petes 2010). The synchronized cells were treated with either γ-radiation in a Shepherd Mark 1 137Cs irradiator at 100 Gy or with UV (254 nm) derived from a TL-2000 Ultraviolet Translinker at a dosage of 10 or 15 J/m2. Following radiation, the cells were plated either on nonselective plates (SD-arg) or plates that lacked arginine and contained 120 μg/ml canavanine. The subsequent growth of the cells and the analysis of sectors were done as described above for the spontaneous selection with the exception that sectored colonies for the UV-treated samples were isolated from nonselective plates grown at 30° instead of room temperature.
SNP microarrays: design and optimization
We designed the SNP arrays on the basis of genomic sequence information available from the Saccharomyces Genome Database (SGD) for S288c (very closely related to W303a) (Winzeler et al. 2003) and YJM789 (Wei et al. 2007). Microarrays that were capable of detecting LOH for SNPs in PG311 were designed on the basis of principles outlined by Gresham et al. (2010). For each SNP represented on the array, four 25-base oligonucleotides were used: one for each strand of the W303a SNP and one for each strand of the YJM789 SNP. The SNP was located in the middle of the 25-base oligonucleotide. Although there are ∼55,000 SNPs in PG311, about three-quarters of these SNPs were not used for our analysis. We excluded most of the SNPs found in repeated genes. We also screened out oligonucleotides that had a melting temperature for the perfectly matched 25-bp duplex that was <55° or >59°. The remaining oligonucleotides (representing ∼15,000 SNPs) that were printed on the microarray are listed in Supporting Information, Table S3. We also included on the array ∼120 oligonucleotides that were not different from W303a and YJM789; these are listed in Table S4. Oligonucleotides were printed onto the microarrays by Agilent Technologies in slides containing ∼105,000 oligonucleotides. Many of the oligonucleotides are represented more than once in the microarrays. Following experiments to determine which oligonucleotides resulted in the most specific hybridization signals (described in File S1), we reduced the number of SNPs used in our analysis to 13,000. This final set of oligonucleotides is presented in Table S5.
Methods used for microarray analysis: sample preparation, hybridization conditions, and data analysis
The methods used for sample preparation, hybridization conditions, and data analysis are similar to those described previously (Lemoine et al. 2005; McCulley and Petes 2010). A detailed description of these procedures is presented in File S1. In brief, genomic DNA from the experimental strain was labeled with Cy5-dUTP, DNA from the control strain (PG311) was labeled with Cy3-dUTP, and the two labeled samples were competitively hybridized to the microarrays. The arrays were then scanned at wavelengths of 635 and 532 nm using a GenePix scanner and GenePix Pro software using settings recommended by the manufacturer. The ratio of the medians (635 nm/532 nm; RM) for each probe was used for analysis, and replicate RM’s were averaged. The data were centered around a value of one by dividing each probe RM by the average of all of the probe RM’s to normalize for differences in the hybridization levels for the control and experimental strain samples.
We calculated 95% confidence intervals on the median sizes of conversion tracts using table B11 of Altman (1990). Comparisons of conversion tract lengths under different experimental conditions were done using the Mann–Whitney test on the VassarStats website (http://faculty.vassar.edu/lowry/VassarStats.html).
Generation and analysis of HTS data
Samples were prepared for HTS as described above for the SNP microarray sample preparation with the exception that genomic DNA was sonicated to 300- to 700-bp fragments. The DNA was then prepared for sequencing using the protocol recommended by Illumina for the Genome Analyzer IIx. The samples were sequenced using an Illumina GAIIx machine, generating 67- to 75-bp paired-end reads. For the eight sequenced samples, coverage varied from 90- to 180-fold.
The details of the HTS data analysis are presented in File S1. In brief, we detected regions of LOH by identifying SNPs in the experimental strains in which at least 90% of the “reads” that were originally heterozygous were now identical to one of the original alleles. We identified new mutations by finding bases that were identical in the original diploid, but had a novel base in at least 40% of the “reads” in the irradiated diploid; we use the 40% criterion because we expect that any new mutation will be heterozygous. Mutations that appeared in more than one independent isolate were not counted as de novo mutations, since such mutations presumably arose in the strain before treatment with the DNA-damaging agent.
Results
As described in the Introduction, we previously selected spontaneous mitotic crossovers, as well as crossovers induced by γ-rays, that occurred between the centromere of chromosome V and the can1-100/SUP4-o markers, an interval of ∼120 kb (Figure 2) (Lee et al. 2009; Lee and Petes 2010). The diploid used in these studies (PG311) was constructed by a cross between two haploids that are isogenic with W303a and YJM789 and is heterozygous for ∼55,000 SNPs. In our previous analysis, the positions of the crossovers and associated gene conversion events were mapped to a resolution of ∼4 kb using a PCR-based strategy that determined whether the SNPs were heterozygous or homozygous. This procedure is impractical for genome-wide mapping of recombination events. Below, we describe the use of SNP arrays to map spontaneous, UV-induced, and γ-ray-induced crossovers selected on chromosome V as well as unselected crossovers and gene conversion events throughout the genome.
SNP arrays have been used previously to map LOH events in tumor cells (Lindblad-Toh et al. 2000), to map meiotic recombination events in Saccharomyces cerevisiae (Mancera et al. 2008), to characterize chromosome rearrangements and chromosome loss in Candida albicans (Abbey et al. 2011), and in a variety of other experiments. Using principles outlined by Gresham et al. (2010), we developed a SNP array to examine LOH events throughout the genome in the diploid PG311. This array has oligonucleotides that distinguish >13,000 SNPs, resulting in an average density of one oligonucleotide per kilobase of genomic DNA. The details of the array design and the specific sequences of the probes are in File S1 and in Table S3, Table S4, and Table S5.
In our experiments, we labeled genomic DNA from a diploid with a recombination event with one fluorescent nucleotide and DNA from the control diploid PG311 with a different fluorescent nucleotide. The samples were mixed and competitively hybridized to the SNP arrays. If SNPs retained heterozygosity in the experimental strain, then the hybridization signal for the oligonucleotides representing that SNP was similar to that of the control strain. LOH was detected by an increase in hybridization to oligonucleotides that had one form of the SNP (e.g., the W303a form) and a decrease in hybridization to oligonucleotides that had the other form (e.g., the YJM789 form).
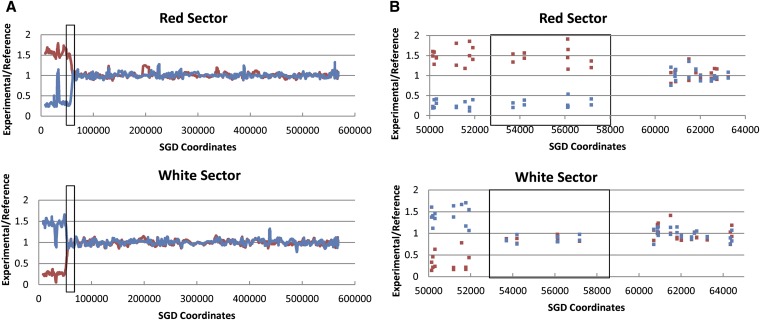
As described above, crossover events between CEN5 and the can1-100/SUP4-o markers produce canavanine-resistant red/white sectored colonies. Thus, all of the samples analyzed had a selected recombination event on chromosome V, and we found that samples treated with UV or γ-rays also had unselected events on other chromosomes. We isolated genomic DNA from both the red and the white sides of the sectors and examined the DNA by SNP arrays for diploids untreated with recombinogenic agents, as well as for diploids treated with UV or γ-rays. An example of the analysis for chromosome V for a selected spontaneous crossover (PG311-2A) is shown on Figure 4. In Figure 4A, a low-resolution depiction of the hybridization levels is shown for both the red (top) and the white (bottom) sectors. In both sectors, the hybridization pattern indicates that the transition from heterozygous SNPs to homozygous SNPs is at approximately SGD coordinate 55000. As expected, the DNA that is centromere-distal to the crossover junction from the red sector hybridizes well to the W303a-specific probes (red line) and poorly to the YJM789-specific probes (blue line), since the red sector is generated by LOH events that include the can1-100 marker that is derived from the W303a-related homolog (Figure 2). Genomic DNA from the white sector shows the reciprocal pattern of hybridization. The “spike” of increased hybridization in the red sector for YJM789 SNPs near SGD coordinate 30000 is an artifact resulting from a deletion of YJM789 sequences during the insertion of the SUP4-o gene into the YJM789-derived chromosome.
Figure 4.
Analysis of a spontaneous reciprocal crossover (PG311-2A) on the left arm of chromosome V by SNP microarrays. Most of the details concerning this figure are explained in the text. In brief, DNA samples isolated from the red and white sectors were labeled with one fluorescent nucleotide and DNA from a heterozygous control strain was labeled with a different fluorescent nucleotide. The samples were competitively hybridized to the SNP array, and we measured the ratio of hybridization of the probes to SNP-specific oligonucleotides. The red and blue colors indicate hybridization to the W303a- and YJM789-specific oligonucleotides, respectively. CEN5 is located approximately at SGD coordinate 152000. (A) Low-resolution depiction of the samples derived from the red and white sectors. In the boxed region, the red sector has a region of LOH whereas the white sector is heterozygous at the same position. This pattern indicates a 3:1 conversion associated with the crossover. Centromere-distal to the conversion event, the red and white sectors are homozygous for the W303a- and YJM789-specific SNPs, respectively. (B) High-resolution depiction of the samples derived from the red and white sectors. Each blue and red square represents hybridization to a single oligonucleotide on the array; the converted region is boxed.
In Figure 4B, we show the same crossover event at higher resolution. Each square in Figure 4B shows the hybridization ratio to a specific oligonucleotide. In the red sector, the transition between the homozygous SNPs and the heterozygous SNPs is between SGD coordinates 57170 and 60701. In the white sector, the transition occurs between 51915 and 53692. Thus, there is a region (boxed in Figure 4B) in which one sector is homozygous for SNPs and the other is heterozygous. This region is a 3W:1Y gene conversion tract (W and Y indicating W303a-derived and YJM789-derived SNPs, respectively), equivalent to the boxed region in Figure 2B; in our subsequent discussions, a 3:1 conversion event indicates a 3W:1Y conversion and 1:3 indicates a 1W:3Y conversion. We estimate the length of the gene conversion tract by averaging the maximal length of the tract (the distance between markers that are not within the gene conversion tract: 8.8 kb) and the minimal length of the tract (the distance between the converted markers: 3.5 kb). For the tract shown in Figure 4B, this length is ∼6.2 kb. It is important to emphasize that the presence and extent of gene conversion tracts can be identified only when the patterns of LOH are analyzed in genomic DNA from both sectors of the sectored colony.
With the genome-wide SNP analysis, we found that the parental diploid PG311 and all of its subsequent derivatives had two LOH events that were unexpected from the sequence of the parental haploids. All strains were homozygous for W303a-derived SNPs centromere-distal to SGD coordinate 685 kb on chromosome XIII and were homozygous for W303a-derived SNPs between coordinates 412715 and 414085 on chromosome X. Since these events, presumably generated during subculturing of PG311, were present in all strains, they were excluded from our analysis.
Analysis of spontaneous selected LOH events on chromosome V by SNP arrays
We examined by SNP arrays genomic DNA from both the red and white sectors from fourteen independent canavanine-resistant colonies. The crossover events in five of the isolates had been mapped previously by PCR-amplifying regions along the left arm of chromosome V that contained polymorphic restriction enzyme sites and testing for homozygosity or heterozygosity by a restriction enzyme digest analysis (Lee et al. 2009). We also mapped an additional nine events solely by SNP arrays. In the previous study, we used 34 markers in the 120 kb CEN5-can1-100/SUP4-o interval. We monitored 172 markers in this same interval by SNP arrays.
In our previous study, we found that most of the spontaneous crossovers were crossovers without conversions (Figure 2A), crossovers with associated 3:1 conversions (Figure 2B), crossovers with 4:0 conversions (Figure 2C), or crossovers with hybrid conversions (3:1/4:0 or 3:1/4:0/3:1 tracts; Figure 3) (Lee et al. 2009). A comparison of the mapping of recombination events by the PCR-based method and SNP arrays for four of the sectored colonies is shown in Table 1. In Table 1, we define the position of the crossovers with only two SGD coordinates: the position of the centromere-proximal heterozygous marker that is closest to the crossover/conversion event and the position of the centromere-distal homozygous markers representing the crossover. Although the agreement between the two methods was reasonably good, as expected, the SNP array mapped events with better resolution and also revealed that some of the conversion events were more complex than previously determined. For example, in PG311-1.4, we previously mapped a crossover between SGD coordinates 125754 and 133080 that appeared to be unassociated with gene conversion. With the SNP arrays, we mapped the transition at higher resolution between SGD coordinates 127038 and 130096, and we detected a SNP in this region that had undergone gene conversion. The complete description of all spontaneous crossovers and associated gene conversion events is given in Table S6.
Table 1. Comparison of mapping methods for four spontaneous crossovers on chromosome V.
| PCR-based method | SNP microarrays | |||||
|---|---|---|---|---|---|---|
| Strain name | Centromere-proximal coordinate | Centromere-distal coordinate | Event description | Centromere-proximal coordinate | Centromere-distal coordinate | Event description |
| PG311-1.4 | 133080 | 125754 | CO, no conversion | 130096 | 127038 | CO + 3:1 tract |
| PG311-1.7 | 151440 | 146855 | CO, no conversion | 151419 | 150291 | CO, no conversion |
| PG311-4.1 | 99267 | 60163 | CO + hybrid tract | 98763 | 62494 | CO + complex tract |
| PG311-4.11 | 133080 | 94329 | CO + hybrid tract | 129511 | 97792 | CO + hybrid tract |
One exception to the generally good agreement between the two mapping methods is isolate PG311-1.6. This event was originally classified as crossover associated with a conversion tract that extended from SGD coordinate 31694 to 63936. SNP analysis demonstrated that the white sector had a terminal deletion on chromosome V, beginning near coordinate 62000. The same sector also had a large terminal duplication on chromosome VII. Although this rearrangement has not been fully characterized, since there are a cluster of δ elements near the breakpoint on chromosome V and Ty elements at the breakpoint on chromosome VII, it is possible that the strain has a chromosome V-VII translocation, similar to those that we have characterized previously (Argueso et al. 2008). No alterations were detected on either chromosome V or VII in the red sector. Sequence analysis indicated that the red sector retained the SUP4-o gene. It is possible that the cell that gave rise to the red sector lost the prion PSI, which affects the efficiency of ochre suppressors (Shkundina and Ter-Avanesyan 2007), although other possibilities cannot be excluded. Whatever the details of the genetic alterations in PG311-1.6, the event does not represent a conventional allelic crossover on chromosome V and, therefore, is excluded from our analysis.
Of the 13 colonies with spontaneous reciprocal recombination events analyzed by SNP arrays, the numbers of colonies of various classes were the following: (1) two crossovers without detectable conversions, (2) two crossovers with 3:1 conversion events, (3) one crossover with a 0:4 conversion, (4) one crossover associated with a hybrid tract (1:3 and 0:4 segments), and (5) seven crossovers with complex conversion tracts (Table 2). The complex conversion tracts will be discussed further below.
Table 2. Summary of all crossovers diagnosed by SNP microarray.
| Spontaneous | γ-ray | UV | ||||
|---|---|---|---|---|---|---|
| Types of reciprocal crossovers | Selected | Unselected | Selected | Unselected | Selected | Unselected |
| No detectable conversions | 2 | 0 | 0 | 0 | 0 | 0 |
| 3:1 or 1:3 conversions | 2 | 0 | 0 | 1 | 0 | 2 |
| 4:0 or 0:4 conversions | 1 | 0 | 1 | 0 | 0 | 0 |
| Hybrid conversionsa | 1 | 0 | 2 | 3 | 2 | 0 |
| Complex conversions | 7 | 0 | 4 | 0 | 1 | 5 |
| Total crossovers | 13 | 0 | 7 | 4 | 3 | 7 |
In this table, we summarize data from selected crossovers and associated conversion events on the left arm of chromosome V as well as unselected crossovers and associated conversions on other chromosomes. For this table, the data obtained with high-throughput DNA sequencing were not used.
3:1/4:0, 3:1/4:0/3:1, 1:3/0:4, or 1:3/0:4/1:3 conversion events.
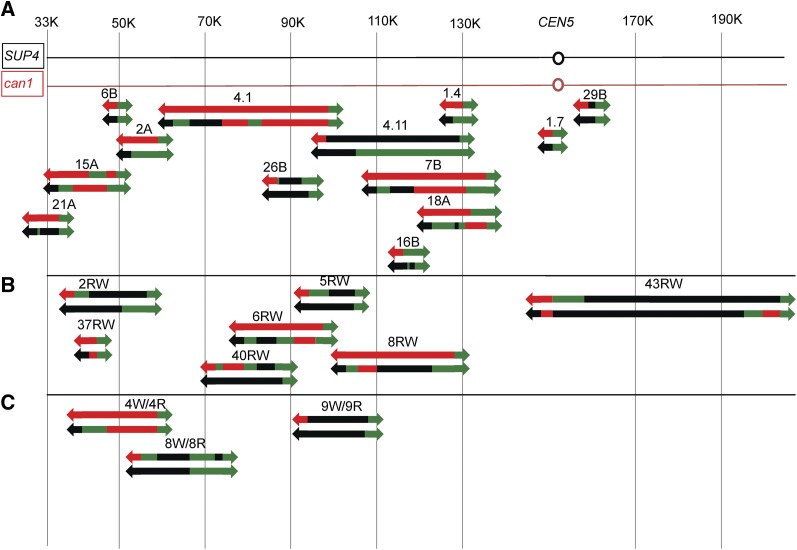
The locations of the spontaneous crossovers and associated conversion tracts are shown in Figure 5A. Each sectored colony is depicted as a pair of lines with the upper line representing the red sector and the lower line representing the white sector. The red and black line segments indicate that the sector is homozygous for the W303a-associated SNPs and the YJM789-related SNPs, respectively. The green line segment indicates that the sector is heterozygous for the SNPs. We show the two chromosomes within each sector as a single line because our analysis does not allow us to determine the coupling relationships for heterozygous SNPs between the two homologs. The median length of all crossover-associated conversion tracts was 6.1 kb, similar to the median observed in our previous study of spontaneous conversion tracts (6.5 kb) (Lee et al. 2009). Only one unselected LOH was observed in unirradiated cells. Both sectors in PG311-7B had a gene conversion event on chromosome VIII unassociated with a crossover (Table S6). Thus, the frequency of spontaneous unselected LOH events/cell is very low (∼0.08) as expected.
Figure 5.
Mapping of crossovers and associated conversion events on the left arm of chromosome V in untreated cells and cells treated with γ-rays or UV by SNP microarrays. Each red/white sectored canavanine-resistant colony is given a number and is depicted as a pair of lines with the upper line representing the red sector and the lower line the white sector. The colored segments signify heterozygosity for the markers (green), homozygosity for the YJM789-derived SNPs (black), or homozygosity for the W303a-derived SNPs (red). The green arrows show that the markers are heterozygous from the position at which the green segment begins to the end of the chromosome, and the red and black arrows indicate that the markers are homozygous for the W303a- or the YJM789-derived SNPs, respectively, from the point at which the segment begins to the telomere of the chromosome. Internal regions of heterozygosity and homozygosity are shown as line segments without arrows and are drawn approximately to scale. The numbers at the top of the figure are SGD coordinates. and the region between CEN5 and the can1-100/SUP4-o markers is ∼120 kb in length. (A) Spontaneous crossovers and conversions. (B) γ-Ray-induced crossovers and conversions. (C) UV-induced crossovers and conversions.
For the events shown in Figure 5A, 3:1 conversion events could reflect an initiating DNA lesion occurring anywhere within the tract, since events can be propagated bidirectionally from the DSB (Tang et al. 2011). For 4:0 or 3:1/4:0 hybrid tracts, the initiating lesion presumably occurs within the 4:0 region of the tract (Figure 3). Although we do not see any strong hotspots for spontaneous events with this limited data set, in a larger sample, we found that the region between SGD coordinates 41000 and 60000 had a significantly elevated level of crossovers and the region near CEN5 had a significantly reduced level of events (Lee et al. 2009).
As shown in Figure 5A, many of the recombination events are associated with multiple transitions between heterozygosity and homozygosity. In Table S6, for each sectored colony, we assigned a letter to represent each transition point; for each transition point, we also show two SGD coordinates, indicating the positions of the closest SNPs on the arrays that flank the transition points. The simplest events (crossovers without gene conversion: 6B and 1.7 in Figure 5A) have a single transition point. The recombination event shown in Figure 4 (which corresponds to event 2A in Figure 5A) has two transition points at different positions, one in the red sector and one in the white sector. In contrast to these relatively simple events, the sectored colony 18A (Figure 5A) has six transitions, one in the red sector and five in the white sector. In our analysis, if the transition point is identical in both sectors, it is counted only once. In Table S6, we also assign a class (A–L) for all events. In Table S1 and Table S2, each class of event is diagrammed using the same approach employed in Figure 5A. In Table S1 and Table S2, we also indicate the number of events in each class in Figures S1–S40 (File S2), which show the pattern of DNA repair events consistent with the specific conversion event. The same methods are used to describe the recombination events induced by DNA damage as were used to describe the spontaneous events. Multiple transitions within conversion tracts could reflect “patchy” repair of mismatches within long heteroduplexes (discussed further below) or template switching between homologs. For the spontaneous conversion events in Figure 5A, we did not find a correlation between SNP density and the number of transitions within the tract (r2 = 0).
Analysis of LOH events by SNP arrays in cells treated with γ-rays
We analyzed PG311 sectors that were generated in a previous study (Lee and Petes 2010) by treatment of G1-arrested cells with 100 Gy of γ-radiation, followed by selection of red/white sectored colonies on canavanine-containing plates; this dose of γ-rays elevated the frequency of sectoring ∼26-fold. All of the colonies examined had a crossover on chromosome V. We analyzed seven of these sectored colonies with SNP microarrays, and two of these were also examined by HTS. The SNP array data are shown in Table S7 with depiction of the recombination events in Table S1 and Table S2.
The positions of the selected crossovers and associated gene conversion events in the CEN5-can1-100/SUP4-o interval are shown in Figure 5B. Our mapping of these events by SNP arrays is in reasonably good agreement with our PCR-based mapping method (Lee and Petes 2010). All of the conversion events had at least one SNP that was homozygous on both sides of the sector (4:0 conversion) as expected if the recombination events were a consequence of repair of two sister chromatids broken at the same position (Figure 2 and Figure 3).
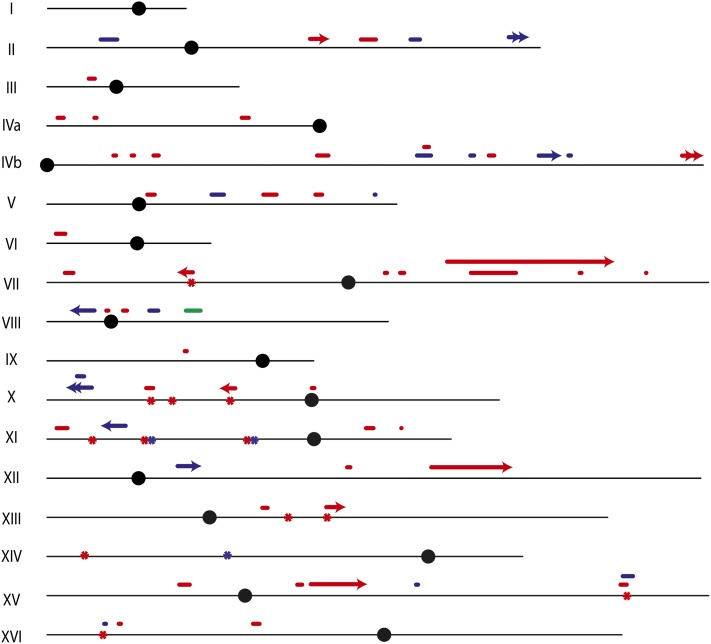
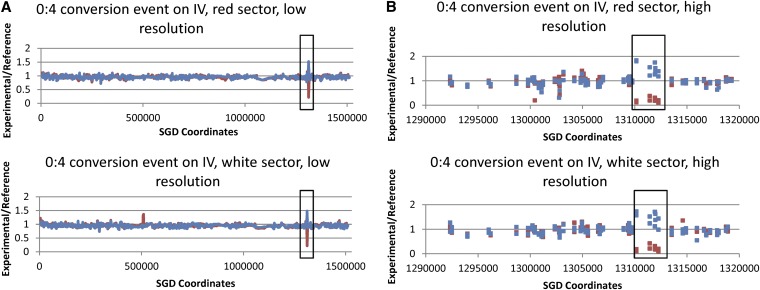
In addition to the selected events, from our genome-wide analysis, we observed 17 unselected events on other chromosomes among the seven colonies: four crossovers associated with conversion (Table 2), 11 conversions that were not associated with crossovers, and two BIR events (Table 3). Since the frequency of unselected crossovers in unirradiated samples is very low (<0.1/cell), it is likely that all of the events in the irradiated cells reflect the repair of γ-ray-induced DNA damage. The locations of these unselected events are shown as blue symbols in Figure 6. The events appear randomly distributed in this small data set. The SNP arrays for radiation-induced unselected crossovers and associated conversions have patterns similar to the selected crossover shown in Figure 4. In addition, we observed many conversion events unassociated with crossovers; Figure 7 shows at low and high resolution a 0:4 conversion event in which both sectors have gained YJM789 SNPs and lost W303a SNPs. This pattern could represent an event that occurred prior to radiation. However, since such events were observed commonly in irradiated cells but not in control diploids, we assume that most (or all) were induced by γ-rays.
Table 3. Summary of all unselected conversion events unassociated with LOH or BIR events as diagnosed by SNP microarrays.
| Type of event | Spontaneous | γ-ray | UV |
|---|---|---|---|
| 3:1 or 1:3 conversions | 0 | 4 | 9 |
| 4:0 or 0:4 conversions | 1 | 4 | 12 |
| Hybrid conversionsa | 0 | 2 | 9 |
| Complex conversions | 0 | 1 | 3 |
| BIR | 0 | 2 | 1 |
| Totals | 1 | 13 | 34 |
3:1/4:0, 3:1/4:0/3:1, 1:3/0:4, or 1:3/0:4/1:3 conversion events.
Figure 6.
Genomic locations of unselected recombination events and de novo mutations in untreated cells and in cells treated with UV or γ-rays as determined by SNP microarrays and HTS. The horizontal black lines depict each chromosome and are proportional to the chromosome length except for chromosome XII. The solid circles depict the centromere of each chromosome. Short horizontal bars above each chromosome depict conversion events unassociated with crossovers and the length of each bar is approximately proportional to the length of the conversion tract. All conversion tracts are shown as single solid lines without regard to the complexity of the event (e.g., transitions between 4:0 and 3:1). Single arrowheads depict reciprocal crossovers and double arrowheads depict BIR events. Asterisks located on the chromosome indicate the approximate positions of mutations induced by UV or γ-rays; two of the mutations (located at SGD coordinates 171529 and 301552 on X) are in regions of LOH. Events observed in untreated cells, in cells treated with UV, and in cells treated with γ-rays are shown in green, red, and blue, respectively. None of the events selected on the left arm of V are shown.
Figure 7.
SNP array analysis of a gene conversion event unassociated with a crossover. In cells treated with γ-rays, one of the canavanine-resistant red/white sectored colonies (43RW) had an unselected gene conversion event on chromosome IV. As shown at low (A) and high (B) resolution, both sectors had an LOH region in which YJM789-derived SNPs became homozygous (0:4 conversion event). The depiction of the SNP array data is the same as in Figure 4. The length of the conversion tract is ∼3 kb. CEN4 is located approximately at SGD coordinate 450000.
Since the red and white sectors are produced by the two daughter cells resulting from the division of radiation treatment of a G1-synchronized cell, the analysis of genomic DNA from both sectors gives valuable mechanistic information even for unselected events. For example, if we observed an interstitial LOH event by examining only the white sector, we would not know whether this event was a consequence of a 3:1 conversion, a 4:0 conversion, or a two-strand double crossover. This ambiguity can be resolved by examining genomic DNA from the red sector.
We observed two sectored colonies that had BIR events. In single BIR events (such as class L2 in Table S1), one sector has an LOH event that extends from an internal site on the chromosome to the telomere, whereas the other sector is heterozygous for the same SNPs. In double-BIR events, both sectors have LOH events extending from an internal site to the telomere (class L1 in Table S1). Interestingly, in the colony with the single BIR event, there is a conversion event on the chromosome that was originally the sister chromatid of the one involved in BIR. This result argues that both sister chromatids had DSBs at approximately the same position. The molecular interactions required to produce classes L1 and L2 are shown in File S2, Figure S39 and Figure S40.
All of the selected and unselected reciprocal crossovers induced by γ-rays were associated with conversion tracts. The median length of all conversion tracts (both associated and unassociated with crossover) was 12.9 kb (95% confidence limits of 5.2–20.4 kb). The median lengths of conversion tracts associated and unassociated with crossovers were 18.4 kb (10.8–25.3) and 8.4 kb (2.6–13.3), respectively. By the Mann–Whitney test, the median lengths of crossover-associated and crossover-unassociated γ-ray-induced conversion tracts were significantly different (P = 0.01).
Analysis of LOH events by SNP arrays in cells treated with UV
G1-synchronized PG311 cells were treated with a UV dose of 10–15 J/m2. This dose resulted in no significant loss of viability but stimulated the frequency of sectors by ∼1000-fold. We examined three sectored colonies by SNP arrays, and two of these colonies were also analyzed by HTS. In addition to the selected crossover on chromosome V, among the three colonies, there were seven unselected crossovers, 33 unselected conversion events, and one BIR event (Table 2 and Table 3). Thus, there were about 14 unselected LOH events per UV-treated cell. The locations of selected chromosome V events and the unselected LOH events are shown in Figure 5 and Figure 6, respectively. The UV-induced LOH events are distributed fairly evenly throughout the genome (Figure 6). As observed for the γ-ray-induced BIR events, the UV-induced BIR event is located close to the telomere. The detailed information about breakpoints in UV-treated cells is shown in Table S1, Table S2, and Table S9.
All of the crossover events had an associated conversion. In most of the conversion events, there was at least one SNP with the 4:0 or 0:4 pattern, suggesting that UV-induced damage in G1 may result in DSBs (Figure 2B). The median length of all UV-induced conversion tracts was 9.2 kb (6.5–10.3), whereas the median lengths of conversion tracts associated and unassociated with crossovers were 10.3 (7.0–18.9) and 7.5 (4.5–10.2) kb, respectively. Although the crossover-associated conversions are longer than the crossover-unassociated conversions, this difference is not significant (P = 0.08 by Mann–Whitney test).
HTS analysis of LOH in γ-ray- and UV-treated cells
Since G1-arrested yeast cells treated with 100 Gy of ionizing radiation have ∼35 DSBs per cell (Lee and Petes 2010), the average number of unselected LOH events per cell (two to three) indicates that most events must be repaired by mechanisms that do not generate LOH. An alternative possibility is that a substantial fraction of the events have short conversion tracts that are not detectable by the SNP arrays. Since HTS can detect LOH events for all of the 55,000 SNPs existing in the diploid strain, rather than the 13,000 SNPs represented on the SNP array, we sequenced genomic DNA samples from both red and white sectors of two sectored colonies of γ-ray-treated samples (PG311-GR-37R/W and PG311-GR-40R/W) and two colonies of UV-treated samples (PG311-UV-8R/W and PG311-UV-9R/W). The details of this analysis are described in File S1. All of the LOH events detected by SNP arrays were also found by HTS. LOH events that had not been previously detected by the SNP arrays were confirmed by resequencing the relevant PCR fragment. The patterns of LOH as identified by HTS in the γ-ray- and UV-treated samples are in Table S8 and Table S10.
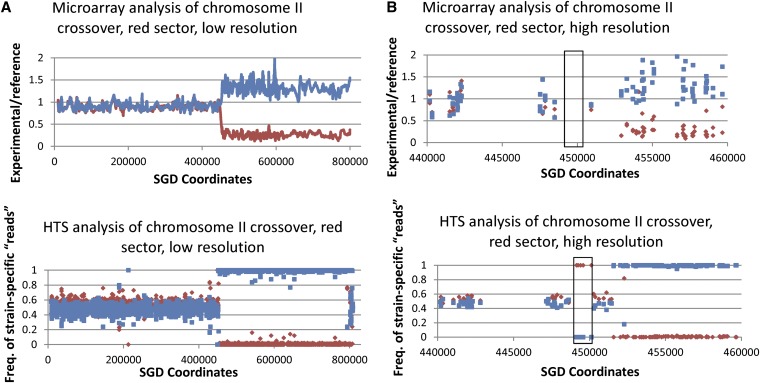
Figure 8 shows a comparison between SNP microarray and HTS data for an LOH event on chromosome II in a UV-treated sample (PG311-UV-8R). The SNP microarray indicates that a transition between heterozygous and homozygous SNPs occurs somewhere between SGD coordinates 450919 and 452926, whereas the HTS data refine the mapping of this transition between SGD coordinates 451337 and 451581. In addition, the HTS data showed that the recombination event was more complex than indicated by the microarray data. By HTS, we found that the region between coordinates 448628 and 450279 had undergone LOH; this event was not detected by microarrays because there were no oligonucleotides between 448488 and 450919 on the microarray. A summary of the comparison of data from SNP arrays and HTS for the same samples is given in Table 4.
Figure 8.
Analysis of the same recombination event by both SNP arrays and HTS. This figure shows the analysis of the unselected recombination event on chromosome II in the red sector of the UV-induced sectored colony 8. Our standard SNP array analysis (top, A and B) showed a single transition between heterozygosity and homozygosity at about SGD coordinate 452000. (Bottom, A and B) HTS data for the same genomic sample. For the HTS data, the y-axis represents the frequency of YJM789 SNP (blue) or W303a SNP (red) “reads” for the experimental sample when assembled to the PSL2 genome. For heterozygous regions, there should be approximately equal frequencies of the two types of SNPs. It is clear from the high-resolution depictions of the HTS data that there is a short LOH region (boxed in B) located near SGD coordinate 450000 that was not detected by the SNP arrays. This region was not detected because oligonucleotides containing these SNPs were not present on the array. In the low-resolution depiction of the HTS data, within the LOH region, there is a small region near SGD coordinate 800000 in which SNPs appear to be heterozygous. These signals are artifacts on the basis of “reads” from the repeated diverged MAL and MPH genes that were incorrectly mapped by the genome analysis software to chromosome II. CEN2 is located near SGD coordinate 238000.
Table 4. Comparison between recombination events detected by high-throughput sequencing or by SNP microarrays.
| γ-ray | UV | |||
|---|---|---|---|---|
| Type of event | Array | HTS | Array | HTS |
| Crossovers with no detectable conversion tracts | 0 | 0 | 0 | 0 |
| Crossovers with 3:1 or 1:3 conversion tracts | 0 | 0 | 2 | 0 |
| Crossovers with 4:0 or 0:4 conversion tracts | 1 | 0 | 0 | 0 |
| Crossovers with hybrid conversion tracts | 0 | 1 | 1 | 1 |
| Crossovers with complex conversion tracts | 1 | 1 | 6 | 8 |
| 3:1 or 1:3 conversion tracts without crossovers | 1 | 0 | 8 | 9 |
| 4:0 or 0:4 conversion tracts without crossovers | 1 | 1 | 5 | 4 |
| Hybrid conversion tracts without crossovers | 0 | 1 | 8 | 7 |
| Complex conversion tracts without crossovers | 1 | 2 | 1 | 5 |
| BIR | 0 | 0 | 1 | 1 |
| Total recombination events | 5 | 6 | 32 | 35 |
This table includes data from two UV-induced sectored colonies and two sectored colonies induced by γ-radiation that were analyzed by both SNP arrays and HTS. Both selected recombination events on chromosome V and unselected events on other chromosomes are included. All conversion events unassociated with LOH were unselected.
Although more LOH events were observed with HTS than with SNP arrays, the difference was not large. For example, in the two UV-treated samples, we observed 32 LOH events by SNP arrays and 35 events by sequencing. In the γ-ray-treated samples, we found five events by SNP arrays and six events by sequencing. Since >80% of the events detected by HTS were also detected by microarrays, it is unlikely that our estimates of LOH events are substantially affected by a high frequency of gene conversion events with short conversion tracts. We cannot rule out, however, the possibility of gene conversion events with very short (<100 bp) tracts. As shown in Table 4, a number of the gene conversion tracts analyzed by HTS were more complex than the same tracts examined by the SNP arrays. The frequencies of complex tracts, as determined by HTS and SNP microarrays, were 0.37 and 0.22, respectively. Despite the differences in the numbers and types of LOH events detected by HTS and the SNP microarrays, it is clear that most of the LOH events are detectable by the SNP microarrays.
HTS analysis of mutations induced in γ-ray- and UV-treated cells
About 99.5% of the bases between the two homologs of PG311 are identical. The HTS data generated for the same colonies examined for LOH events were analyzed for radiation-induced mutations. We analyzed both sectors of two sectored colonies induced by UV and two sectored colonies induced by γ-rays for new mutations (Table S11). There were 3 and 12 de novo point mutations detected in the γ-ray- and UV-treated samples, respectively.
All 3 of the mutations induced by γ-rays and 6 of the 12 mutations induced by UV were in both red and white sectors of the colony. The presence of the mutation in both sectors indicates that the mutation induced in G1 by the radiation was represented in both strands of the duplex prior to replication. The mutations in the UV-treated cells that were present in only one sector could reflect a mutant base in only one of the two strands. These two types of events have been observed previously in UV-treated cells (Eckardt and Haynes 1977; James and Kilbey 1977). Nine of 12 of the UV-induced mutations and 2 of 3 of the γ-ray-induced mutations were transitions. In much more extensive study of spontaneous and UV-induced mutations at the SUP4-o locus (Kunz et al. 1987), spontaneous mutations had a ratio of transitions:transversions of 4:6, whereas UV-induced mutations were biased toward transitions (4:1).
Most (11 of 15) of the induced point mutations were located within genes rather than between genes (Table S11). By chi-square analysis, the distribution of mutations throughout the genome is nonrandom (P = 0.002). Five of the 15 mutations are located on the left arm of XI and, remarkably, two mutations (one induced by γ-rays and one induced by UV) are within the NUP120 gene. By similar methods used to detect new base substitutions, we failed to detect any insertion/deletion (indels) mutations in the eight sequenced samples. It should be pointed out, however, that detection of indels in HTS data with short-paired reads is challenging, particularly in a diploid that is heterozygous for many pre-existing indels.
Discussion
In this study, we mapped both selected and unselected mitotic recombination events in a genome-wide analysis. Most of the events were mapped using only SNP microarrays, but four events were examined by both SNP microarrays and HTS. To our knowledge, this study is the first to examine spontaneous and DNA damage-induced LOH events throughout the yeast genome. The conclusions from this study are the following: (1) the gene conversion tracts analyzed by SNP arrays and HTS were often more complex than inferred from our earlier lower-resolution mapping studies (Lee et al. 2009; Lee and Petes 2010); (2) doses of radiation that result in little or no loss of cell viability in G1-synchronized diploid cells resulted in multiple unselected LOH events; and (3) the same doses of γ-rays and UV used in the LOH study result in very low levels of de novo mutations. In addition, we conclude that, although HTS has fourfold better resolution than SNP microarrays, the SNP arrays detect most of the same events identified by HTS. These conclusions will be further discussed below.
Lengths of gene conversion tracts
The median lengths (95% confidence limits shown in parentheses), as measured by SNP arrays, of gene conversion tracts associated with crossovers for spontaneous, UV-induced, and γ-ray-induced events were 6.1 (1.7 to 25.4), 10.3 (7.0 to 19.0), and 18.5 (10.8 to 25.2) kb, respectively. As we observed previously (Lee et al. 2009), mitotic gene conversion tracts are substantially longer than meiotic conversion tracts (Mancera et al. 2008). In addition, for the γ-ray-induced conversion events, the conversion tracts associated with crossovers were significantly longer than those unassociated with crossovers as has been observed previously (Aguilera and Klein 1989; Mancera et al. 2008). One simple explanation of this observation is that conversion events unassociated with crossovers usually are a consequence of SDSA, and such events might involve limited processing of the broken chromosome ends. In contrast, crossovers likely proceed through formation of a double Holliday junction. Formation of this dHJ intermediate may require more extensive processing of broken DNA ends and/or more extensive DNA synthesis primed from the invading end. It is also possible that branch migration of the dHJ intermediate could extend the length of the heteroduplex associated with the crossover; this possibility will be further discussed below.
Recombinogenic DNA damage
Although it is clear from a variety of studies that DSBs stimulate mitotic recombination, the DNA lesion responsible for initiating spontaneous recombination events is not certain. We previously showed that about half of mitotic crossovers on chromosome V are associated with gene conversion tracts that are exclusively 4:0 or 0:4 or that have a region of 4:0 or 0:4 (hybrid tracts). Such conversion tracts indicate that both sister chromatids have breaks at approximately the same position and one simple mechanism consistent with this property is that these spontaneous conversion events reflect a DSB formed in G1 of the cell cycle. Supporting this conclusion, many (44%) of the conversion events induced by γ-rays in G1 of the cell cycle have regions of 4:0 or 0:4 segregation, whereas none of the conversion events induced by γ-rays in G2 had this pattern (Lee and Petes 2010). Among the mechanisms that could produce the spontaneous lesions required to initiate recombination are (1) closely opposed nicks generated during removal of adducts caused by oxidative DNA damage, (2) DSBs caused by Top2p or other cellular endonucleases, (3) lesions resulting from collisions between replication forks and the transcription machinery, (4) DSBs resulting from the collision of converging replication forks, and (5) nuclease-dependent processing of secondary DNA structures (“hairpins” and cruciforms). Thus far, we have been unable to associate the spontaneous recombination events with any of these mechanisms. For example, the two positions that represent the convergence of replication forks on the left arm of chromosome V (Fachinetti et al. 2010) are not hotspots for recombination in our limited data set. If there are several different mechanisms that can produce recombinogenic DNA lesions, we will need to map many events to detect significant associations.
In our current analysis of γ-ray-induced gene conversion events in G1-synchronized cells by SNP microarrays, we found that 10 of the 11 conversion tracts associated with crossovers had a 4:0 or 0:4 segment, and 8 of the 11 conversion tracts that were unassociated with crossovers had such a segment. This observation is consistent with the possibility that most of the observed recombination events in IR-treated cells reflected a DSB introduced by γ-rays in G1.
The recombinogenic effect of UV-induced DNA damage is less clear. One possibility is that small gaps resulting from the removal of UV-induced dimers are the recombinogenic lesion. Galli and Schiestl (1999) found that UV did not stimulate mitotic recombination between direct repeats (single-strand annealing) in G1-arrested cells unless the cells were allowed to enter the S-period after the UV treatment. In contrast, when G1-arrested cells were treated with IR, stimulation of single-strand annealing was observed without requiring the cells to enter the S-period. If the recombinogenic DNA lesion is a DSB, the likely explanation of the different results is that IR directly creates DSBs whereas the repair of UV lesions results in nicks that result in DSBs when the nicked molecule is replicated (Galli and Schiestl 1999). By this explanation, it is surprising that many of the conversion events induced by UV in G1 in our experiments had 4:0 or 0:4 segments, suggesting that these exchanges were a consequence of a G1-stimulated DSB. Such a DSB could be generated in G1 if the removal of dimers on opposite DNA strands resulted in a very short (<10 bp) duplex region separating the 30-bp gaps. On the basis of previous estimates of the number of dimers induced by 20 J/m2 of UV (Daigaku et al. 2010), we calculate that there are ∼7500 dimers/diploid genome induced by a UV dose of 15 J/m2 and, on the basis of Poisson distribution, there would be ∼35 regions per genome in which 2 dimers are on opposite strands within 75 bp of each other. Since the DSB would require two closely opposed gaps rather than two closely opposed dimers, the kinetics of gap formation and gap repair affect the probability of DSB formation by this mechanism. Another complicating factor is that the frequency of closely opposed dimers is higher than expected if dimer formation is random (Reynolds 1987).
A second possibility is that DSB formation is initiated by gaps on opposite strands that are relatively close together (≤500 bp apart), but too far apart to generate a G1 DSB. If a DNA molecule with such gaps is replicated, the product would be two sister chromatids with DSBs located ≤500 bp apart. Processing of the broken ends to yield single-stranded DNA regions ≥500 bp would preclude formation of a dHJ involving the two sister chromatids. Thus, such molecules would likely be repaired using the intact homolog as the template, generating a 4:0 conversion. This model is consistent with the Galli and Schiestl (1999) interpretation. We calculate that cells irradiated with 15 J/m2 would have ∼234 dimers on opposite strands within 500 bp of each other.
An alternative possibility is that recombination events are a consequence of DSBs occurring at replication forks stalled at unexcised pyrimidine dimers. Unrepaired UV-induced damage has been demonstrated to block replication forks, and replication of such damaged templates promotes sister-chromatid recombination (Branzei and Foiani 2010). Although we cannot exclude this model, the observed UV-induced 4:0 events would require that the replication fork stalled at the dimer result in two broken sister chromatids, perhaps by increasing the probability of a replication fork collision. It should be emphasized that, although UV very efficiently stimulates crossovers between homologs, most of the UV-induced recombination events are likely to represent sister-chromatid interactions (Kadyk and Hartwell 1992).
Relationship between mitotic gene conversion and crossovers
In meiosis in yeast, about half of conversion events are associated with crossovers (Petes et al. 1991; Mancera et al. 2008). In our previous mitotic studies, we selected crossovers and found that most (>80%) of these events were associated with an adjacent tract of gene conversion (Lee et al. 2009; Lee and Petes 2010); conversion events unassociated with crossovers could not be selected with our system. In the current study, for unselected events stimulated by radiation, we can estimate the fraction of conversion events that are associated with crossovers.
For IR-treated cells, including all nonselected events except BIR, we found 4 conversions associated with crossovers and 11 conversions unassociated with crossovers (Table 2 and Table 3). In these experiments, we detect the associated crossover because it generates LOH from the conversion tract to the end of the chromosome. As shown in Figure 2, depending on the pattern of chromosome segregation, we expect that only half of the crossovers will lead to LOH of markers distal to the point of exchange, and this expectation has experimental support (Chua and Jinks-Robertson 1991). In addition, as discussed in File S1 and Table S12, we found preliminary evidence in our experiments for conversion events associated with crossovers that did not result in LOH.
Thus, we calculate that, of the 15 conversion events induced by γ-rays, it is likely that 8 were associated with crossovers (53% association). Similarly, among UV-induced recombinants, since there were 7 unselected conversions associated with crossovers and 33 unselected conversions not associated with LOH (Table 2 and Table 3), we calculate that ∼35% of the UV-induced conversion events are associated with crossovers. Our conclusion that the frequency of crossovers associated with conversions is not very different for mitotic and meiotic conversion events is consistent with other recent studies (Ho et al. 2010). In yeast studies in which conversion events are limited in size, the association between conversion and crossovers is weaker (Pâques and Haber 1999). Also, in Drosophila and mammalian cells, conversion events are only rarely associated with crossovers (Andersen and Sekelsky 2010).
Complex gene conversion tracts and BIR events
Previously, we classified conversion tracts as “simple” if the markers within the tract had one of the following patterns: (1) all markers were 3:1 or 1:3 (not mixtures of 3:1 and 1:3 in same tract); (2) all markers were 4:0 or 0:4; or (3) hybrid tracts of the form 3:1/4:0, 1:3/0:4, 3:1/4:0/3:1, or 1:3/0:4/1:3. All such tracts can be explained as the consequence of the repair of one or two broken chromatids by one of the HR pathways shown in Figure 1. There were, however, conversion tracts that were more complicated (termed “complex tracts”), which will be described below. In the UV-treated samples, 6 of 10 of the crossover-associated conversion tracts were complex, although only 3 of 33 tracts were complex in conversions unassociated with crossovers (Table 2 and Table 3); this difference is significant (P < 0.01) by Fisher’s exact test. In the IR-treated samples, the conversion events associated with crossovers were usually more complex than those that were not (Table 2), although the difference was not significant. Mancera et al. (2008) reported that 11% of meiotic crossovers had complex conversion tracts, whereas the frequency of complex tracts among conversions unassociated with crossovers was 3%. One explanation of this difference could be that crossovers that proceed through the pathway shown in Figure 1B are associated with two regions of heteroduplex, while conversions resulting from SDSA or dHJ dissolution have only a single region of heteroduplex (Figure 1, A and C). Additionally, because gene conversion tracts associated with crossovers are usually longer than those unassociated with crossovers, there may be a greater chance to observe patchy repair of mismatches (as defined below) in tracts associated with crossovers.
Diagrams of all recombination events in our study are shown in Table S1 and Table S2, and the patterns of DSB repair required to produce the recombination events are shown in File S2, Figures S1–S40. Most of the complex conversion tracts could be divided into two categories: tracts that had multiple transitions between 3:1, 4:0, and heterozygosity within the tract and those tracts in which 3:1 and 1:3 or 4:0 and 0:4 segments occurred within one tract. Examples of conversion tracts with multiple transitions are strains 18A (class J9, Table S1) and 4.1 (class J8, Table S1); both 18A and 4.1 are also depicted in Figure 5A. The complex tract in 4.1 is consistent with the repair of two DSBs with “patchy” repair of mismatches in two of the resulting heteroduplexes (File S1, File S2, Figure S29). Heteroduplexes will often contain multiple mismatches that can be repaired to produce either a conversion event or a restoration event (Kirkpatrick et al. 1998). For example, in Figure 1A, repair of the heteroduplex resulting in a duplex with two “red” strands would represent conversion-type repair because this pattern produces 3:1 segregation; repair of the mismatch to produce a duplex with two “blue” strands represents restoration-type repair because this pattern generates two cells that retain heterozygosity at the position of the original heteroduplex. Although multiple mismatches within one heteroduplex are generally converted in a concerted manner, yielding a continuous conversion tract, tracts with mixtures of conversion-type and restoration-type repair have been detected in both meiosis (Symington and Petes 1988; Mancera et al. 2008) and mitosis (Nickoloff et al. 1999; Mitchel et al. 2010).
The pathway of DSB repair shown to explain the pattern of markers in the strain 18A conversion event (File S2, Figure S30) invokes patchy repair and branch migration of the dHJ. During recombination in Escherichia coli, a Holliday junction can be translocated by branch migration, resulting in symmetric heteroduplexes (West 1997). Although genetic evidence argues against the formation of symmetric heteroduplexes during meiotic recombination in S. cerevisiae (Petes et al. 1991), symmetric heteroduplexes have been invoked previously to explain certain classes of mitotic gene conversions (Esposito 1978; Roitgrund et al. 1993; Nickoloff et al. 1999). Branch migration can also generate patterns of repair in which a single DSB can produce both 3:1 and 1:3 or 4:0 and 0:4 events as shown in File S2, Figure S10B.
Our data do not allow us to determine unambiguously the pathways required to generate the observed conversion tracts. However, we can state that many of the complex tracts are inconsistent with the simplest form of the recombination models shown in Figure 1. In particular, it is likely that patchy repair of mismatches is a relatively common feature of mitotic gene conversion tracts. A detailed discussion of all of the conversion tracts in our studies is given in File S1.
There were three unselected BIR events observed in our study (class L, File S2, Figure S39 and Figure S40). For two of the three events, we observed a region of conversion associated with the BIR event. This pattern is consistent with the repair of two DSBs, one by SDSA and one by BIR (File S2, Figure S40). The BIR events were about threefold less frequent than unselected crossovers, as expected from previous studies (McMurray and Gottschling 2003; Ho et al. 2010)
Relationship between the level of DNA damage and the frequency of LOH events
The 100-Gy dose of IR used in our experiments is expected to produce ∼35 DSBs/diploid genome (Lee and Petes 2010). Since we observed only 2.4 LOH events/irradiated cell, most of these DSBs must be repaired by a mechanism that does not produce a detectable LOH event. Since the cells in our experiments were irradiated in G1, the DSBs must have been repaired either by an interaction with the homologous chromosome or by NHEJ. We suggest several possible explanations. First, it is possible that the repair of the DSB frequently involves an interaction with the homolog that is associated with a very short conversion tract. Tracts <50 bp would be rarely detected, even by HTS. Such a repair event would likely involve very limited processing of broken DNA ends as well as short excision repair tracts. A system of short-patch (often <12 bp) mismatch repair that is independent of the classical mismatch repair system in S. cerevisiae was described by Coic et al. (2000), although the genes involved in this type of repair have not been identified. In addition, conversion tracts <53 bp have been detected among HO-induced events (Palmer et al. 2003). Second, a related possibility is that gene conversion events occur nonrandomly in regions of the genome that are not represented on our microarrays (regions that are identical between W303a and YJM789 or regions with repeated genes). A third possibility is that the repair of the DSB is associated with restoration-type repair of mismatches within the heteroduplexes. Since most of the crossovers in our study are associated with detectable gene conversion tracts, we would have to hypothesize that conversion events that are not associated with crossovers are much more prone to restoration-type repair than conversion events that are associated with crossovers. A fourth possibility is that the IR-induced DSBs are frequently (Daley et al. 2005) repaired by NHEJ events. Although NHEJ events are repressed in MATa/MATα diploids, since PG311 lacks the MATα locus, NHEJ events will occur. Although NHEJ events will not produce LOH, depending on the nature of DNA ends (compatible single-strand overhangs or blunt), some NHEJ events would be expected to result in loss or gain of a few base pairs . Although we did not observe indels in our HTS analysis, this observation does not rule out the possibility that some repair events reflect NHEJ. It is also possible, of course, that all four possibilities described above are partly responsible for the “missing” LOH events.
One explanation that we can exclude as a major contributor to the discrepancy between the number of lesions and the number of LOH events is chromosome loss. Chromosome loss can be readily detected by the SNP microarrays, and no losses were observed in cells treated with γ-rays or UV. In experiments in which eightfold higher doses of γ-rays were used, ∼10% of the treated yeast cells had chromosome loss (Argueso et al. 2008).
Although we detected >50 unselected LOH events in cells treated with γ-rays and UV, no duplications or deletions were detected. Thus, SNP arrays that can detect both LOH and changes in copy number are a much more efficient method of detecting recombinogenic DNA lesions than comparative genomic hybridization (CGH) arrays. In our previous analysis of γ-ray-treated diploid cells by CGH (Argueso et al. 2008), we found that most of the irradiated cells had one or more chromosome rearrangements, usually nonreciprocal translocations with retrotransposons at the breakpoints. In these experiments, we treated G2-synchronized cells with doses of radiation that were eightfold higher than the doses used in our current study.
Mutations induced by γ-rays and UV
We found only a few mutations induced by γ-rays and UV, averages of 1.5 and 6 mutations/irradiated cell, respectively. Although there are no genome-wide studies of the frequencies of mutations induced by γ-rays, extrapolating from the frequency of induction of X-ray-induced mutations at the CAN1 locus (Goeke and Manney 1979) and the rate of spontaneous mutations per base pair at CAN1 (Lang and Murray 2008), we calculate that the expected frequency of mutations per genome is about two/diploid cell, close to our observed number. The most direct comparison for the UV-induced mutations is with data obtained from HTS of UV-treated stationary-phase haploid yeast strains (Burch et al. 2011). The strains in these studies had a temperature-sensitive mutation in CDC13. However, three of the sequenced isolates were treated at the permissive temperature. By extrapolating their data to our UV dose, we would expect ∼14 mutations/diploid cell, only twofold different from our observed frequencies. In summary, our HTS data detected roughly the expected number of mutations per irradiated strain.
As described in the Results, the mutations induced by UV and γ-rays are nonrandomly distributed among the yeast chromosomes. Although this nonrandom distribution needs to be verified with a large data set, it is possible that the mutagenic DNA damage is distributed nonrandomly because of the specific position of different chromosomes within the nucleus or chromosome-specific chromatin domains. Since the UV-irradiated strains have ∼7500 DNA lesions (as discussed above), the vast majority of these lesions must be repaired by nucleotide excision repair in a manner that does not result in LOH or mutations.
We assume that most of the UV-induced mutations reflect errors introduced during the bypass of pyrimidine dimers by Rev1p and Polζ, since 90% of UV-induced mutations require these activities (Lawrence 2002). The source of the mutations in the γ-ray-treated samples is less clear. Since the mutations are not associated with regions of LOH, the mutations probably do not reflect errors introduced by DSB repair. It is possible that bases damaged by γ-rays are bypassed by error-prone polymerases by a mechanism similar to that associated with UV-induced DNA damage.
All three of the mutations introduced by γ-irradiation and about half of the mutations caused by UV were found in both sectors of sectored colonies. This result indicates that the introduced mutation was placed into both strands of the duplex before DNA replication. Such events, which have been observed previously for UV-induced DNA damage (Eckardt and Haynes 1977; James and Kilbey 1977), have been termed “two-strand” mutations (Abdulovic et al. 2006). One model for such events is that they reflect the repair of two closely opposed DNA lesions by nucleotide excision repair. During the repair of one lesion, a mutation is introduced. The repair of the second lesion on the opposite strand results in a gap that includes the mutant substitution and the filling in of the gap results in mutant substitutions in both strands of the duplex. Whatever the explanation of two-strand events, both UV and γ-rays efficiently produce this type of mutation.
The repair of DSBs is associated with a 100-fold elevation in the frequency of reversion of a closely linked mutation (Strathern et al. 1995), and ∼1000-fold elevated rates of mutation have been observed during BIR (Deem et al. 2011) and other gene conversion events that result in two newly synthesized strands (Hicks et al. 2010). In addition, the frequency of UV-induced mutagenesis is elevated >100-fold in regions of single-stranded DNA next to DSBs or abnormal telomeres (Yang et al. 2008). On the basis of these observations, we checked whether the de novo mutations were nonrandomly associated with LOH regions associated with gene conversion or BIR. There were 15 base substitutions observed among four sectored colonies resulting from the irradiation of G1 cells. The total lengths of the unselected gene conversion and BIR events among these strains were 163 kb (PG311-UV-8R/W), 271 kb (PG311-UV-9R/W), 22 kb (PG311-IR-37R/W), and 50 kb (PG311-IR-40R/W). The fraction of the genome with these LOH regions was ∼0.01. Two of the 15 (0.13) mutations were in regions of LOH. Although this calculation suggests that the LOH regions may have a significantly elevated frequency of mutations, most of the induced mutations are located outside of the LOH regions.
Comparison among methods of physically mapping recombination events
In our previous studies, we mapped recombination events by a PCR-based technique (described in the Introduction). As employed in our analysis of chromosome V events, this approach was time-consuming and expensive and mapped events with relatively poor resolution (∼4 kb). More importantly, this method could not be easily used to map events throughout the genome. In addition, the PCR-based approach did not allow us to examine changes in gene dosage (deletions or duplications). For example, we found that an event classified as a crossover on chromosome V by the PCR-based method was actually a terminal deletion on V when examined by SNP arrays.
In contrast, both SNP arrays and HTS allow analysis of events throughout the genome. The advantages of SNP arrays compared to HTS are (1) relatively low cost (∼$100/sample), (2) speed of analysis (∼4 hr for SNP arrays vs. 1 week for HTS), and (3) relative ease in detecting changes in gene dosage. The major advantages of HTS are (1) higher resolution (1 kb for SNP arrays vs. 250 bp for HTS) and (2) the ability to detect de novo mutations. In addition, diagnosis of LOH by HTS can be done with any diploid in which the progenitor haploid strains have been sequenced, whereas diagnosis of LOH by SNP arrays requires the construction of strain-specific microarrays. Although SNP arrays are probably a more cost-effective and faster approach for mapping large numbers of recombination events at present, as HTS becomes cheaper and analysis of HTS data becomes faster, HTS is likely to be the method of choice in the future. Neither SNP microarrays nor HTS, however, can map recombination events that do not involve LOH (e.g., sister-chromatid exchanges).
Conclusion
In conclusion, we have used SNP microarrays and HTS to map crossovers and gene conversion events at high resolution throughout the yeast genome. These studies represent the first genome-wide measurement of the number and types of unselected LOH events induced by UV and γ-rays. In G1-synchronized cells treated with either UV or γ-rays, 4:0 conversion events are common, suggesting that many of the LOH events reflect the repair of two sister chromatids broken at approximately the same position. In addition, the high-resolution analysis of recombination events by SNP arrays and HTS reveals that gene conversion tracts, particularly those associated with crossovers, are more complex than was previously recognized by low-resolution studies.
Supplementary Material
Acknowledgments
We thank members of the Petes and Jinks-Robertson laboratories for discussions and Lucas Argueso, Kat Mitchel, Sue Jinks-Robertson, and Wei Song for comments on the manuscript. We also thank members of McCusker’s laboratory for technical help and Argueso and J. Rine for useful suggestions. The research was supported by National Institutes of Health grants GM24110 (T.D.P.), GM52319 (T.D.P. and P.M.), and 5RC1ES18091 (T.D.P. and P.M.) and by funds from the Duke University School of Medicine.
Footnotes
Communicating editor: E. Alani
Literature Cited
- Abbey D., Hickman M., Gresham D., Berman J., 2011. High-resolution SNP/CGH microarrays reveal the accumulation of loss of heterozygosity in commonly used Candida albicans strains. G3: Genes, Genomes, Genetics 1: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulovic A., Kim N., Jinks-Robertson S., 2006. Mutagenesis and the three R’s in yeast. DNA Repair (Amst.) 5: 409–421 [DOI] [PubMed] [Google Scholar]
- Aguilera A., 2002. The connection between transcription and genomic instability. EMBO J. 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Klein H. L., 1989. Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics 123: 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Altman D. G., 1990. Practical Statistics for Medical Research. CRC Press, Boca Raton, FL [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic vs. mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic vs. mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Westmoreland J., Mieczkowski P. A., Gawel M., Petes T. D., et al. , 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. USA 105: 11845–11850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera M. A., Petes T. D., 2006. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 12819–12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Breen A. P., Murphy J. A., 1995. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 18: 1033–1077 [DOI] [PubMed] [Google Scholar]
- Burch L. H., Yang Y., Sterling J. F., Roberts S. A., Chao F. G., et al. , 2011. Damage-induced localized hypermutability. Cell Cycle 10: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P., Jinks-Robertson S., 1991. Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coic E., Gluck L., Fabre F., 2000. Evidence for short-patch mismatch repair in Saccharomyces cerevisae. EMBO J. 19: 3408–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y., Davies A. A., Ulrich H. D., 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley J. M., Palmbos P. L., Wu D., Wilson T. E., 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451 [DOI] [PubMed] [Google Scholar]
- Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., et al. , 2011. Break-induced replication is highly inaccurate. PLoS Biol. 9: e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt F., Haynes R. H., 1977. Induction of pure and sectored mutant clones in excision-proficient and deficient strains of yeast. Mutat. Res. 43: 327–338 [DOI] [PubMed] [Google Scholar]
- Esposito M. S., 1978. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. USA 75: 4436–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F., 1978. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature 272: 795–798 [DOI] [PubMed] [Google Scholar]
- Fachinetti D., Bermejo R., Cocito A., Minardi S., Katou Y., et al. , 2010. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell 39: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin W. A., Doetsch P. W., Haseltine W. A., 1985. Structural determination of the ultraviolet light-induced thymine-cytosine pyrimidine-pyrimidone (6–4) photoproduct. Nucleic Acids Res. 13: 5317–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz A., et al. , 2006. DNA Repair and Mutagenesis. American Society for Microbiology (ASM), Washington, DC [Google Scholar]
- Galli A., Schiestl R. H., 1999. Cell division transforms mutagenic lesions into deletion-recombinogenic lesions in yeast cells. Mutat. Res. 429: 13–26 [DOI] [PubMed] [Google Scholar]
- Goeke E., Manney T. R., 1979. Expression of radiation-nduced mutations at the arginine permease (CAN1) locus in Saccharomyces cerevisiae. Genetics 91: 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Curry B., Ward A., Gordon D. B., Brizuela L., et al. , 2010. Optimized detection of sequence variation in heterozygous genomes using DNA microarrays with isothermal-melting probes. Proc. Natl. Acad. Sci. USA 107: 1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego [Google Scholar]
- Heyer W. D., Ehmsen K. T., Liu J., 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44: 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks W. M., Kim M., Haber J. E., 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329: 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K., Mazon G., Lam A. F., Symington L. S., 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genetic integrity in budding yeast. Mol. Cell 40: 10077–10082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. P., Kilbey B. J., 1977. The timing of UV mutagenesis in yeast: a pedigree analysis of induced recessive mutation. Genetics 87: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Allers T., Lichten M., 2005. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169: 1353–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H., 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D. T., Dominska M., Petes T. D., 1998. Conversion-type and restoration-type repair of DNA mismatches formed during meiotic recombination in Saccharomyces cerevisiae. Genetics 149: 1693–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., Pierce M. K., Mis J. R., Giroux C. N., 1987. DNA sequence analysis of the mutational specificity of u. v. light in the SUP4-o gene of yeast. Mutagenesis 2: 445–453 [DOI] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178: 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., 2002. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst.) 1: 425–435 [DOI] [PubMed] [Google Scholar]
- Lee P. S., Petes T. D., 2010. From the cover: mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc. Natl. Acad. Sci. USA 107: 7383–7388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. S., Greenwell P. W., Dominska M., Gawel M., Hamilton M., et al. , 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5: e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase alpha: a model for chromosome fragile sites. Cell 120: 587–598 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Tanenbaum D. M., Daly M. J., Winchester E., Lui W. O., et al. , 2000. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol. 18: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley J. L., Petes T. D., 2010. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc. Natl. Acad. Sci. USA 107: 11465–11470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M. A., Gottschling D. E., 2003. An age-induced switch to a hyper-recombinational state. Science 301: 1908–1911 [DOI] [PubMed] [Google Scholar]
- Merker J. D., Dominska M., Petes T. D., 2003. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 165: 47–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K., Zhang H., Welz-Voegele C., Jinks-Robertson S., 2010. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol. Cell 38: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S., Mortimer R. K., 1969. Studies of the mechanism of radiation-induced mitotic segregation in yeast. Mol. Gen. Genet. 103: 329–338 [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Sweetser D. B., Clikeman J. A., Khalsa G. J., Wheeler S. L., 1999. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics 153: 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S., Schildkraut E., Lazarin R., Nguyen J., Nickoloff J. A., 2003. Gene conversion tracts in Saccharomyces cerevisiae can be extremely short and highly directional. Nucleic Acids Res. 31: 1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 53: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peak M. J., Peak J. G., 1986. DNA-to-protein crosslinks and backbone breaks caused by far- and near-ultraviolet, and visible light radiations in mammalian cells. Basic Life Sci. 38: 193–202 [DOI] [PubMed] [Google Scholar]
- Petes T. D., Hill C. W., 1988. Recombination between repeated genes in microorganisms. Annu. Rev. Genet. 22: 147–168 [DOI] [PubMed] [Google Scholar]
- Petes T. D., Malone R. E., Symington L. S., 1991. Recombination in yeast, pp. 407–521 The Molecular and Cellular Biology of the Yeast Saccharomyces, edited by Broach J. R., Jones E. W., Pringle J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Reynolds R. J., 1987. Induction and repair of closely opposed pyrimidine dimers in Saccharomyces cerevisiae. Mutat. Res. 184: 197–207 [DOI] [PubMed] [Google Scholar]
- Roitgrund C., Steinlauf R., Kupiec M., 1993. Donation of information to the unbroken chromosome in double-strand break repair. Curr. Genet. 23: 414–422 [DOI] [PubMed] [Google Scholar]
- Setlow R. B., 1966. Cyclobutane-type pyrimidine dimers in polynucleotides. Science 153: 379–386 [DOI] [PubMed] [Google Scholar]
- Shkundina I. S., Ter-Avanesyan M. D., 2007. Prions. Biochemistry (Mosc.) 72: 19–36 [DOI] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L. P., Nickoloff J. A., 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Shafer B. K., McGill C. B., 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Petes T. D., 1988. Expansions and contractions of the genetic map relative to the physical map of yeast chromosome III. Mol. Cell. Biol. 8: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Dominska M., Greenwell P. W., Harvenek J., Lobachev K., et al. , 2011. Friedreich’s ataxia (GAA)/(TTC) repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 7: e1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. F., 1990. The yield of DNA double-strand breaks produced intracellularly by ionizing radiation: a review. Int. J. Radiat. Biol. 57: 1141–1150 [DOI] [PubMed] [Google Scholar]
- Wei W., McCusker J. H., Hyman R. W., Jones T., Ning Y., et al. , 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. USA 104: 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., 1997. Processing of recombination intermediates by the Ruv ABC proteins. Annu. Rev. Genet. 31: 213–244 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Castillo-Davis C. I., Oshiro G., Liang D., Richard D. R., et al. , 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sterling J., Storici F., Resnick M. A., Gordenin D. A., 2008. Hypermutability of damaged single-strand DNA formed by double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 4: e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.