Abstract
We address a conceptual flaw in the backward-time approach to population genetics called coalescent theory as it is applied to diploid biparental organisms. Specifically, the way random models of reproduction are used in coalescent theory is not justified. Instead, the population pedigree for diploid organisms—that is, the set of all family relationships among members of the population—although unknown, should be treated as a fixed parameter, not as a random quantity. Gene genealogical models should describe the outcome of the percolation of genetic lineages through the population pedigree according to Mendelian inheritance. Using simulated pedigrees, some of which are based on family data from 19th century Sweden, we show that in many cases the (conceptually wrong) standard coalescent model is difficult to reject statistically and in this sense may provide a surprisingly accurate description of gene genealogies on a fixed pedigree. We study the differences between the fixed-pedigree coalescent and the standard coalescent by analysis and simulations. Differences are apparent in recent past, within ∼ <log2(N) generations, but then disappear as genetic lineages are traced into the more distant past.
IN the early 1980s, Hudson (1983a,b) and Tajima (1983) reframed population genetics in terms of gene genealogies, which are the ancestral relationships among samples of genetic data from a population. Gene genealogies exist, but they are generally unobservable and random models are used to describe them. Patterns of genetic variation result from mutations along the lineages of the gene genealogy. Today, the theory of gene genealogies is central to both mathematical and empirical population genetics. This theory has been developed for diploid as well as haploid organisms and further extended to include migration, selection, recombination, and other important biological phenomena; see reviews by Hein et al. (2005) and Wakeley (2008).
Kingman (1982a,b,c) gave a mathematical proof of the basic model, his “n-coalescent,” which holds for a sample of size n from a large, well-mixed population of constant size in which all genetic variation is selectively neutral. The gene genealogy at a single locus without recombination is modeled as the outcome of a continuous-time process in which binary mergers between ancestral genetic lineages (coalescent events) occur with rate equal to one independently for each pair of lineages. The process stops at the (n − 1)th coalescent event, that is, when the most recent common ancestor of the entire sample is reached. The result is a random binary tree with associated coalescence times and is interpreted as a pseudosample from a prior distribution of gene genealogies.
Coalescent models are used to describe the distribution of genetic diversity across the genome. For loci that are far enough apart to segregate independently, the coalescent is applied separately at each locus, as in Figure 2 of Garrigan and Hammer (2006), which compares coalescent predictions to the distribution of inferred times to the most recent common ancestor for 51 human loci. A more recent example is Huff et al. (2010), who contrasted the distribution of genetic diversity in samples of size n = 2 among 2432 randomly selected loci to that among 610 loci that each contained a rare insertion sequence in one of the two samples. For linked loci, the coalescent with recombination is used, as in the recent articles by Gronau et al. (2011) and Li and Durbin (2011).
We are concerned with the application of coalescent models to data from diploid organisms. A number of processes conspire to produce such data. Individuals are born, live for some time, and then die. During their lives, they move around their habitat, find potential mates, and mate either successfully or not. Population-genetic models include specific assumptions about these processes. However, with respect to genetic variation, the crucial product of all of them is the population pedigree. The population pedigree is the set of all family relationships among members of the population for every generation. Genetic lineages are transmitted through this pedigree, forward in time, according to Mendel’s laws of independent segregation and, possibly, independent assortment. The validity of Mendel’s laws is well accepted, while the processes by which population pedigrees are laid down, and the role of randomness therein, are poorly known.
All of these processes have already occurred by the time we take a sample of genetic data from the population. Importantly, there is just one population pedigree. Within this single pedigree, to the extent that different loci have assorted independently into gametes, a large number of gene genealogies may exist across the genome. There is in fact no randomness either in the pedigree or in the collection of gene genealogies across the genome. They surely exist, fixed by past events. The only uncertainty about them is our own lack of knowledge. The appropriate statistical analogy for samples of genetic data and their underlying gene genealogies is the framework of survey sampling, in which the experimenter samples randomly from an existing population.
Because the relative frequencies of gene genealogies across the genome result from Mendelian inheritance within a single pedigree, our random model for the distribution of gene genealogies across the genome should include a single pedigree within which genetic lineages percolate to form gene genealogies. The coalescent does not do this. When applied to unlinked multilocus data, the coalescent implicitly generates a new random pedigree for every locus. Even for single-locus or linked genetic data, the predictions of the coalescent do not reflect the effects of pedigree structure because they are obtained by averaging over the process of reproduction within each generation.
The effects of pedigree structure on gene genealogies can be dramatic because the ancestral lineages of a sample are restricted to the pedigree ancestors of the sampled individuals. Samples containing sibs or cousins can be detected—see Huff et al. (2011), for example—and the Kingman coalescent statistically rejected on the basis of the distribution of coalescence times or patterns of haplotype sharing across the genome. In a large population, most samples will not contain sibs, first cousins, or even second cousins. However, the Kingman coalescent is also inappropriate in this case because coalescent events will be impossible in the first, second, and third generations in the past.
The random experiment envisioned in the coalescent is apparent in its mathematical derivation, even for a sample of size n = 2. Starting, as Kingman did, with the haploid exchangeable models of Cannings (1974), the standard derivation of the coalescent proceeds by calculating
| (1) |
in which νi is the number of offspring of (haploid) individual i, one of N possible parents in the immediately previous generation. The sum in Equation 1 adds up the probabilities of coalescence over all of the N possible parents. There is only one expected value on the right in Equation 1 because exchangeability means that E[νi(νi − 1)] = E[νj(νj − 1)] for all i and j. Since E[ν1] = 1 in a population of constant size, expected value E[ν1(ν1 − 1)] is equal to the variance of the number of offspring of a single (haploid) individual, which is often denoted σ2.
Mathematically, the continuous-time Kingman coalescent exists for a sample of finite size n in the limit N → ∞. To obtain the coalescence rate of one per pair of lineages, time must be measured in appropriate units. If the natural unit of time is one generation, then the rescaled unit of time will be cN generations, where c is a constant that depends on the details of demography and reproduction in the population (Sjödin et al. 2005). Following Equation 1, Kingman obtained c = σ−2 for a subset of the haploid exchangeable models of Cannings (1974). Later, Möhle (1998a,b) proved that the same coalescent process holds under the usual diploid, two-sex, Wright–Fisher model if c = 8r(1 − r), where r is the fraction of the population that is female.
The point we wish to emphasize does not lie in the technical details of taking limits and rescaling time, but rather in the fact that all derivations of the coalescent begin, either implicitly or explicitly, by averaging over the random process of reproduction within each generation. The expectations in Equation 1, which in this case are taken over the distribution of haploid offspring numbers (ν1, ν2, … , νN), are precisely such averages. For the diploid monoecious Wright–Fisher model, this averaging is the source of the familiar P(coal) = 1/(2N), or c = 2. The coalescent analysis for a sample from a diploid two-sex population involves additional formalism (Möhle 1998b), but the concept of averaging over all possible outcomes of the process of reproduction, in taking expectations as in Equation 1, is identical. This is true as well for derivations that consider recombination (Hudson 1983b; Hudson and Kaplan 1985, 1988).
Thus the Kingman coalescent and its extensions do not describe the process of coalescence within fixed population pedigrees. We study this process, using simulations to generate pedigrees and also to construct gene genealogies within pedigrees. We consider a number of methods of generating pedigrees, including the canonical diploid two-sex Wright–Fisher model and a novel method of joining families into a population pedigree. In the latter case, we employ family data from 19th century Sweden (Low and Clarke 1991, 1992) and account for known rates of marriages between cousins (Bittles and Egerbladh 2005). We also consider a restricted version of the Wright–Fisher model, in which there is just a single generation of random mating. The motivation for including such a range of pedigrees is to ask whether our results hold only for highly idealized models, such as the Wright–Fisher model, or apply more broadly.
The idea that the population pedigree might constrain gene genealogies is not new, of course. Some of the results we present are foreshadowed (see Discussion) in the simulation studies of Avise and colleagues (Ball et al. 1990; Wollenberg and Avise 1998; Kuo and Avise 2008). In addition, Derrida et al. (2000) and Barton and Etheridge (2011) studied related problems mathematically, providing a basis for our heuristic analyses of coalescence within a fixed pedigree. The foundation of all these works is the fact that, under biparental reproduction, the number of ancestors of each individual increases by a factor of 2 each generation. If N is the population size, then on the order of log2(N) generations ago the numerous ancestors of the sample overlap completely (Chang 1999), and this results in a nearly constant probability of coalescence in each generation.
After having gone to some length in this Introduction to demonstrate that the coalescent is generally misapplied, our results will show that the Kingman coalescent provides a surprisingly accurate description of gene genealogies within fixed pedigrees. More precisely, it can be difficult to reject the Kingman coalescent even with a great deal of data. Of course, this is not true for all pedigrees and samples. Gene genealogies in general depend on the details of pedigree structure. Even when the Kingman coalescent cannot easily be rejected, on average, the distribution of gene genealogies constrained by a population pedigree is different from that predicted by the coalescent. However, in well-mixed populations, these differences are restricted to the most recent log2(N) generations or some small multiple thereof. We describe these differences using simulations and derive heuristic mathematical predictions that fit key aspects of our simulation results.
Methods and Results
We developed several pieces of software. Programs sufficient to reproduce all of the results we present are available at www.oeb.harvard.edu/faculty/wakeley. In general, these programs work by constructing and storing a population pedigree and then following ancestral genetic lineages backward in time within that pedigree. One set of programs simply recorded pairwise times to common ancestry, while another generated full gene genealogies for larger samples. In all programs, individuals are diploid and divided into males and females, so there is no possibility of selfing. We do not explore the consequences of uneven sex ratio, although in the pedigrees generated using the Swedish family data we allowed small variations in male and female population sizes.
Mendel’s law of independent segregation was implemented backward in time by tracing lineages to the mother or father of any individual uniformly (i.e., with a 50:50 chance). When two lineages trace back to the same individual, they coalesce with probability 1/2; otherwise they remain distinct. When two lineages are in a single individual and are distinct, one of them traces back to the mother and the other traces back to the father. Again, which was which was determined randomly and uniformly. Mendel’s law of independent assortment was modeled by performing the above independently for each genetic locus, but on the same pedigree. Finally, we assume that within a locus there is no recombination and between loci there is free recombination.
Pedigrees based on Wright–Fisher reproduction
Wright–Fisher pedigrees were constructed by choosing one female parent and one male parent at random, uniformly among N/2 females and N/2 males, for each of the N individuals in the population. This random sampling was performed independently for 30N generations to decrease the chance that the most recent common genetic ancestor of the sample would not occur within the pedigree. Under the Kingman coalescent as it is applied to the Wright–Fisher model, i.e., with a coalescence rate of one per 2N generations, this corresponds to a probability of exp(−15) ≈ 3 × 10−7 that a single pair of lineages will not coalesce within the pedigree. However, to ensure that a common ancestor was reached at every locus, we reused the same pedigree by letting the individuals in past generation 30N be identical to the individuals in generation zero (i.e., the present), so that a tiny fraction of genealogies actually loop back through the pedigree.
In an effort to isolate the different sources of randomness in the process of coalescence within a fixed pedigree, we considered a “cyclical” Wright–Fisher model. In this model, the parents of the current generation, labeled zero, were chosen by Wright–Fisher sampling. Then these exact parent–offspring relationships were used to specify the parents of the individuals in generation one and again in every generation in the past. To explain, the individuals in each generation are stored as elements in an array of length N. If the individual in position i in generation zero had the individuals in positions j and k as her parents (in generation one), then the individuals in position i in every generation had the individuals in positions j and k in the previous generation as their parents.
The cyclical Wright–Fisher model has no biological interpretation. Our purpose in studying it is that cyclical pedigrees have less randomness of reproduction than standard Wright–Fisher pedigrees. In particular, because relationships in every generation are identical, they very clearly do not conform to the assumptions of the usual derivation of the coalescent, which requires taking an expectation over the process of reproduction as in Equation 1. Note that it is possible in this case for a pedigree to be composed of two or more disjoint pedigrees. We did observe this, but only rarely and only for N = 100. We excluded these disjoint pedigrees from our results.
Pedigrees based on 19th century Swedish families
In addition to the idealized Wright–Fisher models, we constructed pedigrees using the family data from 19th-century Sweden described in Low and Clarke (1991, 1992). These data were gathered from the records of seven parishes—Fleninge, Gullholmen, Locknevi, Nedertorneå, Svinnegarn, Tuna, and Trosa—and contain all men married between 1824 and 1840 and all of their descendants up to 1896 (1922 in the case of Gullholmen). There are 512 of these extended families, which vary in size from 3 individuals (i.e., two parents and their child) to 865 individuals and from two generations to five generations in length.
To facilitate the construction of population pedigrees, we extracted all two-generation families from these data. We define a two-generation family to be a set of all siblings, half siblings, and their parents. To illustrate, an extended family containing a granddaughter, her two parents, and her four grandparents would yield 3 two-generation families, each containing two parents and one child. Applying this to the entire data set resulted in 1884 two-generation families (hereafter “Swedish families”) containing a total of 3856 daughters, 4033 sons, 2200 mothers, and 2251 fathers. These data are available at www.oeb.harvard.edu/faculty/wakeley.
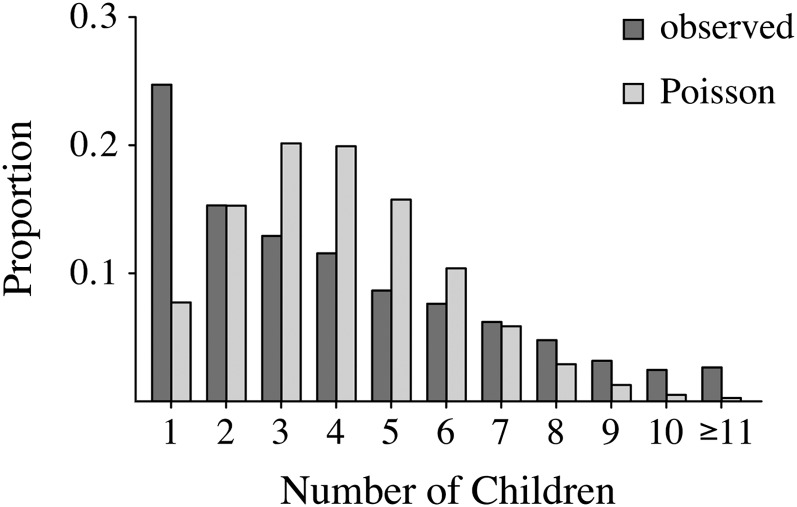
Although we have abstracted the data considerably, note that these Swedish families exhibit a rather different structure from families generated by Wright–Fisher reproduction. For example, 1549 (82.2%) of the Swedish families are monogamous: neither the mother nor the father had children with any other partners. A randomly chosen child has a 77.6% chance of being born to monogamous parents. Such high levels of monogamy are unlikely in all but the smallest Wright–Fisher populations. In addition, the distribution of the number of offspring in the data is very different from the Poisson distribution expected under the Wright–Fisher model (see Figure 1). This is likely due to the differential distribution of wealth, as discussed in Low and Clarke (1991).
Figure 1 .
The observed distribution of the number of children of monogamous parents in the Swedish family data compared to a Poisson distribution with the same mean (∼3.95) and conditional on there being at least one child.
We constructed population pedigrees from the Swedish family data using three different methods, which we designate “random, sibs,” “random, no sibs,” and “cousins, no sibs.” In all cases generations were forced to be nonoverlapping and the population size was held constant over time. In random, sibs, mating occurred at random, allowing the possibility of sib mating. The other two methods reflect aspects of human reproductive behavior. In random, no sibs, mating still occurred at random, but mating between siblings was barred. Finally, in cousins, no sibs, mating was also barred between siblings and the population was structured to reproduce the rates of marriage (here reproduction) between first and second cousins in 19th century Northern Sweden presented in Table 1 of Bittles and Egerbladh (2005).
Consider the simplest method (random, sibs) applied to the entire data set of 1884 families containing 3856 daughters, 4033 sons, 2200 mothers, and 2251 fathers. The population size is fixed at 3856 + 4033 = 7889 and in every generation 2200 females and 2251 males reproduce successfully, precisely according to the structures of the 1884 single-generation families. Thus, this entire set of single-generation families is reused to represent parent–offspring relationships in each past generation. Our three methods differ in the way in which generations are linked together by mapping the children in these families onto the parents in the next (descendant) generation. In random, sibs, children of parents in generation two in the past are equated with parents in generation one in the past by sampling 2200 daughters and 2251 sons at random without replacement.
The method we call random, no sibs is identical to random, sibs except that these random assignments of children to parents are rejected if they would result in reproduction between siblings and are resampled until a nonsib pair is found.
In cousins, no sibs, we made an effort to account for known rates of marriage between cousins. In the period from 1820 to 1899, the numbers in Table 1 of Bittles and Egerbladh (2005) show that 2.56% of marriages were between first cousins and 2.65% were between second cousins. These rates are higher than those obtained under either model of random mating using the full data of 1884 families. We increased the rates of consanguineous matings by subdividing the population into demes between which there was limited migration. For computational simplicity we measured deme size by the number of families in each deme (Fdeme). Because Fdeme generally would not divide the number of families evenly, the remainder of families was added to one of the demes. Once demes were established, a number of parents (M, divided equally into males and females) were reassigned to randomly chosen demes. The mapping of children to parents in the next (descendant) generation occurred as in random, no sibs but separately within each deme.
On the basis of simulated cousin rates obtained for the full data of 1884 families, we chose Fdeme = M = 30. This gave rates of 2.0% and 2.9% for first-cousin matings and second-cousin matings, respectively. It was not possible to match both the first-cousin and the second-cousin rates perfectly. In addition, rates of third-cousin matings in our simulated pedigrees (not shown) were generally higher than those in Bittles and Egerbladh (2005). Our difficulty in fitting these rates using a random model is not surprising, since prospective couples actively respond to a complex set of social factors and very likely know their relationship.
In constructing pedigrees of a given size (with <7889 individuals), we created subsets of the 1884 Swedish families by sampling families randomly without replacement. Because these families are of fixed sizes, we had to allow some variation in the sizes of populations. For each subset, we required that the total number of children be within 2.5% of the desired size. To be able to link generations by mapping children onto parents in each generation, we also required that the number of female (resp., male) children was greater than the number of mothers (resp., fathers). If a subset of families did not meet these criteria, we rejected it and sampled another subset. As in our Wright–Fisher pedigrees, these pedigrees from the Swedish family data were composed of a fixed number of generations, in this case 2000 for computational reasons, and any lineages that did not coalesce by then would loop back through the pedigree.
Since the derivation of the Kingman coalescent is invalid for any given pedigree, we investigated the power of two tests of the coalescent model, using data simulated on fixed Wright–Fisher and Swedish family pedigrees.
A chi-square test of the coalescent
We considered the power to reject the simplest prediction of the Kingman coalescent—that coalescence times for a sample of size n = 2 should follow an exponential distribution—using pairwise coalescence times at independent loci simulated on fixed pedigrees. For each of six different population sizes, from 250 to 8000 on a log2 scale, we generated 2000 pedigrees. For each pedigree, we sampled 2 distinct individuals randomly from the current generation. We then simulated 1000 independent pairwise coalescence times (corresponding to the gene genealogies of 1000 independently segregating loci), each time starting from one gene copy in each of these 2 individuals. The maximum population size of N = 8000 was chosen to be close to the total number of children in our Swedish family data, which again was 7889.
From these 1000 coalescence times, which we assumed were known without error, we first computed the arithmetic mean coalescence time. We then compared the observed distribution of the 1000 coalescence times to an exponential distribution with the same mean. We used a simple chi-square goodness-of-fit test, in which we divided the sample space of the exponential distribution into 50 bins of equal probability (1/50 = 0.02) and counted the number of the 1000 loci that had coalescence times in each bin. For an exponential distribution with mean 1/λ, the first bin will include times between zero and −log(0.98)/λ and the 50th bin will include all times > −log(0.02)/λ. We thus expect to observe 20 loci in each bin. We performed a chi-square test with d.f. = 48 to assess the goodness of fit of the exponential.
As noted in the Introduction, when applied to multiple loci, the Kingman coalescent in effect assumes that each locus comes from an independent population, with its own independent pedigree. It is not feasible to perform this pseudoexperiment using our simulations for any but the smallest population sizes. Therefore, in addition to standard Wright–Fisher and one-generation cyclical Wright–Fisher pedigrees (in both cases with all 1000 single-locus coalescent simulations starting from the same two individuals sampled at random without replacement), we considered a third model that we expected would conform well to the Kingman coalescent. In this model, the 1000 coalescence times were generated on the same Wright–Fisher pedigree, but starting at a newly sampled pair of individuals for each locus.
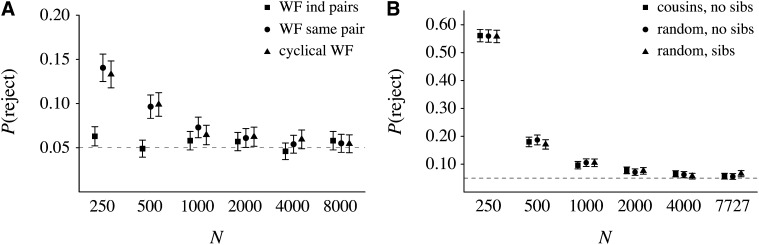
The results of these chi-square tests are displayed in Figure 2. As anticipated, the Kingman coalescent is not rejected for data from Wright–Fisher pedigrees with independently sampled individuals for each locus (Figure 2A, boxes). This shows that our minimum population size of N = 250 is large enough that the coalescent will not necessarily be rejected simply because it is a continuous-time model while time in our pedigrees is discrete. Figure 2A further shows that the Kingman coalescent is not rejected at above the nominal significance level for multilocus data from the same pair of individuals as long as the size of a Wright–Fisher population is a few thousand or larger.
Figure 2 .
Probabilities of rejecting the Kingman coalescent using coalescence times on a fixed pedigree and the chi-square test described in the text. (A and B) The mean and estimated 95% confidence intervals for the probability of rejecting the coalescent using 1000 independent coalescence times, for a series of population sizes, under (A) three different Wright–Fisher pedigree models and (B) three different methods of building pedigrees from the Swedish family data. Dashed lines show the nominal significance level of the tests, which was 5%.
Although our simple chi-square test may not be the most powerful test, the fact that coalescence within pedigrees under the standard Wright–Fisher model leads only to nominal rejection probabilities when the population size is not too small illustrates what Ball et al. (1990, p. 365) noted in their simulations: “Results suggest that gene lineages transmitted through a single organismal pedigree show nearly as much independence as do gene lineages traced through separate organismal pedigrees generated under a common set of demographic conditions.” Probably the most surprising result in Figure 2A is that rejection probabilities for one-generation cyclical Wright–Fisher pedigrees are indistinguishable from those for standard Wright–Fisher pedigrees. Repeated independent realizations of the process of reproduction in every generation are not required for pairwise coalescence times to appear exponential, at least by our approximate measure of the distribution.
Figure 2B shows the chi-square results for the three different methods of constructing pedigrees from the Swedish family data. Rejection probabilities for all three types of Swedish pedigrees are very similar. As with the Wright–Fisher pedigrees, they approach the nominal significance level when the population size is a few thousand or more. However, as a result of differences in the distribution of family sizes and propensity toward monogamy, for small populations the rejection probabilities for pedigrees from the Swedish family data are greater than those for cyclical and standard Wright–Fisher pedigrees.
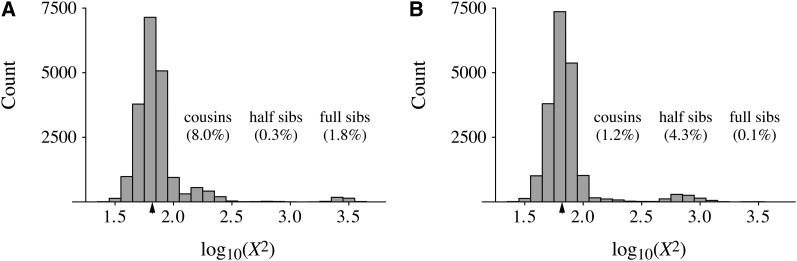
These differences in family structure can be seen in the distribution of the chi-square statistic among samples within a single pedigree. Figure 3 depicts two such distributions, among 20,000 data sets for each of two pedigrees: one with N = 250 individuals (specifically, 129 females and 123 males) constructed from the Swedish family data and one with N = 96 individuals (chosen to yield approximately the same chi-square rejection probability) constructed by Wright–Fisher reproduction. While the bulk of the distributions in Figure 3, A and B, is similar, the right-hand tails are different due to differences in the occurrence of full sibs, half sibs, and cousins in the two pedigrees.
Figure 3 .
(A and B) Distributions of chi-square values (X2 on a log10 scale) among 20,000 randomly sampled pairs of individuals on (A) one pedigree from the Swedish family data containing 129 females and 123 males in each generation and (B) one Wright–Fisher pedigree containing 48 females and 48 males in each generation. The overall rejection probabilities were approximately the same for the two pedigrees: 0.535 vs. 0.529. Peaks in the right-hand tails are labeled by the relationship of the two sampled individuals, and the frequencies of each relationship among the 20,000 samples are given in parentheses. Minor differences in these frequencies occur among pedigrees (results not shown) but the overall patterns are robust. Triangles mark the chi-square cutoff for 5% significance.
Tajima’s D at 10 independent loci
Our application of the chi-square results presented in Figures 2 and 3 is unrealistic. On the one hand, we assumed that coalescence times could be known exactly. In truth, coalescence times can be inferred only with some error from genetic variation that has resulted from the stochastic process of mutation. On the other hand, we restricted ourselves to samples of size 2, while there will necessarily be more information in larger samples. To provide a more realistic picture of the power to reject the Kingman coalescent using data from a single population pedigree, we generated sequence data for samples of n = 20 and n = 100 under the infinite-sites mutation model (Kimura and Crow 1964; Watterson 1975). We modeled 10 independent loci, with mutation rates such that the expected number of pairwise sequence differences was equal to one at each locus.
We computed Tajima’s D from the pseudodata at each locus. Specifically,
where S is the observed number of polymorphic sites in the sample, is the average number of pairwise differences, and c1,n, c2,n, and c3,n are constants defined in Tajima (1989). We implemented a two-tailed test, with upper and lower critical values obtained from simulations of the Kingman coalescent with mutation parameter θ = 1. For n = 20, the cutoffs were −1.886 and 2.316, and for n = 100, they were −1.779 and 2.719. These cutoffs yielded approximately symmetrical tests at approximately the 1% significance level when applied to a single locus under the null model.
As in our chi-square test, we constructed 2000 population pedigrees. We then sampled n individuals from the current generation without replacement. For each of 10 loci, independently, we traced the ancestry of a sample of size n gene copies, one from each individual, backward in time. This resulted in 10 gene genealogies, each typically with its own branching structure and times to common ancestry. Although each population pedigree and sample has its own characteristic times to common ancestry, for Wright–Fisher pedigrees we found that these were generally close to the usual Wright–Fisher expectation of 2N generations. Therefore, if was the total length of the gene genealogy at locus i, then for pedigrees based on Wright–Fisher reproduction, we placed a Poisson-distributed number of mutations, with mean uniformly on the genealogy. For pedigrees from the Swedish family data, preliminary simulations were run for each population size so the mutation rate could be set to give an average number of pairwise differences equal to one.
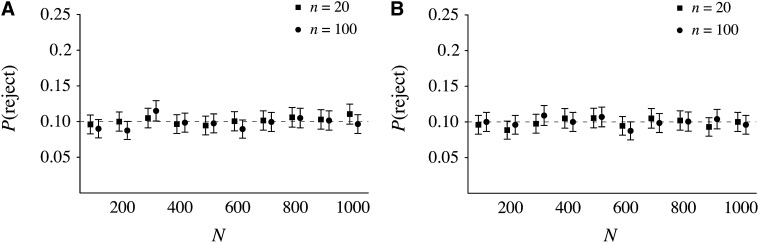
For each pedigree, we asked whether the data from ≥1 of the 10 loci rejected the null model at the 1% significance level. This corresponds to a 10-locus test at the 10% significance level, with a Bonferroni correction. On the basis of our expectation that this test would be less powerful than the chi-square test, we considered a series of 10 different population sizes from N = 100 to N = 1000. The results are displayed in Figure 4A for the cyclical Wright–Fisher model and in Figure 4B for the cousins, no sibs model with Swedish families. The results show zero power to reject the Kingman coalescent, even for a sample size equal to population size (n = 100, N = 100). The same was true for our other pedigree models (results not shown).
Figure 4 .
Probabilities of rejecting the Kingman coalescent using 10-locus data from a fixed pedigree and Tajimas’s D. (A and B) The mean and estimated 95% confidence intervals for the probability of rejecting the coalescent using Tajima’s D at 10 independent loci, for different sample sizes and population sizes, when the expected number of pairwise differences is equal to one, for (A) one-generation cyclical Wright–Fisher pedigrees and (B) pedigrees from the Swedish family data. Dashed lines show the nominal significance level of the test, which was 10%.
This lack of power may be due to several factors. The size and shape of gene genealogies are difficult to discern with random mutations and expected number of pairwise differences equal to one. However, the power to reject the null model remained insensitive to population size in subsequent simulations with mutation rates 10 or 100 times higher (results also not shown). Given the results in Figure 3, A and B, it seems likely that deviations in the size or shape of our simulated gene genealogies differ from those under the Kingman coalescent only very close to the tips, that is, only in the very recent ancestry of the sample. These times are characteristically very short, so regardless of the mutation rate only a small fraction of mutations will fall on this part of the gene genealogy. This basic intuition is borne out by our detailed examination of the distribution of pairwise coalescence times in the following section.
The shapes of coalescence-probability distributions
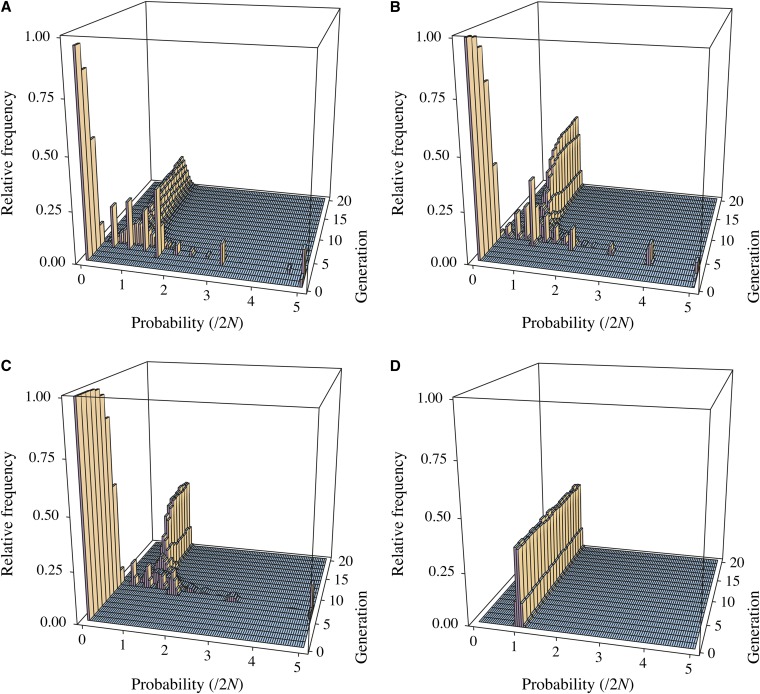
For each of three different population sizes (N = 102, N = 103, and N = 105), Figure 5 shows a series of histograms of the pairwise coalescence probability, one histogram for each of the past 20 generations. These coalescent-probability distributions are different from the usual distributions from the Kingman coalescent, which give the distribution of the time to the coalescent event between a pair of lineages, averaged over all possible pedigrees. Here, instead, we consider the pedigree to be fixed. Conditional on the pedigree and the sampled individuals, there is a fixed probability that two ancestral lineages from those individuals will coalesce in each generation. Figure 5 shows the distributions of these probabilities among pedigrees for each past generation.
Figure 5 .
Three-dimensional histograms of the probability that a sample of size two coalesces in each of the past 20 generations (g) on fixed Wright–Fisher pedigrees, for three different population sizes: (A) N = 102, (B) N = 103, and (C) N = 105. Within each generation, histograms show the relative frequency of the coalescence probability among 10,000 pedigrees. (D) These distributions for the case of independent samples from a geometric distribution with parameter 1/(2N), with N = 105 as in C.
The data for Figure 5 were generated by randomly constructing 10,000 standard Wright–Fisher pedigrees for each population size. The results for the cyclical Wright–Fisher model (not shown) are indistinguishable from those in Figure 5. Likewise, the results for the Swedish family data display the same overall patterns, with differences only in the recent generations (recall Figure 3). For each pedigree, two individuals were sampled randomly without replacement. Starting with this pair of sampled individuals, in the cases N = 102 and N = 103 (also N = 104, not shown), 107 ancestries of two gene copies, one from each individual, were simulated back to their most recent common genetic ancestor. In the case N = 105, 108 ancestries were simulated. The probability of coalescence in generation g for a given pedigree was estimated by the fraction of times (of 107 or 108) that the most recent common ancestor of the pair occurred in generation g.
The histograms for each generation give the relative frequency of these estimated probabilities of coalescence among the 10,000 pedigrees. These were constructed as follows. Estimated probabilities between zero and 5/(2N) were put into one of 100 possible bins, each of equal width 0.05/(2N). Thus the usual Wright–Fisher P(coal) = 1/(2N) is the cutoff between the 20th and the 21st bin. Estimated coalescence probabilities >5/(2N) were placed in an additional 101st bin. For example, two genetic lineages have a chance to coalesce in the immediately previous generation (g = 1 in Figure 5) only if the two sampled individuals share at least one parent. The probability of sharing a parent is inversely related to the population size, so most pedigree-sample cases have a coalescence probability equal to zero for the immediately previous generation. At the same time, if we were to average P(coal at g = 1) over pedigrees, we would obtain 1/(2N).
In considering the pattern of histograms in Figure 5, A–C, it is helpful to imagine how these would appear if coalescence times were drawn directly from a geometric distribution with mean equal to 2N, which is the standard Wright–Fisher prediction (averaged over pedigrees) that produces the Kingman coalescent. Figure 5D shows one such distribution, for N = 105. The distribution traces a sharp ridge along P(coal at g) = p(1 − p)g−1, with P = 1/(2N). This ridge has some width only due to the fact that P(coal at g) has been estimated from a finite number of simulated coalescence times. Note that for later generations the shape of the ridge in Figure 5C becomes indistinguishable from the one in Figure 5D. However, it emerges in Figure 5C only after ∼log2(N) generations in the past, while more recent generations show complicated patterns of coalescence-probability distributions. For reference, log2(102) ≈ 6.6, log2(103) ≈ 10.0, and log2(105) ≈ 16.6.
Consistent with the results in Figure 2A, we also observe a simple ridge like the one in Figure 5D if we obtain the distribution of coalescence times for each pedigree by resampling pairs of individuals, although we do not show these results.
Heuristic analysis of the coalescence-probability distribution within a generation
These complicated distributions and the transition to a simple ridge depend on the overlap in the family trees of the two sampled individuals. Tracing backward in time, common pedigree ancestors accumulate rapidly because the expected number of ancestors of an individual increases twofold each generation. Chang (1999) showed that a common pedigree ancestor of everyone in the current generation would occur at ∼log2(N) generations in past and that all family trees would overlap completely after ∼1.77 log2(N) generations. Derrida et al. (2000) showed that the family trees of a pair of individuals overlap completely over this same time frame. Matsen and Evans (2008) showed that the first occurrence of overlap may involve multiple common pedigree ancestors.
We can gain some intuition about the features of the complicated distributions in Figure 5 from the following approximate calculation, which applies to Wright–Fisher pedigrees with equal numbers of males and females. A single individual has 2g ancestors in generation g in the past. If the first occurrence of shared pedigree ancestry in the family trees of a pair of individuals is a single common ancestor between the two in generation g, then the probability of coalescence is given by
| (2) |
Equation 2 is the product of three probabilities: the probability that the genetic lineage from the first individual traces back to the common pedigree ancestor (1/2g), the probability that the genetic lineage from the second individual also traces back to this individual (1/2g), and the probability the two lineages coalesce when they reach that ancestor (1/2).
We can apply Equation 2 to Figure 5C. In particular, the first nonzero mode in generation 8 of the coalescence-probability distribution in Figure 5C has its tallest bar at histogram bin 31. This bin includes probabilities between 7.50 × 10−6 and 7.75 × 10−6. If we put g = 8 in Equation 2, we obtain a coalescence probability of 7.63 × 10−6. Thus, this first nonzero mode corresponds to the occurrence of a single common pedigree ancestor in the families of the two sampled individuals in generation g = 8. A similar calculation shows that the second nonzero mode in Figure 5C at g = 8 corresponds to there being two common pedigree ancestors.
Numerical calculation of coalescence probabilities on fixed pedigrees
If N is not too large—up to a few thousand—simulations can be replaced by exact numerical calculations. In particular, it is possible to compute the full joint distribution of the locations of a pair of lineages among the individuals in the population in each past generation. The following can be considered a special case of the very general method of Cannings et al. (1978) or an extension (to account for coalescence) of the method of Derrida et al. (2000). Further, the matrix approach of Barton and Etheridge (2011) could in principle be extended to the problem of two lineages and would provide an avenue to rigorous asymptotic analysis of their joint distribution. In the Supporting Information of Barton and Etheridge (2011), an efficient way to compute probabilities of identity by descent on a pedigree is described. Here, we use the full joint distribution of the locations of a pair of lineages among individuals to compute probabilities of coalescence in each generation. Note that computing the entire joint distribution is required for computing probabilities of coalescence exactly.
Let wij(g) be the probability that sampled lineage 1 is in individual i ∈ (1, … , N) and sampled lineage 2 is in individual j ∈ (1, … , N) in past generation g, given that the two lineages did not coalesce in any of the intervening generations: 1, 2, … , g − 1. As in our simulations, we imagine the individuals in any generation lined up in an array of length N. Thus, these wij(g) compose an N × N matrix whose entries sum to one for every g. Initially, in the current generation g = 0, a single entry in the matrix is equal to one, with i and j corresponding to the locations of the two sampled individuals, and all other entries are equal to zero. For a given fixed pedigree, we compute the entries wij(g) backward in time from the entries wij(g − 1), using two intermediate probabilities: is the probability the two lineages trace back to individuals i and j, and is the probability the two lineages trace back to individuals i and j and do not coalesce.
We begin by setting all elements equal to zero. For individuals i and j in generation g − 1, let iM and iF be the mother and father of individual i and let jM and jF be the mother and father of individual j. Considering every i and j, (1) if i = j, then one-half of wij(g − 1) is added to and one-half to , and (2) if i ≠ j, then one-fourth of wij(g − 1) is added to each of , , , and . These rules result from the fact that each lineage is equally likely to trace back to either parent. Now, if two lineages trace back to the same parent, they coalesce with probability 1/2. Therefore, for each i and j, (1) if i = j, , and (2) if i ≠ j, . The overall probability of coalescence is given by
| (3) |
To condition on the absence of a coalescent event in generation g, we let
and this completes the calculation for past generation g.
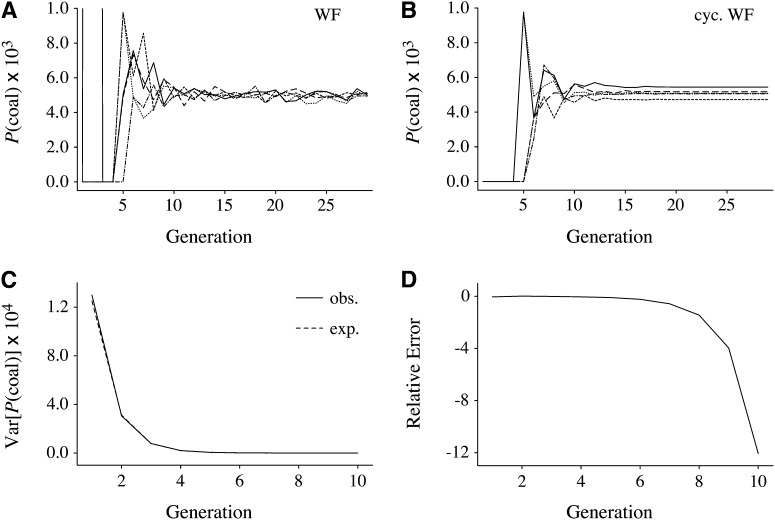
Figure 6 shows the probability of coalescence in each of the past 29 generations for five different pedigrees of N = 1000 individuals (500 males and 500 females). These were computed exactly for each pedigree using Equation 3, beginning with two lineages in a pair of individuals sampled randomly without replacement. The five trajectories in Figure 6, A and B, are for five pedigrees simulated under the standard Wright–Fisher model and under the cyclical Wright–Fisher model, respectively. In contrast to Figure 5, here it is possible to see the particular values for each pedigree in each generation. Another, minor difference is that Figure 6 shows coalescence probabilities conditional on not having coalesced in any intervening generation (1, … , g − 1), while Figure 5 simply depicts the probabilities of coalescence in each generation.
Figure 6 .
Numerical analysis and simulations of pairwise coalescence probabilities in past generations. (A and B) The probabilities of coalescence for a sample of size n = 2 from a population of size 1000 at each generation in the past given no coalescence up to that generation, computed analytically using Equation 3 for five Wright–Fisher and five cyclical Wright–Fisher pedigrees, respectively. (C and D) Comparison of analytical and simulation results for the variance of the probability of coalescence in each generation in the past for a population of size N = 500. C shows both the expectated variances (exp.) given by Equation 4 and the observed variances (obs.) among 100,000 simulated Wright–Fisher pedigrees. D shows the relative error, (obs. – exp.)/exp., in each generation for the same data as in C.
Figure 6A demonstrates that for a given Wright–Fisher pedigree and pair of sampled individuals, the probability of coalescence varies greatly in the recent generations. For example, in generation one this probability will be 1/4 for full sibs, 1/8 for half sibs, and zero for all other relationships. After some small multiple of log2(N) generations, it settles near 1/2N, but with some variation even in the distant past due to stochasticity in the parent–offspring relationships in each generation. Figure 6B shows that a very similar phenomenon holds for cyclical Wright–Fisher pedigrees. However, because now the offspring distribution does not vary from one generation to the next, each cyclical pedigree has its own characteristic probability of coalescence. The trajectories in Figure 6B become completely flat after ∼log2(N) generations.
Recall that the Kingman coalescent assumes a constant probability of coalescence in every generation. The flat lines in Figure 6B explain the result suggested by Figures 2A and 4A. Namely, the Kingman coalescent is a reasonable approximation to the ancestral process on fixed, cyclical Wright–Fisher pedigrees (albeit with an “effective population size” that differs slightly from 2N). For standard Wright–Fisher pedigrees, the Kingman coalescent is also a reasonable approximation, because the fluctuations in the probability of coalescence around 1/2N are small.
Similarly to Equation 2, we can interpret the results in Figure 6, A and B, simply from the initial geometric increase of the number of ancestors of the sampled individuals. Assume that N is large and g small, so that the numbers of ancestors of each individual make up a small fraction of the population (2g ≪ N). In this case, the recent ancestries of two lineages will include either one shared pedigree ancestor or none. Ignoring events in generations 1 through g − 1, we have
The probability of one shared pedigree ancestor results from the fact that each of the first individual’s 2g pedigree ancestors could be among the second individual’s pedigree ancestors, which compose a fraction 2g/N of the population. Also, following Equation 2, we have
Then, the first two moments of P(coal) at generation g are
| (4) |
Under the initial assumption that 2g ≪ N, we have Var[P(coal) at g] ≈ E[P(coal)2 at g].
This approximate derivation shows that, for a randomly sampled Wright–Fisher pedigree, the expected probability of coalescence in each generation is the same and equal to the usual value 1/2N. It further shows that variance of the probability of coalescence in each generation among randomly sampled Wright–Fisher pedigrees should begin at a relatively large value in generation g = 1 and then decrease by a factor of 4 in subsequent generations. We do not expect this level of decrease to continue for very many generations, however, because it depends on the assumption that 2g ≪ N. Figure 6C compares Equation 4 to the observed variances among 100,000 Wright–Fisher pedigrees for a population of size 500. Although the approximation captures the initial behavior well, Equation 4 tends to zero as g grows, and this contradicts the pattern in Figure 6A, which depicts a nonzero, but small, level of variation in P(coal) among pedigrees in later generations. The relative error of Equation 4 is shown in Figure 6D. Results for the cyclical Wright–Fisher model (not shown) are indistinguishable from those in Figure 6, C and D.
Numerical calculations of coalescence probabilities can also be performed on pedigrees constructed from the Swedish family data. The results (not shown) are broadly the same as those in Figure 6, but with differences due to the detailed structure of the Swedish families that are rather unlike the families that result from Wright–Fisher reproduction, as noted previously.
Discussion
We have studied the process of coalescence within single fixed pedigrees. We have mimicked the sampling of gene genealogies underlying multilocus genetic data close to how it actually occurs, rather than as it is conceptualized in the derivation of the Kingman coalescent, which averages over pedigrees. We considered standard Wright–Fisher pedigrees, cyclical Wright–Fisher pedigrees, and pedigrees constructed from a large data set of families from 19th century Sweden. The results reveal that, for these types of pedigrees and for most samples, the Kingman coalescent generally does provide an accurate description of the distribution of gene genealogies among independently segregating loci on a single fixed pedigree, in the sense that the coalescent is not rejected with much power using simple statistical tests.
Considerably greater power could likely be achieved by looking at patterns of haplotypes in recombining sequences. Although we have not considered restricted nonzero recombination, it is important to recognize that the treatment of recombination in coalescent theory involves the same averaging over all possible outcomes of reproduction that occurs in the derivation of the single-locus coalescent process (Hudson 1983b; Hudson and Kaplan 1985, 1988).
The Kingman coalescent has been shown to be a robust model for many sorts of perturbations from the canonical assumptions (Möhle 1998a,b). However, our results do not follow immediately from previous analyses because these begin with finite-state homogeneous Markov chains obtained by averaging over the process of reproduction (i.e., the pedigree) within each generation. Even so, the basic idea behind previous robustness proofs, namely a separation of timescales, is also relevant here. Figures 5 and 6 suggest that the Kingman coalescent is a poor approximation over the short time frame of the recent past, but becomes an accurate approximation after some small multiple of log2(N) generations. Because log2(N) is small relative to the typical coalescent timescale of N generations, excepting loci that coalesce within this very recent past, the distribution of gene genealogies is quite similar to that predicted by the Kingman coalescent.
The heterogeneity of coalescence probabilities shown in Figures 5 and 6 is due to the particular patterns of shared ancestry of the sampled individuals in each pedigree. For example, the solid line in Figure 6A begins at zero in generation 1, then jumps up to 1/32 (which is outside the range of the vertical axis in Figure 6A) in generation 2, and then jumps back down to zero in generation 3. It happened in this simulation that the two sampled individuals had no parents in common, exactly one shared grandparent, and only the two parents of their shared grandparent in common among their great-grandparents. Even this amount of recent shared ancestry is unlikely in all but very small populations, as indicated by the fact that the other nine trajectories in Figure 6, A and B, remain stuck at zero until at least generation 5.
While all sample-pedigree combinations deviate greatly from the coalescent prediction of a constant rate of coalescence in recent generations, if we consider only those genetic ancestries in which two lineages did not coalesce within some small multiple of log2(N) generations, then Figure 6, A and B, demonstrates that the more distant past for any pedigree will conform well to the predictions of the Kingman coalescent. Loosely speaking, an averaging over reproduction reminiscent of that in the derivation of the coalescent occurs on fixed pedigrees because there are so many possible ancestors of each individual: 2g at generation g in the past. Two lineages that do not coalesce in the recent past will traverse the pedigree for many generations before they meet, effectively sampling a large number of different reproduction events.
This remarkable result certainly must depend on the population or pedigree being well mixed in some sense, and we have not explored the stringency of this requirement. There has been one study of pedigrees in a spatially structured population: Kuo and Avise (2008) studied the effects of pedigrees on the branching structure of gene genealogies for samples of size n = 4 from a population arrayed along a circle and found results that pertain to models of isolation by distance. However, they were not concerned with the simple Kingman coalescent.
Our results extend those of previous simulation studies. In particular, Ball et al. (1990) simulated diploid pedigrees for populations of size 100 and then followed a single genetic lineage from each individual through the pedigree backward in time, creating a single-locus gene genealogy with n = N = 100 tips. They then sampled pairs of individuals from the tips of this gene genealogy and compiled the distribution of coalescence times among these pairs. They did not incorporate mutations, but dealt directly with coalescence times. Their study design was to generate 50 single-locus gene genealogies for each of 50 independent population pedigrees. For each of these 50 × 50 = 2500 gene genealogies, they randomly partitioned the N = 100 tips into 50 nonoverlapping pairs and studied the distribution of coalescence times among these pairs.
Ball et al. (1990) found that the distribution of coalescence times among pairs within single pedigrees (at a single locus) seldom fitted coalescent predictions for pairwise times to common ancestry. This stems from the fact that pairwise coalescence times are highly correlated within a single gene genealogy (Slatkin and Hudson 1991). Not surprisingly, Ball et al. (1990) also found that when these single-locus distributions were averaged over independent pedigrees, they conformed well to coalescent predictions. The surprising result of Ball et al. (1990) was that when these single-locus distributions were averaged among independent loci within a single pedigree, they also conformed well to coalescent predictions. We have confirmed this result with extensive simulations and described both the details of deviations from it in the recent past and the subsequent approach to a coalescent-like behavior, even for pedigrees that are not generated by Wright–Fisher reproduction.
Acknowledgments
We thank Nick Barton, Alison Etheridge, and Pleuni Pennings for helpful discussions and comments on the manuscript.
Footnotes
Communicating editor: Y. S. Song
Literature Cited
- Ball M., Neigel J. E., Avise J. C., 1990. Gene genealogies within organismal pedigrees of random-mating populations. Evolution 44: 360–370 [DOI] [PubMed] [Google Scholar]
- Barton N. H., Etheridge A. M., 2011. The relationship between reproductive value and genetic contribution. Genetics 188: 953–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles A. H., Egerbladh I., 2005. The influence of past endogamy and consanguinity on genetic disorders in northern Sweden. Ann. Hum. Genet. 69: 549–558 [DOI] [PubMed] [Google Scholar]
- Cannings C., 1974. The latent roots of certain Markov chains arising in genetics: a new approach. I. Haploid models. Adv. Appl. Probab. 6: 260–290 [Google Scholar]
- Cannings C., Thompson E. A., Skolnick M. H., 1978. Probability functions on complex pedigrees. Adv. Appl. Probab. 10: 26–61 [Google Scholar]
- Chang J. T., 1999. Recent common ancestors of all present-day individuals. Adv. Appl. Probab. 31: 1002–1026 [Google Scholar]
- Derrida B., Manrubia S. C., Zannette D. H., 2000. On the genealogy of a population of biparental individuals. J. Theor. Biol. 203: 303–315 [DOI] [PubMed] [Google Scholar]
- Garrigan D., Hammer M. F., 2006. Reconstructing human origins in the genomic era. Nat. Rev. Genet. 7: 669–680 [DOI] [PubMed] [Google Scholar]
- Gronau I., Hubisz M. J., Gulko B., Danko C. D., Siepel A., 2011. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43: 1031–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein J., Schierup M. H., Wiuf C., 2005. Gene Genealogies, Variation and Evolution: A Primer in Coalescent Theory, Oxford University Press, Oxford [Google Scholar]
- Hudson R. R., 1983a. Testing the constant-rate neutral allele model with protein sequence data. Evolution 37: 203–217 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 1983b. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1988. The coalescent process in models with selection and recombination. Genetics 120: 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff C. D., Xing J., Rogers A. R., Witherspoon D., Jorde L. B., 2010. Mobile elements reveal small population size in the ancient ancestors of Homo sapiens. Proc. Natl. Acad. Sci. USA 107: 2147–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff C. D., Witherspoon D., Simonson T. S., Xing J., Watkins W. S., et al. , 2011. Maximum likelihood estimation of recent shared ancestry. Genome Res. 21: 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Crow J. F., 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982a. The coalescent. Stoch. Proc. Appl. 13: 235–248 [Google Scholar]
- Kingman J. F. C., 1982b. On the genealogy of large populations. J. Appl. Probab. 19A: 27–43 [Google Scholar]
- Kingman J. F. C., 1982c. Exchangeability and the evolution of large populations, pp. 97–112 Exchangeability in Probability and Statistics, edited by Koch G., Spizzichino F. North-Holland, Amsterdam [Google Scholar]
- Kuo C., Avise J. C., 2008. Does organismal pedigree impact the magnitude of topological congruence among gene trees for unlinked loci? Genetica 132: 219–225 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of population history from individual whole-genome sequences. Nature 475: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. S., Clarke A. L., 1991. Family patterns in nineteenth-century Sweden: impact of occupational status and landownership. J. Fam. Hist. 16: 117–138 [Google Scholar]
- Low B. S., Clarke A. L., 1992. Resources and the life course: patterns through the demographic transition. Ethol. Sociobiol. 13: 463–494 [Google Scholar]
- Matsen F. A., Evans S. N., 2008. To what extent does genealogical ancestry imply genetic ancestry. Theor. Popul. Biol. 174: 182–190 [DOI] [PubMed] [Google Scholar]
- Möhle M., 1998a. A convergence theorem for Markov chains arising in population genetics and the coalescent with partial selfing. Adv. Appl. Probab. 30: 493–512 [Google Scholar]
- Möhle M., 1998b. Coalescent results for two-sex population models. Adv. Appl. Probab. 30: 513–520 [Google Scholar]
- Sjödin P., Kaj I., Krone S., Lascoux M., Nordborg M., 2005. On the meaning and existence of an effective population size. Genetics 169: 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Hudson R. R., 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2008. Coalescent Theory: An Introduction, Roberts & Company, Greenwood Village, CO [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Wollenberg K., Avise J. C., 1998. Properties of genealogical pathways underlying population pedigrees. Evolution 52: 957–966 [DOI] [PubMed] [Google Scholar]