Abstract
Hierarchical mixed effects models have been demonstrated to be powerful for predicting genomic merit of livestock and plants, on the basis of high-density single-nucleotide polymorphism (SNP) marker panels, and their use is being increasingly advocated for genomic predictions in human health. Two particularly popular approaches, labeled BayesA and BayesB, are based on specifying all SNP-associated effects to be independent of each other. BayesB extends BayesA by allowing a large proportion of SNP markers to be associated with null effects. We further extend these two models to specify SNP effects as being spatially correlated due to the chromosomally proximal effects of causal variants. These two models, that we respectively dub as ante-BayesA and ante-BayesB, are based on a first-order nonstationary antedependence specification between SNP effects. In a simulation study involving 20 replicate data sets, each analyzed at six different SNP marker densities with average LD levels ranging from r2 = 0.15 to 0.31, the antedependence methods had significantly (P < 0.01) higher accuracies than their corresponding classical counterparts at higher LD levels (r2 > 0. 24) with differences exceeding 3%. A cross-validation study was also conducted on the heterogeneous stock mice data resource (http://mus.well.ox.ac.uk/mouse/HS/) using 6-week body weights as the phenotype. The antedependence methods increased cross-validation prediction accuracies by up to 3.6% compared to their classical counterparts (P < 0.001). Finally, we applied our method to other benchmark data sets and demonstrated that the antedependence methods were more accurate than their classical counterparts for genomic predictions, even for individuals several generations beyond the training data.
WHOLE genome prediction (WGP) using commercially available medium to high density (>50,000) single-nucleotide polymorphism (SNP) panels have transformed livestock and plant breeding. Typically, the allelic substitution effects of all SNP markers are jointly estimated in WGP evaluation models assuming additive inheritance and summed to predict breeding values of each individual animal on the basis of its SNP genotypes (Meuwissen et al. 2001). This technology will not only lead to dramatically increased rates of genetic improvement for economically important traits such as meat and milk production in livestock (Wiggans et al. 2011) or crop production (Lorenz et al. 2011), but would also improve predictions of genetic predisposition to human diseases for personalized medicine (de los Campos et al. 2010).
Currently, the number of available SNP markers (m) is typically much greater than the number of animals having phenotypic records (n). Hence, hierarchical mixed-model or Bayesian approaches have been generally adopted in WGP to efficiently borrow information across these many markers by specifying their corresponding effects to be random. Following Meuwissen et al. (2001), these effects are typically specified to be either Gaussian or Student t-distributed (BayesA), or a mixture of either of these two densities with a point mass on zero (BayesB). When these effects are specified to be Gaussian, then best linear unbiased prediction of these effects is typically pursued because of computational tractability (VanRaden 2008; Hayes et al. 2009); applied to WGP, this procedure is often known as GBLUP (genomic–best linear unbiased prediction). Thus far, the distributional specifications for these various hierarchical modeling approaches have been based on a prior assumption of independence between all such effects.
Gianola et al. (2003) anticipated that some of these effects might be spatially correlated within chromosomes such that greater inference efficiency might be provided by modeling these effects as correlated. Their proposed specifications required equally spaced markers and/or within-chromosome correlations depending strictly on physical/linkage map distance between markers. However, the equally spaced assumption is rather tenuous for most currently available SNP marker panels. Even more importantly, the inferred correlation structure is likely to be nonstationary given that it should be primarily driven by the proximity of SNP markers to quantitative trait loci (QTL) of major effects. In other words, we anticipate that the correlation between the inferred effects of adjacent SNPs distal to major QTL would be substantially smaller than those proximal to these QTL.
Antedependence models have been increasingly advocated for the analysis of repeated measures data (Zimmerman and Nunez-Anton 2010) to parsimoniously account for nonstationary correlations between repeated measurements over time. In this article, we develop first-order antedependence counterparts to BayesA and BayesB. Through simulation, a cross-validation study involving the publicly available heterogeneous stock mice data (Valdar et al. 2006a,b) and journal-provided reference data (Hickey and Gorjanc 2012) to potentially benchmark our proposed methods with others, we demonstrate that, compared to their conventional counterparts, these antedependence-based WGP models improve the accuracy of genomic merit as well as potentially increase the sensitivity of QTL detection, which is the key objective of genome-wide association studies (GWAS).
Materials and Methods
Conventional WGP model
The base linear mixed model used for WGP is generally written as
| (1) |
Here, is a n × 1 vector of phenotypes, β is an p × 1 unknown vector of fixed effects connected to y via a known n × p incidence or covariate matrix X (e.g., environmental effects) is an m × 1 vector of random SNP effects connected to y via a known n × m matrix of SNP genotypes coded as 0, 1, or 2 copies of the minor allele for each SNP (column) and animal (row) in Z. Furthermore, is a q × 1 vector of random polygenic effects connected to y via a known n × q incidence matrix W, and is the residual vector. We assume that , where A denotes the pedigree-derived numerator relationship matrix (Henderson 1976) and is often included in WGP models due to insufficient genome coverage by Z (Calus and Veerkamp 2007). Furthermore, we specify , where and . From a Bayesian perspective, a subjective prior may be also specified on β using with and taken as known (Sorensen and Gianola 2002).
Now the distinction between GBLUP, BayesA, and BayesB in Meuwissen et al. (2001) depends upon the characterization of G. If (i.e., ), then the model is defined to be GBLUP. If, instead, the diagonal elements of G are independent random draws from an scaled inverted chi-square distribution, i.e., such that , then the model is said to be BayesA such that marginally gj is a random draw from a Student t distribution with mean 0, degrees of freedom , and scale parameter (de los Campos et al. 2009; Gianola et al. 2009). Now BayesB further extends BayesA by including a two-component mixture with one component being and the other component being a spike or point mass at 0; i.e.,
| (2) |
That is, () represents the proportion of SNP markers having no associated genetic effects on the trait of interest.
Clear warnings have been provided on how sensitive inferences using BayesA or BayesB may be to specification of the hyperparameters (de los Campos et al. 2009; Gianola et al. 2009). It has not been widely appreciated that and are estimable; this recognition is critical as both hyperparameters help define the genetic architecture in BayesA and BayesB. That is, characterizes the variability of about a typical variance component of . Details on how to estimate and in the context of BayesA were previously provided by Yi and Xu (2008). Furthermore, is estimable in BayesB. For both BayesA and BayesB, we specify the prior distribution , similar to what we have previously adopted in other applications (Kizilkaya and Tempelman 2005; Bello et al. 2010). Furthermore, we specify for BayesB, with values of and chosen to reflect prior uncertainty on . We also specify a proper conjugate prior on in BayesB; i.e., recognizing that the specifications on and become increasingly influential as → 1.
Finally, we specify noninformative priors and , which are congruent with specifying uniform priors on and , respectively, and in line with recommendations for variance components by Gelman (2006). We similarly and confidently specify in BayesA, given that m is generally large enough for stable inference on without the need for more informative priors.
Antedependence extensions of WGP models
We propose a nonstationary first-order antedependence correlation structure for g based on the relative physical location of SNP markers along the chromosome(s):
| (3) |
Here ,, whereas is the marker interval-specific antedependence parameter (Zimmerman and Nunez-Anton 2010) of on in the specified order. We can rewrite the recursive expression in (3) in matrix notation,
| (4) |
where for I being a m × m identity matrix, and T having all null values except for elements tj,j-1 at the corresponding subscript addresses. It can be readily seen using Equation 4 that , where is a lower triangular matrix with diagonal elements equal to 1 and . As further illustrated in supporting information, File S1, from the is a readily determined tridiagonal matrix (Zimmerman and Nunez-Anton 2010), which is important as it facilitates inference on g.
Some of the other developments closely follow the BayesA and BayesB models of Meuwissen et al. (2001). That is, we specify in a model that we label ante-BayesA. Similarly, we propose an ante-BayesB model whereby we specify a mixture similar to Equation 2 except that it is specified on ; i.e., a mixture of point mass on zero with probability and scaled inverted chi-square prior with probability (1 − ). As we suggested earlier for , we believe that is estimable such that ante-BayesA is merely a special case of ante-BayesB. In turn, BayesA is merely a special case of ante-BayesA, as is BayesB of ante-BayesB, when ; i.e., tj,j-1 = 0 .
These antedependence extensions, nevertheless, do require inference on the unknown m − 1 nonzero elements of T. Borrowing from Daniels and Pourahmadi (2002) and Bello et al. (2010), we specify as a conjugate prior in both ante-BayesA and ante-BayesB, thereby allowing flexible inference on the nonstationary correlation structure in G. However, it should be further noted that if interval j, j − 1 specifies that between the last SNP of one particular linkage group or chromosome and the first SNP in the arbitrarily subsequent linkage group, then we set the corresponding . The remaining prior specifications are specified on the hyperparameters that essentially characterize the hypothesized genetic architecture of the trait and are virtually identical to those previously prescribed for BayesA and BayesB; i.e., , , with , , and again all specified as known. Similarly, we also estimate and by placing subjective priors, and on these key hyperparameters, where , are specified to be known.
As in Meuwissen et al. (2001) and subsequent work, our implementation strategy is based on the use of Markov chain Monte Carlo (MCMC) methods; however, we also additionally infer key hyperparameters, i.e., , , and , that characterize the genetic architecture of the trait, as alluded to earlier. Further details on the full conditional densities and any necessary Metropolis–Hastings strategies to sample from the joint posterior density of all unknown parameters using MCMC are provided in File S1.
Simulation study
We compare the performance of BayesA and BayesB with their antedependence counterparts, ante-BayesA and ante-BayesB, in a simulation study. Twenty replicated data sets were each generated from a base population containing 50 unrelated males and 50 unrelated females. Each data set underwent random mating while maintaining constant population size for 6001 generations beyond the base population. The entire genome was composed of one chromosome of length 1 M. All of 20,001 potential SNP markers were equally spaced on this genome with a potential QTL placed directly in the middle of each interval of adjacent markers. In the base population, all 20,000 QTL and 20,001 SNP marker alleles were coded as monomorphic. The number of simulated crossover events per meiosis was generated from a Poisson (mean 1) distribution with the location of the crossover events uniformly distributed throughout the chromosome in accordance with the Haldane mapping function. The mutation rate for both QTL and SNP markers was specified to be 10−4 per locus per generation and to be recurrent, that is, switching between one of two alternative allelic states 0 and 1 whenever mutation occurred so as to ensure biallelic loci (e.g., Coster et al. 2010; Daetwyler et al. 2010).
In Generation 6001, all SNP markers and QTL with a minor allele frequency (MAF) <0.05 were discarded. We then randomly selected only 30 of the remaining QTL and their corresponding allelic substitution effects. For each of these k = 1, 2, …, 30 QTL, an allelic substitution effect () was drawn from a reflected gamma distribution with shape parameter 0.4 and scale parameter 1.66, with a positive or negative sign on sampled with equal probability. The genetic variance at QTL k was determined to be , where is the MAF at QTL k. The total genetic variance was subsequently determined to be the summation of these terms across the 30 selected QTL, i.e., as . Now the true breeding values (TBV) were defined to be a genotype-based linear function of the 30 generated QTL effects, which, because these QTL were located between various SNP, are not subsets of g. These TBV were further scaled such that the total genetic variance was 1 as per Meuwissen and Goddard (2010). Residual effects were, in turn, sampled from a standard normal distribution, such that the heritability was 0.50. That is, each phenotypic record was generated by adding the TBV for that animal plus its corresponding residual. Hence, 100 animals with known phenotypes and genotypes in Generation 6001 were simulated for inferring upon the SNP effects, using each of the competing methods. Genotypes and the TBV for each of 100 offspring were also generated in Generation 6002, on the basis of randomly mating animals in Generation 6001.
For each of the 20 replicated data sets, the effect of six different marker densities on the comparison between the competing methods were investigated by selecting every 1, 4, 7, 10, 15, and 20 SNP markers from those with MAF > 0.05. That is, the data sets were used as a blocking factor in comparing different marker densities for the accuracy of predicting genetic merit in Generation 6002, using each of the four different methods: BayesA, BayesB, ante-BayesA, and ante-BayesB. Accuracy was defined as the correlation between estimated breeding values (EBV) for Generation 6002, using just Generation 6001 phenotypes and genotypes data, and the corresponding TBV of Generation 6002. These EBV are based on the posterior mean () of g; i.e., EBV are elements of . Comparisons between the BayesA/BayesB procedures and their antedependence counterparts were also drawn for inference on the key hyperparameters that characterize genetic architecture. This was conducted using multifactorial ANOVA on the posterior means using replicate as the blocking factor for assessing the importance of model and marker density and their interaction across the 20 replicates. Furthermore, an assessment of the relative ability of ante-BayesB compared to BayesB to identify the top QTL by genetic variance was based on the difference in posterior probabilities of and , respectively, of adjacent SNP markers being nonzero. As QTL were placed between SNP markers and never on top of SNP markers, we calculated this probability of association by determining the proportion of MCMC cycles that either or both of the two markers adjacent to the known QTL were chosen to be nonzero within each analysis.
All comparisons were based on the linear mixed model in Equation 2 with X being a column vector of ones, except that polygenic effects (u) were ignored for simplicity and computational tractability.
Application to heterogeneous stock mice data set
We used a data set publicly available from the Wellcome Trust (http://gscan.well.ox.ac.uk/), which includes phenotypic records on 2296 mice, each genotyped for 12,147 SNP markers. This data resource, which also includes pedigree information, was based on an advanced intercross mating among eight inbred strains after 50 generations of random mating (Valdar et al. 2006a). The average linkage disequilibrium (LD), as measured by r2 between adjacent markers is 0.62 (Legarra et al. 2008), which is high compared to commonly used SNP panels available for livestock populations. For example, the average r2 between adjacent markers in most commercially available livestock SNP panels ranges from 0.10 to 0.37 for markers that are generally around 100 kb apart (Du et al. 2007; de Roos et al. 2008; Abasht et al. 2009; Bohmanova et al. 2010).
Given this high pairwise LD, we considered only a random subset of all markers from this data set to ensure adjacent marker LD levels that are representative of livestock populations. We first excluded SNP markers if the percentage of missing genotypes across samples was >10% or if the MAF was <2.5%. We also discarded animals having greater than 20% missing SNP genotypes. We then randomly selected 50 SNP markers from each of the 19 autosomes, leading to an average LD of r2 of 0.35 between adjacent markers. The resulting data set then involved records on 1917 animals with genotypes on 950 SNPs. The phenotypes and pedigrees are available as File S2 and the SNP genotypes as File S3 based on the chosen SNP markers are provided in File S4.
As in Legarra et al. (2008), we also added the random effect of cage in the WGP model of [2]; i.e., , where and S is the corresponding incidence matrix with all other terms defined as before. Furthermore, we specified Gelman's prior on in addition to all previously provided prior specifications. Also, as per Legarra et al. (2008), we chose to use the data provided on body weight at 6 weeks that was already precorrected for fixed effects such that X was a column vector of ones and β consisted of just an overall mean. Missing SNP genotypes were simply imputed from binary distributions on the basis of their corresponding allelic frequencies in the data set following Legarra et al. (2008). We adopted the same within-family cross-validation technique as described in Legarra et al. (2008) by randomly partitioning each family into two. This partitioning was replicated 20 times to obtain 20 different nearly equal-sized partitions of training and validation data subsets. Also, as in Legarra et al. (2008), we compared the various methods using predictive abilities, defined as the correlation between phenotypes in the validation subset and their corresponding predictions on the basis of their inferences from the training data subset.
Application on simulated genomic data from Hickey and Gorjanc
To provide a benchmark comparison of our proposed methods with competing methods in other articles in this issue, we analyze simulated data sets provided by and described in detail by Hickey and Gorjanc (2012). They generated 10 replicated data sets for each of four different traits whereby 9000 QTL effects were generated for trait 1 and 900 QTL effects were generated for trait 2. Traits 3 and 4 mirrored traits 1 and 2, respectively, with the further requirement that the MAF for these QTL was <0.30. Since we were permitted to simultaneously run 144 jobs on the high-performance computing cluster at MSU (hpcc.msu.edu), we chose to compare the four methods for each of the four traits on each of the first nine data sets (4 × 4 × 9 = 144). For all analyses, training data were based on 2000 animals in generations 4 and 5 whereas TBV were provided on 500 animals within generations 6, 8, and 10. To facilitate computing tractability, we saved every tenth SNP marker that had a MAF >0.20. This led to a range of 2884–2952 SNP markers and an average LD between adjacent markers from 0.16 to 0.17 across the nine replicates. The actual SNP markers chosen from each of the nine data sets are provided in File S5 based on the original data (File S6) as provided by Hickey and Gorjanc (2012). All four models also included polygenic effects. Antedependence methods were directly compared with their classical counterparts for accuracy (correlation of EBV with TBV) and bias (deviation of slope from 1 from regressing TBV on EBV) in these latter validatation generations using a Wilcoxon signed rank test.
Bayesian inference
For each of the four methods, BayesA, BayesB, ante-BayesA, and ante-BayesB in both our simulation study and the heterogeneous stock mice application, we ran MCMC for 50,000 cycles of burn-in followed by an additional 300,000 cycles; for the benchmark data from Hickey and Gorjanc (2012), the corresponding numbers were 80,000 and 1,000,000, respectively. Every tenth MCMC cycle was subsequently saved for inference post burn-in. We monitored MCMC convergence via inspection of trace plots and determined the effective sample size (ESS) for number of random draws from the joint posterior density for all key hyperparameters using the R package CODA (Plummer et al. 2006). The larger number of MCMC cycles for the Hickey–Gorjanc data were based on ensuring that ESS for all hyperparameters at least exceeded 100. Inferences were primarily based on the posterior means and posterior standard deviations for key parameters, including those hyperparameters that characterize genetic architecture.
Prior specifications
For all analyses in this article, we chose and in both BayesB and ante-BayesB to reflect the prior belief that most of the markers will not be associated with any genetic effects; however, the dispersion of this corresponding beta distribution is still large enough such that values of () close to 0.70 are plausible. On the basis of preliminary runs, we also found that this prior specification led to superior mixing properties of the MCMC chains over a naïve Uniform(0,1) prior, yet facilitated domination of data over prior information since << m.
For BayesB and ante-BayesB, we always specified for the Gamma prior on (). For the antedependence-based models, we specified , , −1, and 0, i.e., a standard normal prior on and Gelman's prior on . We also always specified a flat prior on β by defining = 0. Prior specifications for all other parameters (e.g., variance components) were based on those previously recommended in this article.
Results
Simulation study
For the six different marker densities, the average distances between adjacent markers ranged from 0.046 to 0.918 cM over the 20 replicates whereas the average LD between adjacent markers, measured by r2 values, ranged from 0.15 to 0.31, as shown in Table 1. Among the 30 chosen QTL within each of the 20 replicates, anywhere between 6 and 11 of the QTL had variances >2% of the total genetic variance.
Table 1.
Summary statistics for six different marker densities in the simulation study over 20 replicates
| Marker density levela | Average no. of markers per replicate | Average distance between adjacent marker loci (cM) per replicate | Average r2 between adjacent marker loci per replicate |
| 1 | 108 | 0.918 | 0.15 |
| 2 | 145 | 0.689 | 0.18 |
| 3 | 217 | 0.459 | 0.21 |
| 4 | 311 | 0.321 | 0.24 |
| 5 | 545 | 0.184 | 0.27 |
| 6 | 2182 | 0.046 | 0.31 |
Marker density levels 1–6 pertain to saving every 20th, 15th, 10th, 7th, and 4th and every single SNP marker from a single 1-M chromosome within each data replicate.
Inferences on key hyperparameters
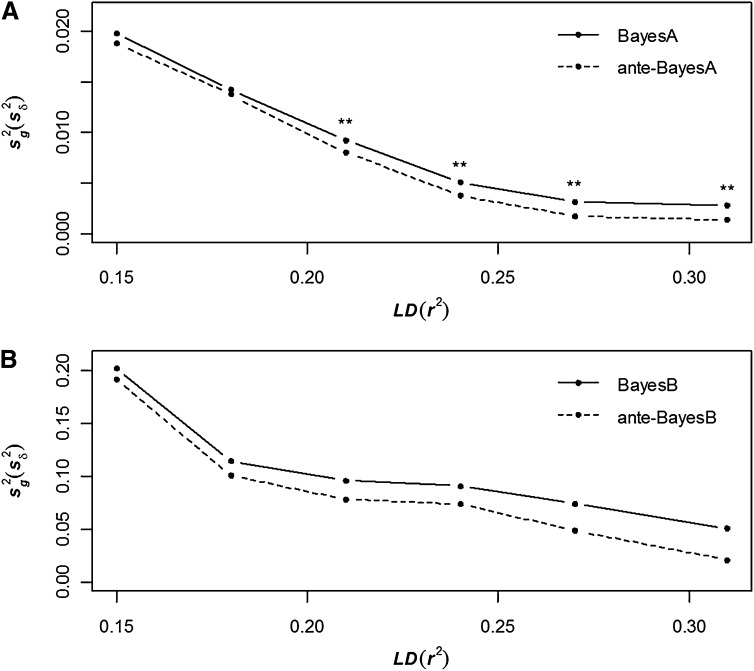
It is important to recognize that none of the modeling assumptions behind BayesA, BayesB, anteBayesA, or anteBayesB truly match the data generation model based on thousands of generations of LD created between markers and QTL, even for simulated data. This goes beyond the fact that the QTL effects were drawn from reflected Gamma distributions in our simulation study as typically done (e.g., Meuwissen et al. 2001; Meuwissen and Goddard 2010). That is, the process of recombination over thousands of generations in terms of how it generates LD between QTL and SNP markers is not explicitly captured in any known WGP model, including any of the competing models, especially when the effects of neighboring SNP markers rather than the causal QTL effects are being estimated. Hence, there is no way to surmise the “true” values of key hyperparameters, whether for , , or in BayesA or BayesB or for ,, , , or in anteBayesA or anteBayesB. However, one should anticipate that estimates of or should be inversely related to marker density, since they closely represent the mean value of the variance components or , respectively, accounted for by each SNP. Indeed, we observe this phenomenon in the comparison between BayesA and anteBayesA in Figure 1A. We also note a similar comparison between and for BayesB vs. anteBayesB in Figure 1B, but further recognize that the corresponding estimates of and are roughly 1 order of magnitude greater than those seen in Figure 1A. That is, and specify a typical value for and , respectively, over many more loci in (ante)BayesA than their (ante)BayesB counterparts. In spite of the lower values observed in Figure 1A, however, there was a significant difference (P < 0.01) between and when r2 ≥ 0.21.
Figure 1 .
Average posterior means of (BayesA, BayesB) and (ante-BayesA, ante-BayesB) across 20 replicates for six different levels of LD comparing BayesA and ante-BayesA (A) and BayesB vs. ante-BayesB (B). Significant differences in posterior means between competing methods at each LD level are indicated: (**) P < 0.001.
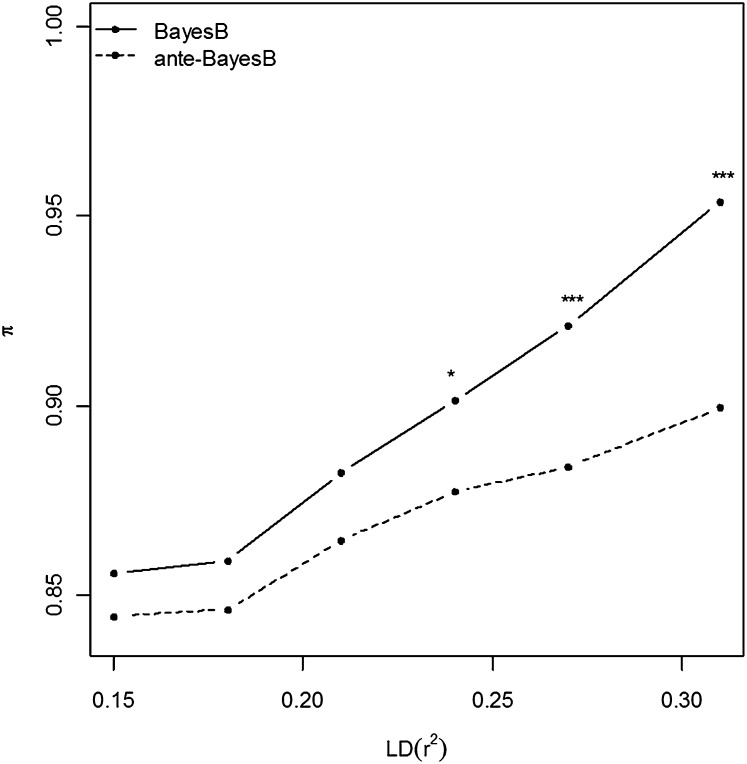
As marker density increased, we also expected that the estimates of or should increase as well; that is, it becomes increasingly unlikely that individual SNP markers become associated with a particular QTL with greater marker density. Indeed we observed this in Figure 2. It was particularly interesting that the posterior means of were generally lower than that of , with differences widening with increasing marker density (i.e., LD level) such that the differences were significant beyond r2 = 0.24 (P < 0.01). Note the subtle difference in interpretation between and as pertains to the probability of nonassociation for the corresponding SNP whereas pertains to the probability of nonassociation conditional on a neighboring SNP.
Figure 2 .
Average posterior means of (BayesB) vs. (ante-BayesB) across 20 replicates as a function of six different LD levels. Significant differences in posterior means between competing methods at each LD level are indicated by (*) P < 0.01, or (***) P < 0.0001.
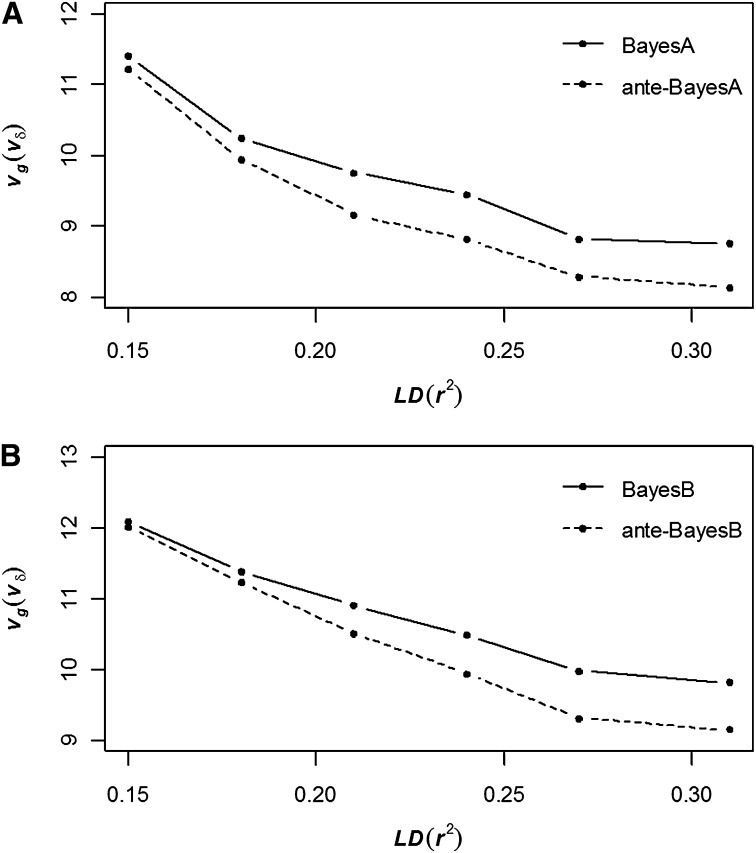
The estimates of and also changed as a function of marker density for anteBayesA vs. BayesA in Figure 3A and for anteBayesB vs. BayesB in Figure 3B. Specifically, the posterior means of and particularly of both decrease with increasing marker intensity. Since these parameters, respectively, characterize the heterogeneity of and across SNP or, alternatively, the heaviness of the tails for the resulting marginal Student t distribution on and across SNP, our results imply that these hierarchical methods, and particularly those based on nonstationary first-order antedependence correlation structures, identify SNP with large effects as being more outlying relative to a normal distribution when marker density increases. However, these differences between and were not seen to be statistically significant at any marker density.
Figure 3 .
Average posterior means of (BayesA, BayesB) and (ante-BayesA, ante-BayesB) across 20 replicates for six different levels of LD comparing BayesA and ante-BayesA (A) and BayesB vs. ante-BayesB (B). No significant differences (P > 0.01) were determined between the two sets of competing procedures at each LD level.
Figures S1 and S2 in supporting information File S1 shows the average posterior means for and against LD level across the 20 replicates under both ante-BayesA and ante-BayesB. There was no evidence (P > 0.01) across these 20 replicates that the posterior means of were different from zero at any LD level; however, at higher LD levels, the posterior means tended to converge to zero as anticipated. Similarly, the posterior estimates for were also lower at higher LD levels. Again, this was somewhat anticipated since there should be less disparity in different values of the antedependence parameters () between adjacent markers with increasing marker intensity.
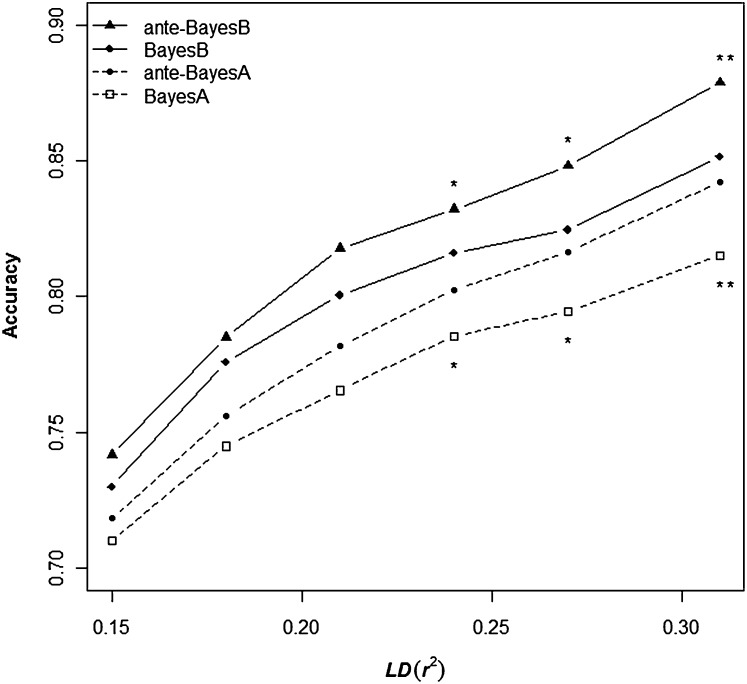
Comparison between true and estimated total breeding values
The average accuracies of the EBV over the 20 replicated data sets are plotted as a function of the average r2 (i.e., the different marker densities) between adjacent markers for the four different methods in Figure 4. As anticipated, given the simulated genetic architecture of few QTL, the accuracies for the BayesB methods were consistently greater than their corresponding BayesA counterparts at all marker densities. Also, ante-BayesA and ante-BayesB outperformed their classical counterparts with differences increasing with LD level. Specifically, anteBayesA had significantly greater accuracies compared to conventional BayesA, as did anteBayesB compared to BayesB (P < 0.01), when average LD levels exceeded r2 = 0.24.
Figure 4 .
Average accuracies of estimated breeding value across 20 replicates for analyses based on each of six LD levels. Differences in accuracy between BayesA and ante-BayesA (circle and square) and between BayesB with ante-BayesB (triangle and diamond) indicated as significant by (*) P < 0.01 or (**) P < 0.001.
We anticipated that the antedependence parameters 's would have greater importance at higher marker densities. To demonstrate this, we standardized the posterior means of these parameters as a ratio over their posterior standard deviations, i.e., , for each analysis. We then determined the proportion of these whose absolute value exceeded an arbitrary value of 2 for each data replicate and marker density analysis to indicate the relative importance of these antedependence parameters. We present boxplots of these proportions across the 20 replicates for ante-BayesA and for ante-BayesB in Figure S3 in File S1. We anticipated and noted that a higher proportion of exceeded 2 in data sets characterized by higher marker densities, thereby indicating that, in general, nonstationary serial correlation between adjacent markers becomes increasingly more important with higher levels of LD. We believe this phenomenon is responsible for driving the differences in accuracies between ante-BayesA (ante-BayesB) and BayesA (BayesB) with increasing LD levels as seen earlier in Figure 4.
Potential merit for GWAS
Hierarchical methods that are similar to BayesB, in that they jointly infer upon all SNP effects, have been increasingly advocated as tools for GWAS (Hoggart et al. 2008; Lee et al. 2008; Logsdon et al. 2010). Figure S4 in File S1 shows the average (across 20 replicates) posterior mean probabilities of identifying the largest QTL by genetic variance within each replicate using BayesB and ante-BayesB, respectively. These estimated posterior probabilities increased with LD level for both models but were significantly greater for ante-BayesB than for BayesB with statistical significance also increasing with LD or marker density. That is, the precision for detecting QTL was increasingly greater for ante-BayesB than for BayesB at higher LD levels. We observed this consistently across data replicates with the ability of ante-BayesB to better track causal variants increasing with marker density (Figure S5 in File S1).
Application to heterogeneous stock mice data
We summarize posterior inferences of key parameters using BayesA and BayesB in Table S1 and for their antedependence counterparts in Table S2 (see File S1) for the heterogeneous stock mice data. Inferences on , , and were consistent with results previously reported by Legarra et al. (2008). As expected from our simulation study, the estimates for () and () were substantially greater for BayesB (ante-BayesB) than for BayesA (ante-BayesA). Although the posterior mean for of 0.81 (BayesB) was only slightly larger than 0.80 (ante-BayesB), the posterior mean of was substantially larger in ante-BayesB than in BayesB. The average estimates ±empirical standard errors of predictive ability correlations over the 10 cross-validation partitions of training and validation data subsets were 0.57 ± 0.01, 0.62 ± 0.01, 0.60 ± 0.01, and 0.66 ± 0.01 for BayesA, BayesB, ante-BayesA, and ante-BayesB, respectively. The differences between BayesA with ante-BayesA and BayesB with ante-BayesB were both determined to be statistically significant (P < 0.005), indicating the relative advantage of the antedependence methods. Furthermore, BayesB and ante-BayesB had significantly greater predictive abilities than BayesA and ante-BayesA, respectively (P < 0.001).
Application to Hickey and Gorjanc data
Average posterior means for key hyperparameters for each of the four methods across the nine replicates are provided in Table S3 in File S1, whereas the corresponding average ESS are provided in Table S4 in File S1. Estimates of () and () were lower whereas estimates of () are higher for traits with higher numbers of QTL (traits 1 and 3) compared to those with lower numbers of QTL (traits 2 and 4), relative to the same number of markers.
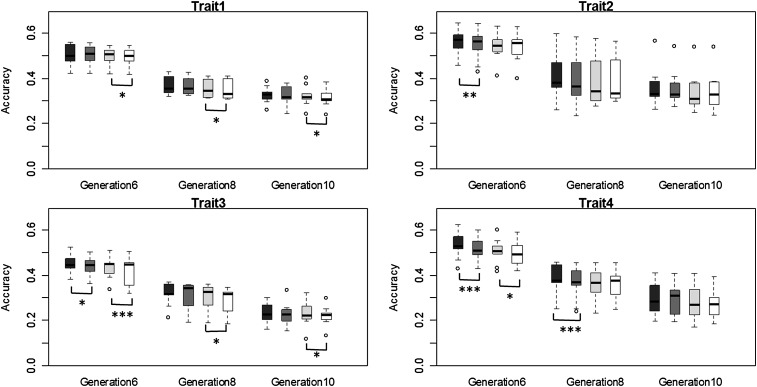
A side-by-side comparison of the accuracies of the four methods across the validation generations (6, 8, and 10) is provided in Figure 5. It is remarkable to note that ante-BayesA had generally significantly greater accuracies than BayesA for traits 1 and 3 (larger numbers of QTL) that was still maintained until generation 10, whereas ante-BayesB had generally significantly greater accuracies than BayesB for traits 2 and 4 (lower numbers of QTL) but only in generations 6 and 8. An assessment of bias of the four procedures based on regressing TBV on EBV is provided in Figure S6 in File S1. For all traits, all four methods had some significant bias in generation 6 but not in later generations.
Figure 5 .
Boxplots of average accuracies of estimated breeding value across nine replicates for four traits in generations 6, 8, and 10 for benchmark data from Hickey and Gorjanc (2012). Differences in accuracy between anteBayesB (solid bars) and BayesB (dark shading) and between anteBayesA (light shading) with BayesA (open bars) indicated as significant by (*) 0.05 < P < 0.10, (**) 0.01< P < 0.05, or (***) P < 0.01.
Discussion
In this article, we extend two very popular Bayesian methods, BayesA and BayesB, for WGP to model potential nonstationary correlations between SNP effects in close proximity to QTL. We demonstrated using a small-scale simulation study that the accuracies of our proposed first-order antedependence extensions, labeled ante-BayesA and ante-BayesB, were greater than their classical counterparts with differences increasing with marker density. This result was anticipated given that the magnitude and importance of the antedependence parameters in T should increase as marker densities increase. To further illustrate the importance of modeling nonstationary correlations, rather than basing correlations between SNP effects purely as a function of distance (Gianola et al. 2003), we observed the magnitude of the posterior means of at the various locations through the chromosome for the first four replicates using ante-BayesA and ante-BayesB (see Figure S7 in File S1). These posterior means tended to be rather large in absolute value in the general vicinity of the major QTL. This result was anticipated since each QTL is likely tracked by several SNP, each in incomplete LD with the QTL (Goddard and Hayes 2009). Interestingly, there appeared to be a greater spread in these posterior means around a greater number of QTL using ante-BayesB compared to ante-BayesA.
We realize that the order in which Equation 3 is specified for the antedependence methods is rather arbitrary; i.e., one might specify Equation 3 from the end of the p-arm to the end of the q-arm of a chromosome or vice versa. For instance, instead of specifying Equation 3 from , we might have also modeled antedependence in the opposite direction, i.e., from . It has been demonstrated by Zimmerman and Nunez-Anton (2010) in the context of longitudinal data analysis that the (co)variances based on a first-order antedependence model are invariant to directionality as long as the relative order is correctly specified. To illustrate this, we reanalyzed the first four replicates on the basis of the highest average marker density (r2 = 0.31), again using ante-BayesA and ante-BayesB but this time specifying Equation 3 in the opposite direction from what was used previously. We plotted the posterior means () of g on the basis of the analysis in the original direction again the same estimates on the basis of the analysis in the opposite direction (Figure S8 in File S1), further demonstrating that inferences on EBV are invariant to direction. Noting that the EBV are linear functions of , i.e., Z, we noted even greater consistency for EBV between the two directions for these same four replicates in Figure S9 in File S1.
Given that the accuracy of EBV was greater using the antedependence-based procedures compared to their classical counterparts, we examined the elements of for each marker between the two different classes of models within each of the first four replicates and the highest average marker density (Figure S10 in File S1). It was interesting to note that these elements were more shrunk to zero using the antedependence-based procedures than were the conventional counterparts. Given the specification of nonstationary correlations between effects of adjacent SNP markers, the effective number of SNP may be considered to be somewhat lower using the antedependence-based procedures, particularly in regions containing a major QTL. That is, larger elements of using the antedependence-based models will tend to be more highly correlated and hence shrunk closer to zero than elements of derived from their conventional counterparts. Goddard et al. (2009) have previously described the optimal statistical properties of when g is specified as a set of “random effects” having an exchangeable distribution, such as the normal or Student's t. However, these properties are better realized when a more appropriate correlation structure is specified, as would be true, for example, modeling polygenic effects with a classical animal model (Henderson 1984). The more optimal properties of using the antedependence-based models was also partly reflected in the earlier EBV comparisons with their classical counterparts.
In spite of the limited scale of our simulation studies, it has been demonstrated that inferences on accuracy based on 100 individuals and a genome length of 1 M is roughly equivalent to inferences derived from 3000 individuals and a genome length of 30 M (Meuwissen and Goddard 2010), the latter of which might depict a more common scenario in livestock populations. We also based all of our simulation work on a heritability of 50%; for situations with lower or higher heritabilities, we would naturally expect the accuracies to, respectively, decrease or increase accordingly in concert with previous simulation results (Calus and Veerkamp 2007) and/or analytical derivations (Meuwissen and Goddard 2010); however, we believe that there is no reason to believe that the antedependence methods would not outperform the conventional Bayesian WGP methods in these situations as well. This was further substantiated by our analyses of the data from Hickey and Gorjanc (2012), which were based on a heritability of 25%. In our own simulation study, there were around 2200 markers per the single 1-M chromosome using the highest average LD level of r2 = 0.31, whereas there were around 100 markers per the single chromosome with the lowest average LD level of r2 = 0.15. Using Meuwissen and Goddard (2010), these two specifications are, respectively, analogous to a panel of 60,000 SNP markers and to a panel of 3000 SNP markers for a 30-M genome; commercially developed panels having roughly these same numbers of SNP markers are now widely available for cattle (Wiggans et al. 2011). On the basis of the results of our work, we anticipate that the antedependence-based methods, compared to their classical counterparts, would lead to even greater accuracies with higher density SNP marker panels (m > 500,000) that are being developed for livestock or for situations in which there is sequence data (Meuwissen and Goddard 2010). Along those lines, we anticipate that these methods would also perform better in populations where LD is greater between markers due to other phenomena, e.g., selection history.
Our simulation studies were also based on a particular genetic architecture; i.e., 30 QTL that were randomly distributed throughout a 1-M chromosome (or equivalently, 900 QTL for a 30-M genome). Although this is not the focus of our article, we realize that genetic architecture (i.e., number of QTL, average QTL substitution effect, marker density, etc.) can affect the relative merit of BayesA, BayesB, and GBLUP on the basis of other studies in which key hyperparameters such as , , and are arbitrarily specified to be known (Daetwyler et al. 2010; Meuwissen and Goddard 2010). That is, the greater the number of QTL, each with small effects, relative to the number of SNP markers, the more likely the genetic architecture reflects the GBLUP assumptions (, such that ). Conversely, BayesB would be favored in the situation in which SNP marker density is high relative to the number of QTL (). However, we believe that formal comparisons in data fit between BayesA, BayesB, and GBLUP, along with ante-BayesA and ante-BayesB, are not entirely necessary since ante-BayesB represents the most general model. As previously noted, ante-BayesA is a special case of BayesA, as is ante-BayesB of BayesB, when T = 0 such that then , , and . Furthermore, BayesB becomes BayesA as , whereas BayesA becomes GBLUP as . Nevertheless, our claim that one needs only to fit ante-BayesB, rather than any of the other three competing submodels, vitally depends upon reliable inferences being provided on these key hyperparameters defining genetic architecture, rather than arbitrarily specifying them (Daetwyler et al. 2010; Meuwissen and Goddard 2010) or estimating a subset thereof (Habier et al. 2011). We provide details on MCMC inference strategies on these and other unknown parameters in File S1. We are currently pursuing more suitable inferential strategies for variable selection (O'Hara and Sillanpaa 2009) when inferring upon or . Also, although our proposed antedependence methods seem to work well under additive genetic model assumptions, it is not clear how well they may perform in the presence, for example, of extensive nonadditive gene action where nonparametric approaches may be warranted (Gianola et al. 2010). Nevertheless, even in the extensive presence of such phenomena, genetic variance is still considered to be primarily additive (Hill et al. 2008).
Although the scope of this work was focused on the potential merit of these antedependence models for WGP, we suggested earlier that there may be also merit in using these models in GWAS in both livestock and human populations. It has become increasingly recognized that GWAS procedures based on joint analyses of all SNP markers are more powerful than the conventional series of single SNP analyses. Our results suggest that modeling nonstationary correlations between SNP effects will further augment this power. At any rate, we recognize that for reasonably accurate GWAS, a greater marker density (m) per chromosome and sample size (n) should be considered (e.g., Meuwissen and Goddard 2010) than those studied in this article; i.e., most of the posterior probabilities reported in Figures S4 and S5 in File S1 are too low to be of practical benefit in current applications.
We also acknowledge that our ante-BayesA and ante-BayesB models increase the computational load relative to their conventional counterparts. Since m is typically large, the computing time for the proposed antedependence models is bottlenecked primarily by the m elements of δ, the m diagonal elements of Δ, and the m − 1 nonzero elements of T. Similarly, computing time for the two conventional methods, BayesA and BayesB, is primarily restricted by the dimension of δ and Δ, i.e., roughly two-thirds as many variables for the antedependence-based models, ignoring the remaining parameters such as variance components and hyperparameters. Hence, the computing time for the antedependence-based procedures should be somewhat less than one-third greater than for their conventional counterparts. Indeed, we discovered from our simulation study that computing time for all four competing models were linear in m with the antedependence-based models taking <30% greater computing time than the conventional counterparts for the wide range of values of m considered in this article. We recognize for much larger number of SNP markers than those pursued in this study that alternative algorithmic adaptations already developed for models similar to conventional BayesA or BayesB, such as those based on the EM algorithm (Shepherd et al. 2010) or variational Bayes (Logsdon et al. 2010), would be worth exploring.
We believe the proposed antedependence models provide opportunities for further study and extension. For example, it has been previously recognized that basing inferences on allelic effects on the use of multiple marker haplotypes rather than single markers increases accuracy of WGP (Calus et al. 2008; Villumsen et al. 2008) or GWAS (Grapes et al. 2004). Given the difficulty in how to appropriately specify these haplotypes, we believe our antedependence-based methods may help bridge these two different strategies as the effects of adjacent SNP markers connected by large values of may somewhat determine “effective haplotype” effects. We also think that our antedependence specifications might facilitate multiple breed inference if, for example, genomic effect differences between breeds is primarily due to differences in SNP associations with QTL, as partly manifested in T.
Supplementary Material
Acknowledgments
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2011-67015-30338 from the U.S. Department of Agriculture National Institute of Food and Agriculture. Additional support from the Quantitative Biology Program, the Department of Animal Science, and AgBioResearch at Michigan State University is also gratefully acknowledged.
Footnotes
Edited by Dirk-Jan de Koning, Lauren M. McIntyre, and Matias Kirst
Literature Cited
- Abasht B., Sandford E., Arango J., Settar P., Fulton J. E., et al. , 2009. Extent and consistency of linkage disequilibrium and identification of DNA markers for production and egg quality traits in commercial layer chicken populations. BMC Genomics 10: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello N. M., Steibel J. P., Tempelman R. J., 2010. Hierarchial Bayesian modeling of random and residual variance-covariance matrices in mixed effects models. Biom. J. 52: 297–313 [DOI] [PubMed] [Google Scholar]
- Bohmanova J., Sargolzaei M., Schenkel F. S., 2010. Characteristics of linkage disequilibrium in North American Holsteins. BMC Genomics 11: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calus M. P. L., Veerkamp R. F., 2007. Accuracy of breeding values when using and ignoring the polygenic effect in genomic breeding value estimation with a marker density of one SNP per cM. J. Anim. Breed. Genet. 124: 362–368 [DOI] [PubMed] [Google Scholar]
- Calus M. P. L., Meuwissen T. H. E., de Roos A. P. W., Veerkamp R. F., 2008. Accuracy of genomic selection using different methods to define haplotypes. Genetics 178: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster A., Bastiaansen J. W. M., Calus M. P. L., van Arendonk J. A. M., Bovenhuis H., 2010. Sensitivity of methods for estimating breeding values using genetic markers to the number of QTL and distribution of QTL variance. Genet. Sel. Evol. 42: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M. J., Pourahmadi M., 2002. Bayesian analysis of covariance matrices and dynamic models for longitudinal data. Biometrika 89: 553–566 [Google Scholar]
- de los Campos G., Naya H., Gianola D., Crossa J., Legarra A., et al. , 2009. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182: 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886 [DOI] [PubMed] [Google Scholar]
- de Roos A. P. W., Hayes B. J., Spelman R., Goddard M. E., 2008. Linkage disequilibrium and persistence of phase in Holstein, Friesian, Jersey and Angus cattle. Genetics 179: 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F. X., Clutter A. C., Lohuis M. M., 2007. Characterizing linkage disequilibrium in pig populations. Int. J. Biol. Sci. 3: 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., 2006. Prior distributions for variance parameters in hierarchical models. Bayesian Anal. 1: 515–533 [Google Scholar]
- Gianola D., Perez-Enciso M., Toro M. A., 2003. On marker-assisted prediction of genetic value: beyond the ridge. Genetics 163: 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola D., de los Campos G., Hill W. G., Manfredi E., Fernando R., 2009. Additive genetic variability and the Bayesian alphabet. Genetics 183: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola D., Wu X.-L., Manfredi E., Simianer H., 2010. A non-parametric mixture model for genome-enabled prediction of genetic value for a quantitative trait. Genetica 138: 959–977 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10: 381–391 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Wray N. R., Verbyla K., Visscher P.M., 2009. Estimating effects and making predictions from genome-wide marker data. Stat. Sci. 24: 517–529 [Google Scholar]
- Grapes L., Dekkers J. C. M., Rothschild M. F., Fernando R. L., 2004. Comparing linkage disequilibrium-based methods for fine mapping quantitative trait loci. Genetics 166: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A.J., Goddard M. E., 2009. Invited review: genomic selection in dairy cattle: progress and challenges. J. Dairy Sci. 92: 433–443 [DOI] [PubMed] [Google Scholar]
- Henderson C. R., 1976. A simple method for computing the inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 32: 69–83 [Google Scholar]
- Henderson C. R., 1984. Applications of Linear Models in Animal Breeding. University of Guelph, Guelph, Canada [Google Scholar]
- Hickey J. M., Gorjanc G., 2012. Simulated data for genomic selection and GWAS using a combination of coalescent and gene drop methods. G3: Genes, Genomes, Genetics 2: 425–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Goddard M. E., Visscher P. M., 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4: e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart C. J., Whittaker J. C., De Iorio M., Balding D. J., 2008. Simultaneous analysis of all snps in genome-wide and re-sequencing association studies. PLoS Genet. 4: e1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilkaya K., Tempelman R. J., 2005. A general approach to mixed effects modeling of residual variances in generalized linear mixed models. Genet. Sel. Evol. 37: 31–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., van der Werf J. H., Hayes B. J., Goddard M. E., Visscher P. M., 2008. Predicting unobserved phenotypes for complex traits from whole-genome SNP data. PLoS Genet. 4: e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legarra A., Robert-Granie C., Manfredi E., Elsen J.-M., 2008. Performance of genomic selection in mice. Genetics 180: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon B., Hoffman G., Mezey J., 2010. A variational Bayes algorithm for fast and accurate multiple locus genome-wide association analysis. BMC Bioinformatics 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A. J., Chao S. M., Asoro F. G., Heffner E. L., Hayashi T., et al. , 2011. Genomic selection in plant breeding: knowledge and prospects, pp. 77–123 in Advances in Agronomy, vol. 110. Elsevier–Academic Press, San Diego
- Meuwissen T., Goddard M., 2010. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185: 623–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara R. B., Sillanpaa M. J., 2009. A review of Bayesian variable selection methods: what, how and which. Bayesian Anal. 4: 85–117 [Google Scholar]
- Plummer M., Best N., Cowles K., Vines K., 2006. CODA: convergence diagnostics and output analysis for MCMC. R News 6: 7–11 [Google Scholar]
- Shepherd R. K., Meuwissen T. H., Woolliams J. A., 2010. Genomic selection and complex trait prediction using a fast EM algorithm applied to genome-wide markers. BMC Bioinformatics 11: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen D., Gianola D., 2002. Likelihood, Bayesian, and MCMC Methods in Quantitative Genetics, Springer-Verlag, New York [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., et al. , 2006a. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887 [DOI] [PubMed] [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Cookson W. O., Rawlins J. N. P., et al. , 2006b. Genetic and environmental effects on complex traits in mice. Genetics 174: 959–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423 [DOI] [PubMed] [Google Scholar]
- Villumsen T. M., Janss L., Lund M. S., 2008. The importance of haplotype length and heritability using genomic selection in dairy cattle. J. Anim. Breed. Genet. 126: 3–13 [DOI] [PubMed] [Google Scholar]
- Wiggans G. R., VanRaden P. M., Cooper T. A., 2011. The genomic evaluation system in the United States: past, present, future. J. Dairy Sci. 94: 3202–3211 [DOI] [PubMed] [Google Scholar]
- Yi N., Xu S., 2008. Bayesian Lasso for quantitative trait loci mapping. Genetics 179: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman D. L., Nunez-Anton V. A., 2010. Antedependence Models for Longitudinal Data. Chapman & Hall, London/New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.