Abstract
The genome sequence of the paleohexaploid Brassica rapa shows that fractionation is biased among the three subgenomes and that the least fractionated subgenome has approximately twice as many orthologs as its close (and relatively unduplicated) relative Arabidopsis than had either of the other two subgenomes. One evolutionary scenario is that the two subgenomes with heavy gene losses (I and II) were in the same nucleus for a longer period of time than the third subgenome (III) with the fewest gene losses. This “two-step” hypothesis is essentially the same as that proposed previously for the eudicot paleohexaploidy; however, the more recent nature of the B. rapa paleohexaploidy makes this model more testable. We found that subgenome II suffered recent small deletions within exons more frequently than subgenome I, as would be expected if the genes in subgenome I had already been near maximally fractionated before subgenome III was introduced. We observed that some sequences, before these deletions, were flanked by short direct repeats, a unique signature of intrachromosomal illegitimate recombination. We also found, through simulations, that short—single or two-gene—deletions appear to dominate the fractionation patterns in B. rapa. We conclude that the observed patterns of the triplicated regions in the Brassica genome are best explained by a two-step fractionation model. The triplication and subsequent mode of fractionation could influence the potential to generate morphological diversity—a hallmark of the Brassica genus.
ANCIENT polyploidies are prevalent in most eukaryotic lineages, including plants (Van De Peer et al. 2009; Jiao et al. 2011; Proost et al. 2011), fungi (Kellis et al. 2004), and animals (Jaillon et al. 2004; Aury et al. 2006). Much progress has been made in dating these evolutionary events and quantifying the retention and loss of gene duplicates after them. Gene content influences the potential for diversification and specialization of biological functions (Force et al. 1999) and the potential for increases in morphological complexity (Thomas et al. 2006). In both the eudicot and the monocot clades of flowering plants, there have been multiple rounds of polyploidy followed by selective gene losses (Tang et al. 2008, 2010), leaving the gene repertoire of many angiosperm species greatly expanded from an estimated ancestral (i.e., in the last common ancestor) gene number of 12,000–14,000 loci (Sterck et al. 2007; Tang et al. 2008).
Despite the initial expansion of gene numbers immediately following genome duplications, most lineages have since experienced drastic gene loss, genome downsizing (Bennett and Leitch 2005; Leitch and Leitch 2008), and ultimately genetic “diploidization” at many loci (Wolfe 2001). A number of mechanisms could lead to the diploidization, among which the “fractionation” of duplicate genes is a major force (Langham et al. 2004; Thomas et al. 2006). During the fractionation process, many gene copies with redundant functions and with product levels not under stringent control [“gene dosage” theory (Birchler and Veitia 2010)] tend to be lost, resulting in a reduction of gene complement that offsets the initial expansion from genome mergers. In the paleotetraploid maize, using the sorghum genome as an outgroup, the fractionation mechanism was shown to be predominantly short deletions, probably via intrachromosomal recombination, and is certainly not randomization by nucleotide substitutions (Woodhouse et al. 2010). By whatever mechanism, the initially near-identical subgenomes generated by whole-genome duplication events do not fractionate equally—one subgenome consistently has more genes retained on it than the other; this holds true for eukaryotes ranging from Paramecium to flowering plants and fish (Sankoff et al. 2010). This phenomenon, called “fractionation bias,” was first described in the Arabidopsis genome (Thomas et al. 2006) and later generalized throughout major eukaryote lineages with paleopolyploidies (Sankoff et al. 2010).
In plants, if not all eukaryotes, when two genomes find themselves in the same nucleus, one subgenome—as defined by fractionation bias—expresses its genes to a higher mRNA level than does the other subgenome. This is the phenomenon of genome dominance (Schnable et al. 2011). Since the 12-million-year-old maize paleotetraploid displays substantial genome dominance (Schnable et al. 2011), the result that Brassica rapa subgenome III expresses its genes to a higher level than does either subgenome I or II was more affirming than surprising (Wang et al. 2011). However, genome dominance is most evident when tetraploidy was recent: in synthetic and natural hybrids and allotetraploids of cotton (Flagel and Wendel 2010), wide hybrids of Arabidopsis species (Wang et al. 2006; Chang et al. 2010), allotetraploids of Tragopogon species (Buggs et al. 2010a,b), and synthesized Brassica lines (Gaeta et al. 2007; Xiong et al. 2011). It is not yet fully understood why genome dominance persists until today after tens of millions of years of evolution.
The diploid Brassica species were first hypothesized to have been triplicated on the basis of comparative mapping studies (Lagercrantz and Lydiate 1996; Lagercrantz 1998; Parkin et al. 2003, 2005). There was some skepticism on the basis of the observation that most loci were not triplicated; however, subsequent BAC-FISH (Lysak et al. 2005) and comparative BAC sequencing studies (Yang et al. 2006) further supported the triplication hypothesis. The recent sequencing of B. rapa has confirmed the genome triplication event that occurred in the common ancestor of all Brassica species (Wang et al. 2011).
It was demonstrated that B. rapa underwent biased fractionation—subgenome III has retained almost two-thirds of Arabidopsis thaliana orthologous genes, while subgenomes I and II have retained significantly fewer genes (Wang et al. 2011). On the basis of biased fractionation results much like those in B. rapa, the eudicot paleohexaploidy, known as the gamma event, was proposed to have happened by a two-step fractionation process (Lyons et al. 2008). Fortunately, the relatively recent paleohexaploidy in B. rapa and the position of the Arabidopsis genome as an outgroup provide a phylogenetic system with superior analytical power. The two-step fractionation hypothesis was suggested for Brassica’s biased fractionation, to explain the fact that subgenome I is the most fractionated genome and subgenome III the least fractionated genome (Wang et al. 2011). However, this hypothesis was not formally tested.
Herein, we test the “two-step fractionation” hypothesis by examining short, exonic deletions in retained Brassica genes, using Arabidopsis as the outgroup. Such deletions were associated with recent, ongoing biased fractionation in maize (Woodhouse et al. 2010). We found that subgenome II had more deletions than subgenome I or subgenome III, suggesting that a two-step process of genome fractionation did indeed occur. We also show that deletions tend to accumulate in multicopy retained genes rather than in genes retained as a single copy, a phenomenon best explained by relaxed selection in duplicate genes.
Methods
Partitioning of subgenomes according to number of retained genes
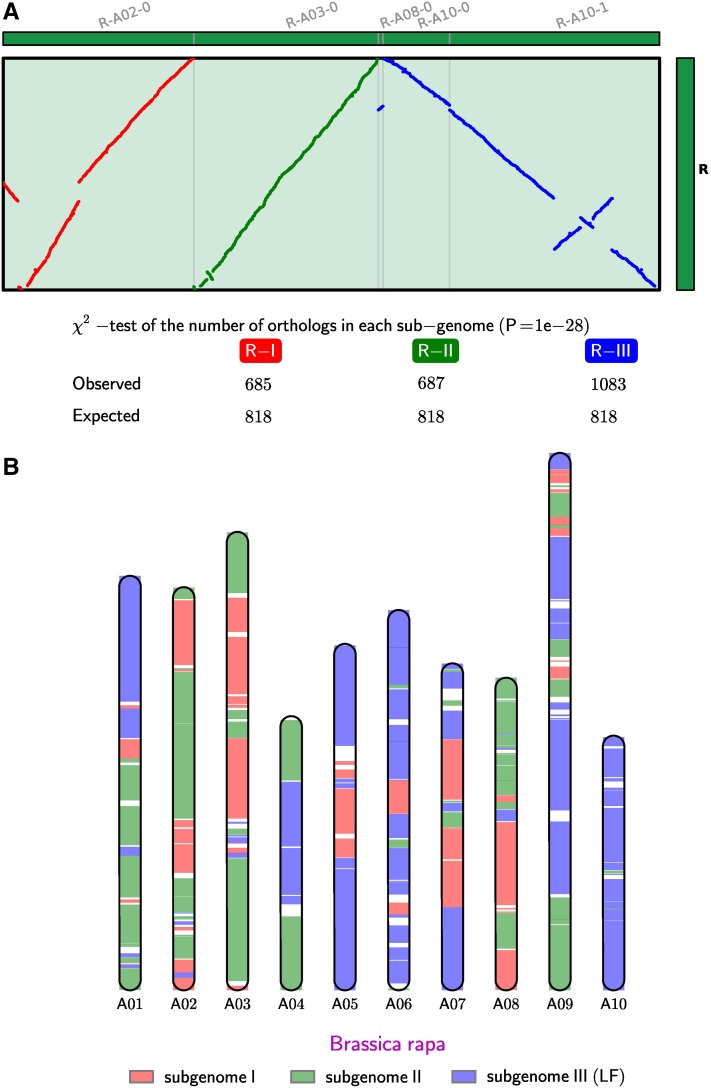
The identification of orthologous regions and partitioning into subgenomes follow the method described in supporting information, File S1, in the B. rapa release (Wang et al. 2011). Briefly, multiple chromosomal segments in B. rapa that are orthologous to the same A. thaliana segment are numbered accordingly, using the established “A to X” numbering system (Wang et al. 2011). All B. rapa segments that match to the same A. thaliana segment are partitioned into three subgenomes (for example, segments matching A. thaliana segment R are partitioned into R-I, R-II, and R-III) (Figure 1A). We exhaustively enumerated all partitions and evaluated each partition on the basis of heuristic rules that were detailed in Wang et al. (2011). After the partitioning, we counted the number of syntenic orthologs within each subgenome. According to the number of retained orthologous genes in each subgenome, each segment was classified and named I, II, and III for “most fractionated,” “moderately fractionated,” and “least fractionated,” respectively, for each A to X segment (Figure 1B). We then examined the number of orthologous genes in each block; nearly all blocks showed a significant difference (with P-value cutoff = 0.01) in gene numbers between the three subgenomes, with the only exception being block T. Finally, we concatenated each set of most fractionated (A-I, B-I . . . , to X-I), moderately fractionated (A-II, B-II . . . , to X-II) and least fractionated blocks (A-III, B-III . . . , to X-III), respectively, for downstream analyses.
Figure 1 .
(A) Dot plot between B. rapa and A. thaliana, with B. rapa segments that are derived from the same A. thaliana origin grouped together, to illustrate the partitioning and test of nonrandom fractionation among B. rapa triplicated regions. This shows only one of the 24 sets of blocks (block R). The table under the dot plot contains the counts of A. thaliana–B. rapa orthologs in the respective subgenomes. Gene losses are not equally distributed in most of the duplicated blocks, as tested by a χ2-test (P = 1 × 10−28 in the case of block R). (B) The partitioning of B. rapa chromosomes into three inferred subgenomes following the partitioning algorithm in Wang et al. (2011).
Determining the sequence divergence among B. rapa homeologs
For paired genes inferred from syntenic alignments, we aligned the protein sequences using CLUSTALW (Larkin et al. 2007) and used the protein alignments to guide coding sequence alignments by PAL2NAL (Suyama et al. 2006). To calculate Ks, we used the Nei–Gojobori method implemented in the yn00 program in the PAML package (Yang 2007). A Python script was used to create a pipeline for all the calculations and is available at http://github.com/tanghaibao/bio-pipeline/tree/master/synonymous_calculation/. The actual distribution of Ks values is modeled and fitted as a log-transformed normal distribution (Tang et al. 2008).
Automated cataloging of internal deletion sites within the B. rapa genes
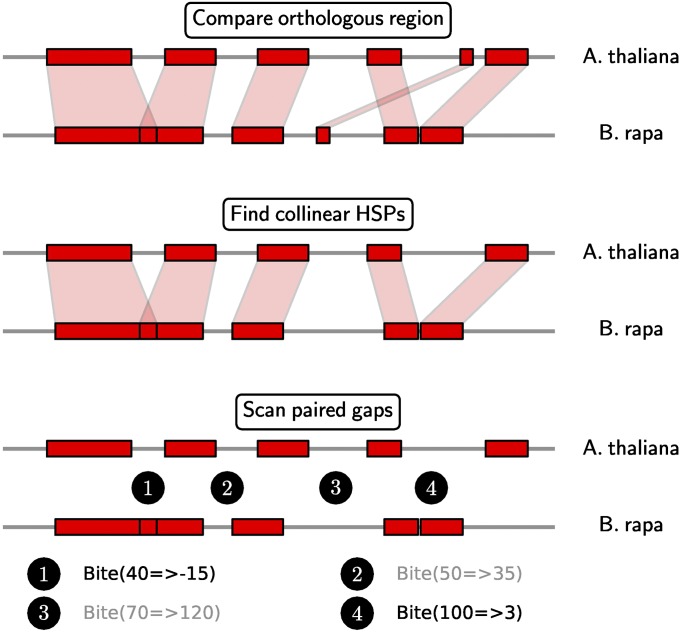
The sites of deletions were identified by an automated pipeline, illustrated in Figure 2. Using the A. thaliana orthologs as reference, we aligned one, two, or three homeologs in B. rapa. For each of 3648 (A. thaliana, B. rapa) pairs, we detected deletions of various sizes in the B. rapa gene compared to the A. thaliana gene. The DNA sequences of the complete genes (containing all exons and introns) were extracted for the BLASTN comparisons. For each gene pair, we used BLASTN with parameters favoring short, strong sequence matches (word size 7, spike length 15 bp, low-complexity filter off). We identified all collinear high-scoring segment pairs (HSP) through the “heaviest increasing subsequence” algorithm (Kurtz et al. 2004). There are unmatching sequences (gaps) between adjacent HSPs. For each gap pair, we noted the size in A. thaliana, as well as in B. rapa, and identified all the sites that were smaller in the B. rapa genes. Links to the GEvo (Lyons and Freeling 2008) URL were configured in the spreadsheet to assist manual proofing.
Figure 2 .
The pipeline for automated deletion discovery. We first compared between A. thaliana and B. rapa orthologous genes using BLASTN. From the initial BLASTN HSPs, we computed a set of collinear HSPs. The unmatching regions in A. thaliana and B. rapa are compared in a pairwise fashion, recording the sizes of the corresponding gaps in A. thaliana and B. rapa, in a notation of “Bite (A → B)”. We selected only the deletions that have A > 30 bp and B < 10 bp, to screen for substantial downsizing in the B. rapa sequence. As examples, the bites in black color are selected on the basis of these criteria whereas the gray ones are ignored.
The changes in the sizes of the sequences are documented in the notation “Bite (A → B)”, which means there are A bases in A. thaliana, but B bases in B. rapa (Figure 2). For example, “Bite (81 → 0)” means 81 A. thaliana bases were removed in the B. rapa gene. A very useful effect of this notation is that it is also possible for “B” to be negative. For example, “Bite (82 → −7)” means 82 A. thaliana bases were removed and adjacent HSPs overlap by 7 bases. This is an indication of 7 bases of flanking direct repeats (as proposed in Woodhouse et al. 2010). We applied cutoffs of A > 30 bp and B < 10 bp, to select DNA chunks that decreased in size from A. thaliana to B. rapa. Exonic deletions were further identified for the deletion locations that intersect A. thaliana exon locations, using the tool INTERSECTBED (Quinlan and Hall 2010).
The full catalog containing a total of 4539 deletion sites along with their locations, gene identifiers, deleted bases, and GEvo links is available in File S1.
Simulation of deletions of homeologs and likelihood-ratio test for model selection
On the basis of the initial hypothesis of a deletion mechanism that independently eliminates one gene at a time, a simulation of gene loss was carried out. Starting with a length equal to the number of all genes, genes were deleted at random until the simulated number of deletions was equal to the true observed number. The distribution of apparent deletion lengths for the run was then saved, and the preceding steps were repeated 1000 times. This gives a distribution of deletion lengths.
A genetic algorithm (GA) using 20 character states, each representing a deletion length of various lengths, was used to determine, given the region length and the distribution of observed deletion lengths, the most likely deletion model to achieve the best match between simulated and observed data. The fitness values of solutions in the genetic algorithm were scored after each step with the fittest solutions being those where the simulated number of deletion runs was least different from the observed number of runs. The components for our deletion size simulation include the following:
Simulate under “model 1 (with only deletion size of one gene)” and then report counts for various deletion sizes.
Simulate under “model 1+2 (with deletion size up to two genes)” and then report counts for various deletion sizes.
Continue the simulation. Add one more deletion size for each new model.
Likelihood-ratio test to see which model gives the best likelihood while keeping the model as simple as possible (based on Occam’s razor): The likelihood function is defined as , where Ci is the simulated count and pi is the actual frequency of deletion size i.
Scripts that perform the simulations and likelihood calculation are available at http://github.com/tanghaibao/bio-pipeline/blob/master/gap_simulations.
Results
One subgenome has retained significantly more genes than the other two
As noted by Wang et al. (2011), subgenomes I, II, and III have retained 5966, 7679, and 11,536 genes, respectively (ignoring genes that do not show conserved synteny with A. thaliana, e.g., those that are unique to B. rapa or have transposed) (Table 1). We find a similar trend in number of nucleotides per subgenome and number of genes per subgenome (both retained and nonretained) (Table 1). The difference in size among all three subgenomes (subgenome I is the smallest and subgenome III the largest) is primarily due to the level of biased fractionation among the three subgenomes. Conversely, whole-gene deletions are 2.1 times more frequent in I than in III (10,423 deletions in subgenome I vs. 4853 deletions in subgenome III). There are also significant differences in numbers of singletons (no whole genome duplicates) in the three subgenomes. The genes that exist only on I, II, and III total 1592, 2449, and 5211, respectively, which suggests that most single-copy genes are retained in the least fractionated subgenome III. In contrast, the differences in gene densities of the three subgenomes are less dramatic than the sheer counts (Table 1). We conclude that (1) the observed gene retention bias cannot be explained by uneven gene density (for example, varied level of heterochromatic vs. euchromatic sequences) and (2) the sequence removal mechanism that has shaped the retention bias did not exclusively target gene-rich regions.
Table 1 . The number of genes and retained genes in each of the three subgenomes in B. rapa when compared to A. thaliana.
| Subgenome I | Subgenome II | Subgenome III | Arabidopsis | |
|---|---|---|---|---|
| Genome span (Mb) | 56.9 | 78.1 | 104.6 | 119.1 |
| No. genes | 8,890 | 11,957 | 16,838 | 27,134 |
| Gene density (genes/Mb) | 156.3 | 153.0 | 160.9 | 227.8 |
| No. retained genes | 5,966 | 7,679 | 11,536 | 16,423 |
| Retained gene density (genes/Mb) | 104.9 | 98.3 | 110.3 | 137.8 |
| % genes retained (compared to Arabidopsis) | 36 | 46 | 70 | 100 |
The “number of retained genes” in each subgenome is taken from Wang et al. (2011).
Genetic distances to A. thaliana orthologs cannot distinguish among B. rapa subgenomes
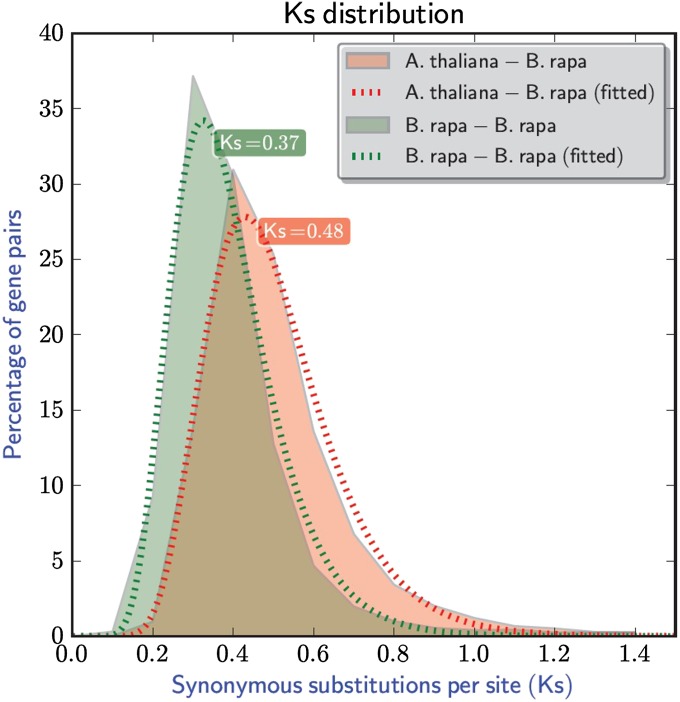
The median Ks value between A. thaliana–B. rapa orthologs is 0.48, while the median Ks value between B. rapa–B. rapa homeologs is 0.37 (Figure 3), supporting the conclusion that Brassica hexaploidy occurred after its divergence from Arabidopsis (Wang et al. 2011). Both A. thaliana–B. rapa gene pairs and the B. rapa–B. rapa gene pairs show a unimodal peak in the Ks distribution (Figure 3).
Figure 3 .
Ks distribution between A. thaliana–B. rapa orthologs and B. rapa–B. rapa homeologs. Solid lines are the observed distribution, and dashed lines are the fitted distribution based on log-normal distribution (Tang et al. 2008).
We collected A. thaliana genes that were represented in B. rapa by two or three orthologs. For each of these orthologs, we noted their subgenome assignment and determined their Ks value in comparison with their single A. thaliana ortholog. The A. thaliana–B. rapa Ks values were compared in a pairwise fashion with the “winner” subgenome inferred (Table 2). The distance between A. thaliana–subgenome III appears to be slightly larger than the distance between A. thaliana–subgenome II (χ2-test, P = 0.004), while the other two pairwise comparisons are not significant at the α = 0.01 level. This suggests that although Ks is able to clearly differentiate between the time of Arabidopsis–Brassica divergence and the hexaploidy, it fails to differentiate the three subgenomes within the hexaploidy.
Table 2 . “Horse race” Ks comparisons.
| “Horse race” Ks comparisons | Counts | P-value (χ2-test) |
|---|---|---|
| I-At > II-At | 1191 | 0.489 |
| I-At < II-At | 1225 | |
| II-At > III-At | 2256 | 0.004* |
| II-At < III-At | 2067 | |
| I-At > III-At | 1678 | 0.027 |
| I-At < III-At | 1809 |
The distances of two B. rapa genes to the A. thaliana reference gene can be compared to each other. For example, 1191 of “I-At > II-At” means that among all the I, II comparisons, 1191 of them showed a higher Ks value of the gene in subgenome I than the gene in subgenome II. *P-value significant at α < 0.01. At, A. thaliana.
Additionally, we employed a tree-based method to attempt to differentiate the B. rapa triplets. We used PhyML (Guindon and Gascuel 2003) to construct the phylogeny of the 3 B. rapa genes, using the single A. thaliana gene as an outgroup. We evaluated a total of 1655 trees with B. rapa triplets. A total of 952 (58%) trees had poor branch support (aLRT value ≤ 0.8), suggesting that in most cases, the relationships among the B. rapa triplets are poorly resolved. Even among the trees that have good resolution on the splitting of the triplets, 243 trees have the “(I, II), III” topology, 203 trees have the “(I, III), II” topology, and 257 trees show the “(II, III), I” topology. These counts do not favor a dominant topology (P = 0.04, χ2-test, significance level = 0.01).
In general, our findings are in agreement with previous results (Wang et al. 2011). On the sequence level, all three subgenomes appear equally diverged from A. thaliana, as would be expected if the divergence between the Arabidopsis and Brassica genomes predated the triplication.
Deletions in B. rapa genes through comparison to the A. thaliana ortholog
To understand the mechanism underlying biased whole-gene removals in the three subgenomes, we asked whether there are differences in the rate of sequence removals within the gene sequences. We cataloged a list of sequence removal events on the basis of pairwise comparisons between each B. rapa gene and its A. thaliana orthologs through an automated pipeline. Briefly, we listed the intervening gaps between the adjacent matching regions (HSPs) and checked whether the corresponding gap in B. rapa was substantially smaller than the corresponding gap in A. thaliana (see Methods) (Figure 2). In this study, we focused only on the deletions that are >30 bases. Shorter deletions are likely affected by the artifacts of sequence alignments, so this arbitrary cutoff is a result of our favoring accuracy over sensitivity.
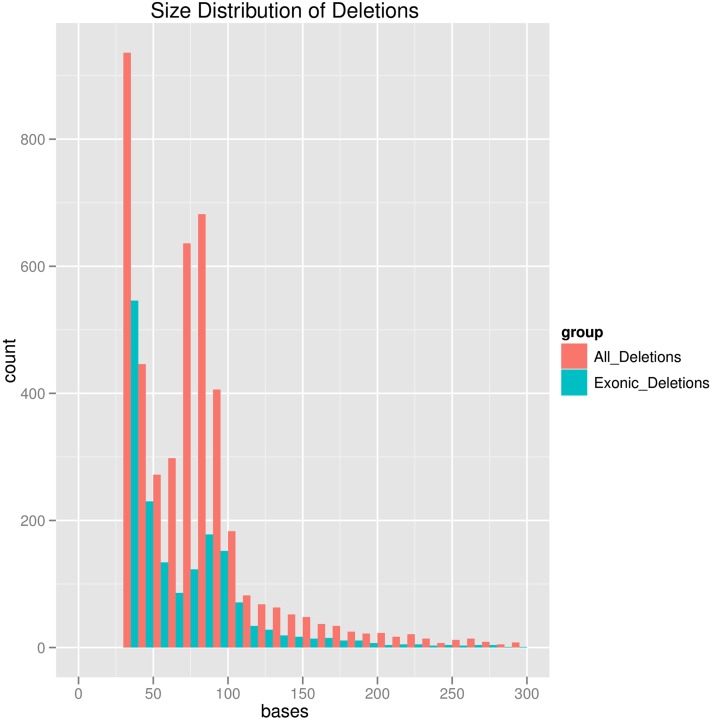
Using our automated deletion discovery pipeline, we identified a total of 4539 deletion sites of 3648 B. rapa genes examined (14.5% of B. rapa genes inspected in this study) (all deletion sites identified are available in File S1). Some B. rapa genes have experienced more than one deletion. Gap sizes ranged from 31 bases (just above the computational cutoff) to 1363 bases, with the size distribution shown in Figure 4. There is an apparent excess of deletion sizes between 70 and 80 bases, in addition to the peak at smaller deletion size ranges.
Figure 4 .
Size distribution of the deletions in B. rapa genes (sequences present in A. thaliana but removed in B. rapa) that we cataloged in this study. The distribution stops at 30 since we focus only on the deletions that are >30 bases (a computational cutoff).
Different parts of the genes have experienced different rates of deletion. Deletions in the intronic sequences were approximately eight times more likely than in exonic sequences (1.63% of total intronic bases vs. 0.24% of exonic bases; Table 3). 5-′ or 3-′ untranslated regions (UTRs) have incurred the fewest deletions, even fewer than in exons (0.04% of UTRs vs. 0.24% of exonic bases), suggesting that some UTRs have functional roles and are under strong purifying selection. Inferred deletions that fall within sequences corresponding to Arabidopsis exons are likely to be the most reliable and therefore are the types of deletions we used to investigate the mechanism of biased gene deletion.
Table 3 . The locations of the deletion sites in A. thaliana genes identified through A. thaliana–B. rapa comparisons.
| No. deletions | No. bases | No. in A. thaliana genome | % | |
|---|---|---|---|---|
| Coding sequence (CDS) | 1,863 | 78,125 | 33,050,356 | 0.24 |
| Introns | 3,345 | 319,220 | 19,590,057 | 1.63 |
| 5′-UTR | 27 | 1,563 | 2,610,978 | 0.06 |
| 3′-UTR | 49 | 2,604 | 4,442,021 | 0.06 |
Note that it is possible for some deletions to be situated across exon–intron boundaries.
Distribution of deletion sites among B. rapa homeologs
We anticipated that exonic deletions would be rarest in genes within subgenome III and that genes in subgenome I would be the most likely to have gaps. Unexpectedly, we found that a higher proportion of genes in subgenome II had deletions vs. those in subgenome I (7.9% vs. 7.1%; Table 4). This is true whether we count the number of deletion sites or count the number of deleted bases. These data track observation of whole-gene fractionation bias among the three subgenomes, in that subgenomes I and II had more numbers of genes with deletions than subgenome III. However, subgenome II still had more genes with exonic deletions than expected, given the overall genome fractionation bias as discussed earlier.
Table 4 . Number of deletions and exonic deletions within B. rapa genes, grouped on the basis of their subgenome assignments and copy numbers.
| No. exonic deletions | No. deleted exonic bases | No. genes | % genes with exonic deletions | |
|---|---|---|---|---|
| Subgenome I | 423 | 18,155 | 5,966 | 7.1 |
| Subgenome II | 609 | 27,096 | 7,679 | 7.9 |
| Subgenome III | 714 | 32,874 | 11,537 | 6.2 |
| Singlet | 594 | 28,131 | 9,252 | 6.4 |
| Doublet | 799 | 33,752 | 10,962 | 7.3 |
| Triplet | 353 | 16,242 | 4,968 | 7.1 |
We also observed differences of deletion frequencies in singlet, doublet, or triplet genes in B. rapa. Singlet genes contain significantly fewer deletions than doublets or triplets. A total of 6.4% vs. 7.1% and 7.3% of the singlet, doublet, and triplet genes contain deletions, respectively (Table 4). This is consistent with different selection regimes on single-copy genes relative to genes with duplicate copies. Single-copy genes are expected to be under stronger purifying selection than genes with duplicate copies that can potentially buffer their functions. We further note that the real differences of the strength of purifying selection on singlet and duplicate genes might be larger than we have observed. The deletions we have counted include the selectively neutral as well as deleterious deletions. Indeed, there is a background rate for neutral deletions, which are expected to be the same between singlet and duplicate genes. This background component in our deletion counts dilutes the signal, reflecting only the purifying selection.
Direct repeats flanking the removed sequences
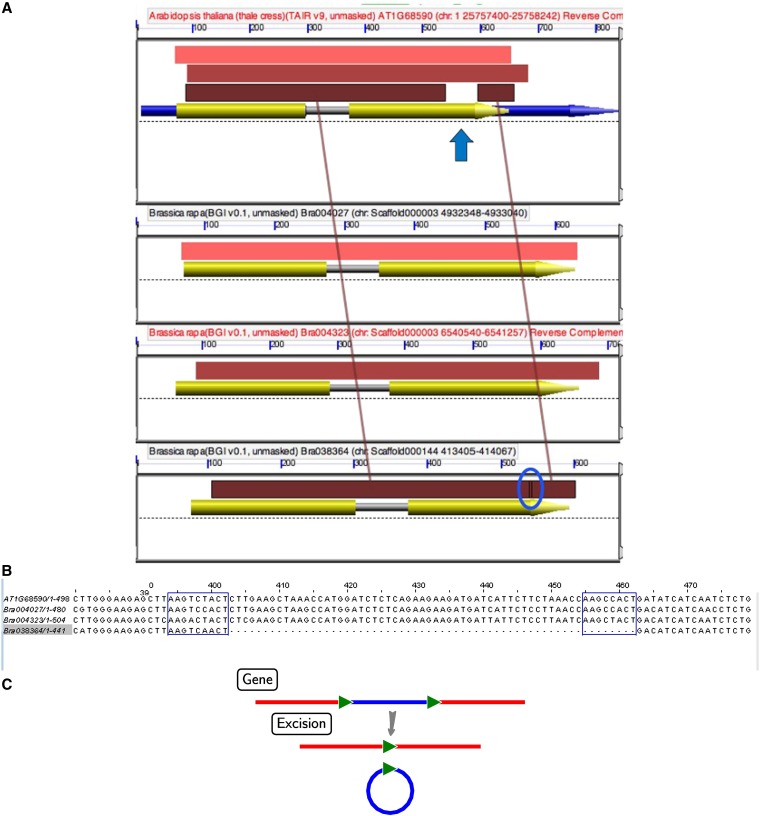
In maize, sequence deletions flanked by direct repeats (Woodhouse et al. 2010) are associated with the biased fractionation among homeologous regions. In B. rapa, about one-third of exonic deletions were flanked by direct repeats, with length up to 19 bp (see Methods). Only one copy of the two original repeat units remained at the deletion site, probably as a direct result of the deletion mechanism (Figure 5). These data suggest that fractionation via small deletions occurs in B. rapa as it does in maize (Woodhouse et al. 2010) and may be a phenomenon general to plants.
Figure 5 .
(A) A GEvo graphic of BLASTN output between orthologous genes in A. thaliana and three B. rapa homeologs. The top panel is a region in A. thaliana and used as the reference, and the following three panels are three B. rapa regions that were derived from the recent hexaploidy event. Arrows represent gene models and colored rectangles show the extents of BLASTN matches (high-scoring sequence pairs, HSPs). The colored rectangles (pink, tan, and brown) represent HSPs or regions with high sequence similarity to each other. A. thaliana is the reference sequence (top panel). As can be seen, a gap is evident (blue arrow) when comparing the HSPs of AT1G68590 and Bra038364 (bottom panel). The deleted sequence (circled in blue) is evident in comparison to other B. rapa homeologs. An overlap between the HSP blocks that flank the deletion can be seen (blue circle); this indicates that the predeleted sequence was flanked by direct repeats. To reproduce this analysis, go to (http://genomevolution.org/r/rmi). (B) ClustalW alignment of the A. thaliana and the three B. rapa sequences from A. The sequences in the blue box in the whole B. rapa homeologs indicate the locations of the direct repeat sequence that originally flanked the deletion in the homeolog containing the deletion (Bra038364). (C) Proposed mechanism for the within-gene deletion via intrachromosomal illegitimate recombination.
Several instances of the flanking repeats are given in Table 5. Some repeats are low-complexity simple sequence repeats (SSRs), e.g., trinucleotide repeats (GAT)n, (TTC)n (Table 5). SSRs have been shown to have high potential for illegitimate recombination of genes (Rocha et al. 2002). Other repeat instances with higher nucleotide complexity are also present. Direct repeats are known to be hotspots of homologous recombination between the repeat units, making the intervening sequences more easily removed (Figure 5C).
Table 5 . Partial list of instances of internal deletions within B. rapa genes that are flanked by direct repeats.
| Deletion ID | Left flank | Right flank |
|---|---|---|
| AT1G10570_Bra019923_Bite(32 → −19) | ggagaatttagtgtattga | agagaatttagtgtattga |
| AT1G47970_Bra018696_Bite(119 → −19) | gacgatgacgatgatgatg | gacgatgatgatgaggatg |
| AT1G52870_Bra018995_Bite(104 → −19) | tttgatgttgcttgagtga | tttgatgttgcttgagtga |
| AT2G21560_Bra030293_Bite(44 → −19) | cttttcttcagtgctctgt | cttatcttcaatgctctgt |
| AT2G44160_Bra004810_Bite(44 → −19) | ttaatgtagataccagctg | ttaatgtagataccagttg |
| AT3G03590_Bra031985_Bite(298 → −19) | tgtaaagactctaagcaaa | tgtgaagactctaaacaaa |
| AT3G49140_Bra018005_Bite(81 → −19) | aacctcagtcattctcttt | aacctcagtcattctcttt |
| AT3G51260_Bra036824_Bite(43 → −19) | catgttctataactaaacc | aatgttctataactaaacc |
| AT4G02880_Bra018525_Bite(50 → −19) | tgaaaatagtgatgccgag | tgaaaatggtgatccagag |
| AT4G18430_Bra012597_Bite(99 → −19) | atctagtcaaatattatat | atctagttaatattatatt |
| AT5G40120_Bra025619_Bite(89 → −19) | gttaatgcagcaggagctt | gttgatgcagcatgaactt |
| AT5G46740_Bra025000_Bite(62 → −19) | cagcaaatggcttctcaga | cagcaaatggtttctcaga |
| AT5G61150_Bra029332_Bite(101 → −19) | ttcctcttcttcatcttca | ttcttcctcttcttcttcc |
This list only shows the flanking sequences that are 19 bases in length (an arbitrary number). See Methods for the notation of deletion identifiers.
Distribution of transposable-element–related sequences
Any sort of mechanism that removes DNA in the genome could potentially be “induced” by a transposon bloom. Since we are testing the two-step model for paleohexaploid fractionation, a past transposon bloom could affect each subgenome in different ways. The deletion mechanism in plants has been hypothesized as an adaptation to fight “genetic obesity” (Devos et al. 2002).
Identification of the B. rapa interspersed repetitive elements followed published methods (Wang et al. 2011). Elements were categorized into classes, with long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), long terminal repeats (LTRs), and DNA transposons being the largest classes (Table 6). Among all major classes of transposable elements (TEs), the distributions are not biased toward any subgenome, suggesting that the “background” insertion and removal rates are equal across the three subgenomes, at least when viewed as they exist today. None of the most abundant TE families with the large counts (>500 copies) across the genome showed any preference for a single subgenome (P-value cutoff = 0.01, χ2-test).
Table 6 . The number of major classes of transposable elements in three subgenomes.
| Frequency in subgenomes (counts/Mb) | ||||
|---|---|---|---|---|
| Type | Total counts | I | II | III |
| LTR | 62,510 | 239 | 248 | 241 |
| LINE/L1 | 28,215 | 109 | 112 | 108 |
| LTR/Copia | 24,809 | 94 | 100 | 95 |
| LTR/Gypsy | 17,004 | 69 | 70 | 61 |
| DNA/hAT-Ac | 13,715 | 54 | 54 | 52 |
| DNA/En-Spm | 10,438 | 41 | 42 | 39 |
| SINE | 10,232 | 40 | 39 | 40 |
| DNA/MuDR | 8,950 | 35 | 35 | 34 |
| DNA/Harbinger | 4,990 | 18 | 21 | 19 |
| DNA/TcMar-Pogo | 4,983 | 18 | 20 | 19 |
| DNA | 4,701 | 18 | 19 | 18 |
| DNA/hAT | 4,542 | 18 | 18 | 17 |
| RC/Helitron | 3,190 | 11 | 13 | 12 |
| LINE/Penelope | 3,096 | 11 | 12 | 12 |
| DNA/TcMar-Stowaway | 2,674 | 11 | 10 | 10 |
| DNA/hAT-Tag1 | 2,262 | 8 | 10 | 9 |
Discussion
Two-step genome merger model could explain the retention bias
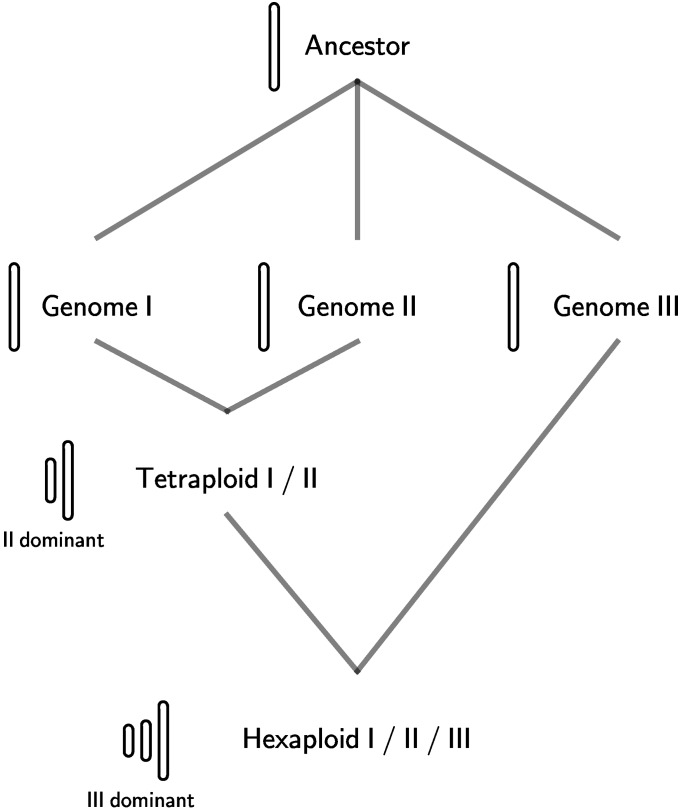
The two-step model for paleohexaploidy formation and fractionation (Lyons et al. 2008) suggests that two of the genomes came together first, and then the third genome was added some time later (Figure 6). The common way to form a hexaploid is between a diploid (2N) and a tetraploid (4N) cross, resulting in a triploid, which on doubling produces a hexaploid. If this were the case, two subgenomes could be in the same nucleus for a longer period of time (as a viable tetraploid) than the third, which is then relatively less fractionated than the first two. Additional support comes from the gene loss pattern between subgenomes I and II, where low-density regions of one of the two more fractionated genomes are compensated by less loss on the other, which indicates that the tetraploid genome (I + II) could be viable since most genes tend to have at least one copy (Wang et al. 2011).
Figure 6 .
The proposed “two-step” model of genome mergers. First, genome I and genome II form a tetraploid, and subsequent addition of genome III forms the hexaploid. Such stepwise genome additions involve shifting roles of the “dominant” genome: in the formation of the tetraploid, subgenome II was the dominant genome, whereas in the hexaploid, subgenome III became the new dominant genome.
We devised an experiment to test for the B. rapa genes that recently underwent fractionation. We reasoned that, if subgenomes I and II had been “at war” for a long time, then perhaps the nondominant genome, I, had already lost nearly all the genes it could lose, so that, when III entered the fray, subgenome II would still have removable genes to be deleted. This is indeed what we observe: subgenome II is the one that has incurred the most exonic deletions, rather than subgenome I.
While the two-step model is the general expectation for the formation of a hexaploid, our model does require fractionation to also have occurred in two distinct steps. There are other alternatives that we have to rule out. For example, the two genome mergers might have occurred sequentially so there was no time to fractionate after the first step. While this alternative is technically two-step formation, in terms of fractionation it is one step. We rendered this hypothesis unlikely.
Rates of recent deletions are actively influenced by selection on gene functions
We found that deletions in the intronic sequences have occurred approximately eight times more frequently than deletions in the exonic sequences (Table 3). Intuitively, corresponding exons are usually much more conserved in genome comparisons than the sequences of the introns, when exons are under strong selective pressure to retain coding capacity. Although the majority of the deletions occurred in the intronic sequences, there are still deletions in the exonic sequences that are tolerated by the B. rapa genes in our comparisons. Both functionally and in terms of technical reliability, exonic deletions are the more relevant to our study.
We consider the deletions in exonic sequences to be recent events, following the assumption that once a gene undergoes short deletions within exons, it is increasingly likely to be rendered nonfunctional, and more deletions will eventually accumulate until the gene sequence is no longer identifiable. Therefore, the presence of one or a few small deletions in the exonic sequences of any gene suggests that the deletion event was relatively recent.
On the whole-gene level, we found that singly retained genes contain fewer sequence removals than the doublets or triplets. In general, genes with duplicate copies show a higher likelihood of functional compensation than single-copy genes (Gu et al. 2003). However, there are many exceptions to this rule. Many duplicate genes that have survived the sequence removal processes following polyploidy have diversified in their regulatory roles (Tang et al. 2010) and have acted as the hub or bottleneck enzymes of metabolic pathways (Wu and Qi 2010). One well-studied example is the FLC gene, for which all three copies are retained in the B. rapa genome (Schranz et al. 2002). All three B. rapa homeologs are additive, in a dosage-dependent manner. We failed to find exonic deletions in any one of the three copies of the FLC homeologs in B. rapa, suggesting that the structures of the three copies are relatively intact. There is likely selective advantage in retaining multiple copies of those genes critical to the adaptation to the environment, e.g., flowering time, so that the actual dosage of the protein can be finely modulated.
Recent deletions in B. rapa homeologs are affected by a complex interplay of genome dominance
Our initial expectation was that the ratios of small deletion events per subgenome should match the gene fractionation ratio per subgenome. In maize, the deletion bias matched the fractionation bias between homeologs (Woodhouse et al. 2010). However, in B. rapa, the ongoing deletions do not closely reflect the pattern of gene loss: among the three subgenomes of B. rapa, subgenome II appears to have undergone more deletions than either subgenome I or subgenome III (Table 4).
Our proposed two-step fractionation hypothesis is capable of explaining this unexpected deviation of subgenome II from the general trend of fractionations. During the first genome merger, fractionation bias favored the retention of genes from one subgenome vs. another, and then the more fractionated genome may have reached its level of saturation for fractionation (subgenome I). When a new genome was introduced, the earlier, more retained genome (subgenome II) may have undergone more fractionation in comparison to the new genome as well as the older, greatly fractionated genome (Figure 6). In this case, one would expect to see more deletions in the genome undergoing more recent fractionation. We would then hypothesize that the genome now undergoing the most fractionation is subgenome II. The two-step process involves a shifting role for subgenome II: after the first step, it is the dominant genome, but only until the second step, when subgenome III dominates both other subgenomes (I and II veteran subgenomes) (Figure 6).
Deletions, not point mutations, are the major mechanism for gene inactivation
We failed to observe consistent relationships among the three subgenomes using both the synonymous substitutions (Ks) and the gene tree method, indicating that base substitutions do not account for the fractionation biases or the three parental species are approximately equally diverged. In contrast, the deletion rates are distinctly different among the three subgenomes, suggesting that the deletions contribute more to the fractionation process than point mutations.
In mammals, the mode of gene inactivation appears to be a pseudogene pathway, which mostly involves point mutations that over time accumulate to render gene products nonfunctional (Schrider et al. 2009). In the case of B. rapa, we favor the sequence removal model as we have observed the presence of small deletions within exons that track the fractionation biases (more exonic deletions in subgenomes I and II than in subgenome III), while in comparison, frequencies of point mutations among the three subgenomes are similar.
Sequence removals are likely facilitated by illegitimate recombination via direct repeats
We argue that fractionation in B. rapa appears to be due in part to a short deletion mechanism via illegitimate recombination, similar to previous observations in maize (Woodhouse et al. 2010). Direct repeats exhibit high levels of recombination intensity (Rocha 2003). Bzymek and Lovett (2001) proposed three major mechanisms for illegitimate recombination: simple replication slippage, sister-chromosome exchange-associated slippage, and single-strand annealing. Presence of the genomic repeats in proximity, e.g., simple di- and trinucleotide repeats, increases the likelihood of illegitimate recombination (Table 5).
Illegitimate recombination is not the only avenue for sequence removals. Interspersed repeats also tend to carry and transpose DNA segments. We could not find such a bias on the basis of our scan for major types of repeat elements (Table 6), suggesting that the sequence removals—especially the “biased” removal patterns—are unlikely to be caused by interspersed repetitive elements, e.g., transposons or retrotransposons.
Cumulative small deletions set the ground for whole-gene removals
We simulated the process of generating the observed gap patterns. Our likelihood-ratio test combined with simulations (see Methods) suggested that small deletion sizes consisting of mostly single-gene and a small number of two-gene deletions are sufficient to generate the observed patterns. The pattern of predominantly one- or two-gene removals at a time is in concordance with the observation of small, internal deletion of genes, in which small chunks of sequences are removed per deletion event. However, when enough sequence removals have accumulated and/or removal of sequences containing critical functional domains has taken place, the entire gene function is compromised and thus more mutations will follow, since there is no purifying selection in place to protect against deleterious mutations. We therefore view the short deletion mechanisms as cumulative mutations that eventually resulted in whole-gene removals that have shaped the gene loss patterns we observe.
Alternative hypotheses that might explain genome dominance in B. rapa
Although we have support for the “two-stage” scenario through the observation that subgenome II contains the most exonic deletions, we do not exclude other possibilities that might also contribute to the current gene loss pattern among B. rapa subgenomes. For example, paleohexaploidy could have quickly taken place with no initial differences in fractionation, but the three genomes may instead have acquired different epigenetic marks, and these epigenetic differences included the fractionation differences observed in B. rapa today. “Genomic dominance” of subgenome D over subgenome A is clear in the allotetraploid cotton genome (Flagel and Wendel 2010). Differential epigenetic marks resulting in the differential expression strengths of duplicate genes might correlate with gene retention favoring the homeolog with higher expression levels. Work in A. suecica showed that homeologous gene loss is certainly correlated with levels of expression and perhaps histone modifications as well (Chang et al. 2010). Genomic and epigenomic changes directed toward one parental genome have also been observed in B. napus (in which the C subgenome tended to be preferentially modified) (Gaeta et al. 2007) and in Triticale polyploids (in which the rye subgenome tended to be preferentially modified) (Ma and Gustafson 2006). Biases in the strength and patterns of epigenetic modifications can lead to different selective constraints on the subgenomes. Such biases may be still ongoing in the modern-day Arabidopsis and maize genomes, and the bias is associated with differential epigenetic marks that result in differential expression levels between homeologs. Biased gene loss is the result of selection against the loss of the homeolog copy that has a higher expression value (which is more likely to compromise the biological function) (Schnable and Freeling 2011).
In any case, there is nothing equal about the behavior of the three different genomes in B. rapa. The competing model that involves differential epigenetic marking might have impact on the subgenome differences we observe, which we hope to evaluate by studying the patterns of gene expression or histone modifications in B. rapa using high-throughput RNA-seq or CHIP-seq data.
Supplementary Material
Acknowledgments
We appreciate financial support from the U.S. National Science Foundation (MCB-0820821 to M.F.).
Footnotes
Communicating editor: M. Kirst
Literature Cited
- Aury J. M., Jaillon O., Duret L., Noel B., Jubin C., et al. , 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 [DOI] [PubMed] [Google Scholar]
- Bennett M. D., Leitch I. J., 2005. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann. Bot. 95: 45–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Veitia R. A., 2010. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs R. J., Chamala S., Wu W., Gao L., May G. D., et al. , 2010a Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 19(Suppl. 1): 132–146 [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Elliott N. M., Zhang L., Koh J., Viccini L. F., et al. , 2010b Tissue-specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 186: 175–183 [DOI] [PubMed] [Google Scholar]
- Bzymek M., Lovett S. T., 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98: 8319–8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. L., Dilkes B. P., McMahon M., Comai L., Nuzhdin S. V., 2010. Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 11: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K. M., Brown J. K., Bennetzen J. L., 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F., 2010. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186: 184–193 [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. L., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., et al. , 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Brunet F., Petit J. L., Stange-Thomann N., et al. , 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B. W., Lander E. S., 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., et al. , 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U., 1998. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U., Lydiate D. J., 1996. Comparative genome mapping in Brassica. Genetics 144: 1903–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham R. J., Walsh J., Dunn M., Ko C., Goff S. A., et al. , 2004. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Leitch A. R., Leitch I. J., 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483 [DOI] [PubMed] [Google Scholar]
- Lyons E., Freeling M., 2008. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53: 661–673 [DOI] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Freeling M., 2008. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates the rosids. Trop. Plant Biol. 1: 181–190 [Google Scholar]
- Lysak M. A., Koch M. A., Pecinka A., Schubert I., 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. F., Gustafson J. P., 2006. Timing and rate of genome variation in triticale following allopolyploidization. Genome 49: 950–958 [DOI] [PubMed] [Google Scholar]
- Parkin I. A., Sharpe A. G., Lydiate D. J., 2003. Patterns of genome duplication within the Brassica napus genome. Genome 46: 291–303 [DOI] [PubMed] [Google Scholar]
- Parkin I. A., Gulden S. M., Sharpe A. G., Lukens L., Trick M., et al. , 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., Pattyn P., Gerats T., Van de Peer Y., 2011. Journey through the past: 150 million years of plant genome evolution. Plant J. 66: 58–65 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P., 2003. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 13: 1123–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P., Matic I., Taddei F., 2002. Over-representation of repeats in stress response genes: a strategy to increase versatility under stressful conditions? Nucleic Acids Res. 30: 1886–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D., Zheng C., Zhu Q., 2010. The collapse of gene complement following whole genome duplication. BMC Genomics 11: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J. C., Freeling M., 2011. Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS ONE 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J. C., Springer N. M., Freeling M., 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M. E., Quijada P., Sung S. B., Lukens L., Amasino R., et al. , 2002. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Costello J. C., Hahn M. W., 2009. All human-specific gene losses are present in the genome as pseudogenes. J. Comput. Biol. 16: 1419–1427 [DOI] [PubMed] [Google Scholar]
- Sterck L., Rombauts S., Vandepoele K., Rouzé P., Van de Peer Y., 2007. How many genes are there in plants (... and why are they there)? Curr. Opin. Plant Biol. 10: 199–203 [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P., 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang X., Bowers J. E., Ming R., Alam M., et al. , 2008. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 18: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Bowers J. E., Wang X., Paterson A. H., 2010. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 107: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. C., Pedersen B., Freeling M., 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Fawcett J. A., Proost S., Sterck L., Vandepoele K., 2009. The flowering world: a tale of duplications. Trends Plant Sci. 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Wei N. E., Jiang H., et al. , 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., et al. , 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., 2001. Yesterday’s polyploids and the mystery of diploidization. Nat. Rev. Genet. 2: 333–341 [DOI] [PubMed] [Google Scholar]
- Woodhouse M. R., Schnable J. C., Pedersen B. S., Lyons E., Lisch D., et al. , 2010. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 8: e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Qi X., 2010. Genes encoding hub and bottleneck enzymes of the Arabidopsis metabolic network preferentially retain homeologs through whole genome duplication. BMC Evol. Biol. 10: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R. T., Pires J. C., 2011. Because shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. J., Kim J. S., Kwon S. J., Lim K. B., Choi B. S., et al. , 2006. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.