Abstract
Yeast remains an important model for systems biology and for evaluating proteomics strategies. In-depth shotgun proteomics studies have reached nearly comprehensive coverage, and rapid, targeted approaches have been developed for this organism. Recently, we demonstrated that single LC-MS/MS analysis using long columns and gradients coupled to a linear ion trap Orbitrap instrument had an unexpectedly large dynamic range of protein identification (Thakur, S. S., Geiger, T., Chatterjee, B., Bandilla, P., Frohlich, F., Cox, J., and Mann, M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell Proteomics 10, 10.1074/mcp.M110.003699). Here we couple an ultra high pressure liquid chromatography system to a novel bench top Orbitrap mass spectrometer (Q Exactive) with the goal of nearly complete, rapid, and robust analysis of the yeast proteome. Single runs of filter-aided sample preparation (FASP)-prepared and LysC-digested yeast cell lysates identified an average of 3923 proteins. Combined analysis of six single runs improved these values to more than 4000 identified proteins/run, close to the total number of proteins expressed under standard conditions, with median sequence coverage of 23%. Because of the absence of fractionation steps, only minuscule amounts of sample are required. Thus the yeast model proteome can now largely be covered within a few hours of measurement time and at high sensitivity. Median coverage of proteins in Kyoto Encyclopedia of Genes and Genomes pathways with at least 10 members was 88%, and pathways not covered were not expected to be active under the conditions used. To study perturbations of the yeast proteome, we developed an external, heavy lysine-labeled SILAC yeast standard representing different proteome states. This spike-in standard was employed to measure the heat shock response of the yeast proteome. Bioinformatic analysis of the heat shock response revealed that translation-related functions were down-regulated prominently, including nucleolar processes. Conversely, stress-related pathways were up-regulated. The proteomic technology described here is straightforward, rapid, and robust, potentially enabling widespread use in the yeast and other biological research communities.
Yeast is one of the most well established model systems in molecular biology. It is used to study a large range of conserved cellular processes, including the cell cycle, metabolism, and stress responses. Yeast was the first organism whose genome was sequenced completely (1), and many other systems-wide biology screens were first carried out in the yeast model (2–6). Large scale proteomics has also been pioneered in yeast, identifying first hundreds and then thousands of proteins (7–13). Using three different analytical strategies, including one with subcellular fractionation and two involving peptide separation into 24 fractions, our group has reported a substantially complete proteome of yeast as judged against genome-wide tagging experiments (14). However, the expertise and analysis times associated with in-depth proteome measurements have so far precluded the widespread adoption of in-depth proteomics in the yeast research community. Targeted proteomics, in the form of multiple reaction monitoring, offers a possible solution to this problem and has recently been used to detect proteins throughout the dynamic range of the yeast proteome, as well as to quantify changes in key proteins after metabolic shift (15). However, targeted proteomics aims at the characterization of relatively few key proteins across many conditions, and it is therefore less well suited to the discovery of biological responses on a global scale.
Both the multiple reaction monitoring experiments and analyses of the total features detectable in the MS retention time contour plots suggest that a very large number of peptides are present in LC-MS runs of total proteome digests (16, 17). We recently investigated the dynamic range of single LC-MS/MS runs and found that even very low-abundance proteins could be detected in this mode (18). Furthermore, direct analysis without prefractionation implies high sensitivity because only a few micrograms of peptides are required to load the column to capacity. However, our previous study was performed with a dedicated chromatographic setup and would not be straightforward to adopt for nonspecialized groups.
A novel mass spectrometer, the Q Exactive, couples a mass selective quadrupole to the Orbitrap analyzer (19). In this bench top instrument, precursor ions are selected by the quadrupole, fragmented by higher energy collisional dissociation (20), and measured at high resolution and mass accuracy in the Orbitrap analyzer. Cycle times for a top10 method (survey scan followed by up to 10 MS/MS scans) are ∼1 s, more than twice as fast as with previous instruments of the Orbitrap family. Thus the Q Exactive offers the potential to analyze many more peptides in a given time, with very high MS/MS data quality. We wanted to combine these benefits with ultra HPLC (UHPLC),1 which was not available to us in the previous single-run analyses. Taking advantage of a newly developed compact UHPLC system termed the EASY-nLC 1000, we achieved higher chromatographic performance with relatively long columns and small particle diameters. Here, we describe this simple but powerful bench top platform and evaluate its capability to characterize the yeast proteome in high throughput but also in-depth fashion.
To quantify proteome states in yeast, SILAC labeling can be employed in the standard format, which requires labeling both the control and the experimental conditions (21). To enable even more streamlined systems analysis of perturbations of the yeast proteome, we further wanted to decouple the SILAC metabolic labeling step from the actual experiments by using a “spike-in” SILAC strategy (22). Here we developed such a standard, taking into account several proteome states of yeast. We then used this standard to quantify yeast proteome changes upon heat shock, an important perturbation frequently encountered with temperature-sensitive mutant strains and synchronization experiments (23, 24).
EXPERIMENTAL PROCEDURES
Yeast Culture and Lysis
The yeast strain W303 MATα was grown in YPD medium until early- to mid-log phase and was harvested by centrifugation at 4000 × g for 5 min at 4 °C. The cell pellet was resuspended in 100 mm Tris, pH 7.6, containing 100 mm dithiothreitol and 5% SDS. The lysates were heated to 95 °C for 5 min followed by sonication using a Bioruptor Sonicator (20 kHz, 320 W, 60 s cycles) for 15 min at the maximum power to achieve complete lysis. The lysate was centrifuged at 16,000 × g for 5 min to clarify the protein extract.
Yeast Spike-in Standard
The W303 MATα strain for heavy lysine labeling was constructed by deletion of the Lys2 gene using the pYM-natNT2 plasmid according to Janke et al. (25). The cells were labeled only with heavy lysine, and not heavy arginine, to reduce sample complexity and avoid arginine to proline conversion. The spike-in standard was used to compare expression levels across different conditions. We cultured 250 ml to log phase (A600 = 0.9) in SCD medium containing [13C6/15N2]l-lysine. To represent further biological conditions in the spike-in mix, we also cultured cells with 2% ethanol as the carbon source as well as at higher temperature (37 °C for 30 min after previous culture at 24 °C). These three conditions were mixed in equal proportions to produce the spike-in mix. This quantity of cultured cells would be sufficient for thousands of spike-in experiments in single-shot measurements (at a few μg/analysis) and hundreds of experiments with an up front pipette-based strong anion exchange fractionation step (26).
Yeast Heat Shock Treatment
Yeast was cultured to mid-log phase to obtain an A600 of 2.5 for cells at 24 °C in the YPD medium and was subsequently shifted to 37 °C via water bath incubation to achieve uniform and efficient heat transfer. Samples were collected at t = 0 and 30 min after incubation at 37 °C to analyze the proteome changes upon heat shock. The samples were lysed as described above.
Protein Digestion
Proteins were digested using the FASP method (27). Briefly, 140 μg of protein was loaded on the filter, and SDS was completely replaced by washing two to three times with buffer containing 8 m urea. The proteins were then alkylated using iodoacetamide, and the excess reagent was washed through the filters. The reduced and alkylated proteins were digested using endoproteinase LysC, which cleaves at the C terminus of lysine residues, with an enzyme to protein ratio of 1:50. Peptides obtained by FASP were desalted using C18 StageTips (28).
Ultra High Pressure Easy LC
The Thermo Scientific EASY-nLC 1000 (Thermo Fisher Scientific, Odense, Denmark) is a split-free, nano-flow LC designed to operate at ultra high pressures up to 1000 bars (15,000 p.s.i.). The system employs two direct-drive syringe pumps to generate binary gradients with minimum stable flow down to ∼50 nL/min. Flow and pressure sensors (one set for each mobile phase) are placed immediately upstream from the high pressure mixing Tee such that sensor output can accurately control the gradient. The LC system is preconfigured, requiring only two liquid connections by which the user connects the column(s) to the eluent flow line and a waste/venting line. This simplicity facilitates daily use, and further ease-of use is obtained by a finger-tight fitting, named NanoViper (Thermo Fisher Scientific), that ensures zero dead volume seals up to 1200 bars. This compact LC instrument, with its maximum pressure limit of 1000 bars, enables the use of long columns with linear velocity of 250 nl/min in the temperature range of 35 °C, rather than the relatively high temperatures of up to 60 °C required in our previous setup without ultra high pressure (18).
LC-MS/MS
Peptides were loaded on a 50-cm column with 75-μm inner diameter, packed in-house with 1.8-μm C18 particles (Dr Maisch GmbH, Germany). Reversed phase chromatography was performed using the Thermo EASY-nLC 1000 with a binary buffer system consisting of 0.5% acetic acid (buffer A) and 80% acetonitrile in 0.5% acetic acid (buffer B). The peptides were separated by a linear gradient of buffer B up to 40% in 240 min for a 4-h gradient run with a flow rate of 250 nl/min in the EASY-nLC 1000 system. The column was operated at a constant temperature of 35 °C regulated by an in-house designed oven with a Peltier element (18). The LC was coupled to a Q Exactive mass spectrometer (19) (Thermo Fisher Scientific) via the nanoelectrospray source (Proxeon Biosystems, now Thermo Fisher Scientific). The Q Exactive was operated in the data-dependent mode with survey scans acquired at a resolution of 50,000 at m/z 400 (transient time = 256 ms). Up to the top 10 most abundant isotope patterns with charge ≥2 from the survey scan were selected with an isolation window of 1.6 Thomsons and fragmented by higher energy collisional dissociation (20) with normalized collision energies of 25. The maximum ion injection times for the survey scan and the MS/MS scans were 20 and 60 ms, respectively, and the ion target value for both scan modes were set to 1E6. Repeat sequencing of peptides was kept to a minimum by dynamic exclusion of the sequenced peptides for 40 s.
Data Analysis
The raw files were processed using the MaxQuant computational proteomics platform (29) version 1.2.0.34. The fragmentation spectra were searched against the yeast ORF database (release date of February 3, 2011; 6752 entries) using the Andromeda search engine (30) with the initial precursor and fragment mass tolerances set to 7 and 20 ppm, respectively, and with up to two missed cleavages. Carabamidomethlyation of cysteine was set as a fixed modification, and oxidation of methionine and protein N-terminal acetylation were chosen as variable modifications for database searching. Both peptide and protein identifications were filtered at 1% false discovery rate and thus were not dependent on the peptide score. Bioinformatics analysis was performed using the Perseus tools available in the MaxQuant environment. All enrichment analysis and analysis of variance tests were performed with Benjamini-Hochberg correction at a false discovery rate of 2%. The raw data are available from the Tranche proteome repository with the following access code: Bz9hlKJ5EaEq/rgoVH0+fHehRgTSaCcD2 + 879Q1JnJm3d9sFaCpNgFnPPZT9WFu5K5mXKz8o1B9qaK7WBFxdFPu2ThkAAAAAAAAPmA = =.
RESULTS AND DISCUSSION
The Single-shot LC-MS/MS System
We aimed to devise a shotgun proteomics workflow with the lowest possible number of processing and analysis steps and consequently high robustness (Fig. 1). Yeast cells were lysed in the presence of SDS, ensuring efficient denaturation and solubilization of all protein classes (“Experimental Procedures”). The proteins were reduced to peptides by LysC digestion using the FASP method (27), and the resulting peptides were purified on StageTips (28). These procedures only involve pipette-based operations, and they can be performed in several hours and in parallel for several conditions. Peptide mixtures were then loaded onto the autosampler of the UHPLC system (EASY-nLC 1000) and analyzed in an automated manner by LC-MS/MS on the bench top quadrupole Orbitrap mass spectrometer (Q Exactive) (19). The LC setup does not use precolumns or flow splitting, avoiding sample loss and reducing solvent consumption. The UHPLC system itself is designed for compactness and simplicity (“Experimental Procedures”).
Fig. 1.
Minimalistic proteomics setup. Yeast samples were lysed and prepared by the FASP method. Peptides were purified on StageTips and placed in an autosampler, which loads them directly on to a relatively long column (50 cm). The binary gradient system is provided by an UHPLC system (EASY nLC 1000) system coupled to a bench top quadrupole Orbitrap mass spectrometer (Q Exactive) via a nanoelectrospray source. The data obtained were analyzed in the MaxQuant computational proteomics platform, and bioinformatics analyses were performed using the Perseus tool.
To facilitate deep sampling of the proteome, we employed relatively long columns and small particle sizes (50 cm, 1.8 μm). This was readily accommodated by the UHPLC pump, which produced a stable flow of 250 nL/min at 500 bars. Another advantage of the UHPLC system is its ability to load samples at a higher flow rate and to equilibrate columns more quickly, leading to a shortening of overhead times. We found the combination of a 50-cm column and 4-h gradients to be a good combination for standard use.
Depth of Analysis of the Yeast Proteome
Having established the single-shot workflow, we next measured six yeast cell lysates, which simulates an experiment with triplicate control and triplicate perturbation. Approximately 4 μg of peptide material was loaded onto the 50-cm column and separated with the 4-h gradients. Joint analysis of the six LC-MS/MS files in MaxQuant resulted in an average of 26,173 ± 286 peptide identifications with unique amino acid sequence for the single runs. Transferring identifications between the runs based on their mass precision and retention time (“match between runs” feature in MaxQuant) led to 33,122 ± 405 sequence-unique peptide identifications per single run (Fig. 2A). Together, 41,035 peptides were identified from this experiment, which took ∼24 h of total measurement time. Even though LysC peptides are on average larger than tryptic peptides and therefore more difficult to identify, the identification rates for runs were above 51%. This is presumably due to the high mass accuracy enabled by the high resolution higher energy collisional dissociation spectra.
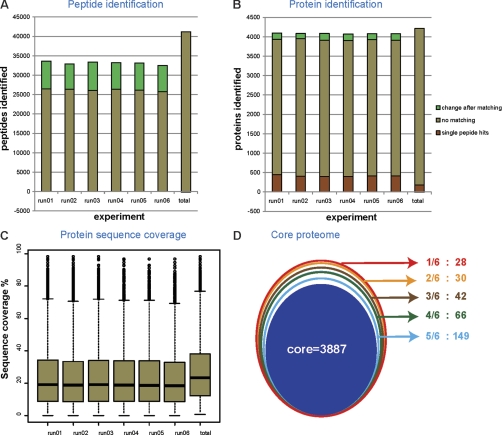
Fig. 2.
In-depth coverage of the yeast proteome. A, number of peptides identified in individual runs with and without matching between the runs. Peptides identified by matching are indicated in green. B, proteins identified in individual runs with the gain from matching between the runs indicated in green. Proteins identified with single peptide hits are shown in red. C, the median sequence coverage of individual runs after matching was ∼17%. The median sequence coverage from the combined run for 4099 proteins was 22.9% as shown. D, the conjoint circles represent the frequency of identification of proteins in the six runs. Proteins identified in all six runs were designated as core proteome in the innermost circle.
When matching between the runs, 4084 ± 8 proteins were identified per run. In the combined data set, 4206 proteins were identified (not counting contaminants such as keratins), and only 180 of these had a single peptide (Fig. 2B and supplemental Tables I and II and other supplemental materials containing the spectra of all the proteins identified with single peptides). We repeated the database search with an arbitrary Andromeda peptide score threshold of 60, which is high for a database with the size of the yeast proteome, and still identified 4137 proteins. This further demonstrates that our data do not rely on low scoring peptides. Our previous study using 8-h gradients, a custom LC setup, and the previous generation Orbitrap instrument identified just under 3000 proteins in a triplicate experiment (18). Here we achieved dramatically increased performance—close to the complete expressed proteome (see below)—with a very streamlined and minimalistic proteomic system.
Median sequence coverage of identified proteins was 23.4% with a median of seven peptide sequences (Fig. 2C). Many more peptides can be detected in LC MS plots than are sequenced and identified by tandem mass spectrometry. In our data set, the median intensity of the fragmented isotope patterns was ∼10-fold higher than that of the nonfragmented isotope patterns (supplemental Fig. 1). This suggests that many more yeast peptides are present in the single-runs than are fragmented and identified, although they may not be accessible to data-driven LC-MS/MS (19).
A key challenge in shotgun proteomics is the “missing value” problem, which refers to the absence of data on particular proteins or peptides in some of the measurements of a series and which is caused by the semi-random nature of peak selection for fragmentation. Remarkably, when comparing identifications in different subsets of the single-shot analyses, we found that a full 3887 of the 4206 proteins (92%) were identified in all six runs (termed core in Fig. 2D), and 96% were identified in at least five of the six data sets. This indicates that for the vast majority of the proteins, there is no or very little “missing value problem.” At the peptide level, naturally, overlap is not as high, but 75% of the peptides are still identified in at least five of the six runs (supplemental Fig. 2). High reproducibility between the single runs is presumably a consequence of the very high sequencing speed of the Q Exactive, combined with the efficient matching of peptides between runs by MaxQuant.
To assess the completeness of our data set, we compared it against our previous in-depth study (14). Despite differences in the yeast background (W303 versus S288C), somewhat different conditions and slight reannotation of the yeast genome in the past 4 years, 95% of the 4206 genes found here were contained in our previous data set. Of the 217 proteins not reported there, 133 were identified in six of six runs (core in Fig. 2D). Yeast has 809 ORFs that are classified as “dubious” by the Saccharomyces Genome Database, and these ORFs are thought not to encode a corresponding protein (Table I). As described before (14), this set of genes provides a useful independent test of false positive identification rates. The combined single-shot data set only identified two dubious ORFs (“majority” protein column in supplemental Table II), whereas on the basis of a 1% false positive rate, we would have expected five false positives hits in this subset (1% of 809 dubious ORFs given our coverage of the 6717 yeast ORFs; Table I). Furthermore, one of the two dubious ORF hits was also found in our previous study because one of only four hits in this subset (YBR126W-A), which suggests that it may not in fact be a false positive. These data provide independent evidence that our false positive rate is below 1%.
Table I. Coverage of Saccharomyces Genome Database annotations and GO biological process terms that are de-enriched.
| Yeast ORFs | Proteins identified | Proteins identified − core proteome | |
|---|---|---|---|
| Total | 6717 | 4206 (63%) | 3887 (58%) |
| Saccharomyces Genome Database | |||
| Verified | 4941 | 3856 (87%) | 3587 (73%) |
| Uncharacterized | 857 | 335 (39%) | 287 (33%) |
| Dubious | 809 | 2 (0%) | 2 (0%) |
| Transposable Elements | 89 | 17 (19%) | 13 (15%) |
| Pseudogenes | 21 | 1 (5%) | 1 (5%) |
| Silenced | 4 | 1 (25%) | 1 (5%) |
| GO biological process | |||
| Maltose metabolic process | 11 | 1 (9%) | 0 (0%) |
| Synapsis | 10 | 2 (18%) | 2 (18%) |
| Multidrug transport | 11 | 0 (0%) | 0 (0%) |
Pathway Analysis of the Detected Proteome
Table I indicates that the six single-shot runs together identified 78% of the ORFs verified as genuine gene products by the Saccharomyces Genome Database; therefore at least this number is expressed as proteins in laboratory yeast. Many pathways and functions are not needed under laboratory conditions, and the corresponding proteins may not be expressed. At 88%, coverage of the proteins in the Kyoto Encyclopedia of Genes and Genomes database was very high in the single shot yeast proteome, as was the coverage of the three gene ontology (GO) categories (GOCC, 85%; GOMF, 82%; and GOBP, 85% CC-cell component, MF-molecular function, BP-biological process). (Because some pathways consist of only a few proteins, we restricted the analysis to pathways with 10 proteins or more; coverage would be even higher without this filter.) Interestingly, the pathways with most missing proteins belong to sugar metabolism and meiosis (Table I), functions that are not expected to be active in haploid yeast growing in glucose media.
Dynamic Range of the Single-shot Proteome
Given the number of identified proteins, we expected the single-shot proteome to have a large dynamic range of protein expression. Indeed, the integrated peptide signals for all the identified proteins spanned approximately 5 orders of magnitude in the single-shot measurements (Fig. 3). A recent multiple reaction monitoring study examined the detectability of 127 proteins chosen to represent the full range of the yeast protein expression from most abundant to least abundant protein classes (15). Our single shot proteome included 121 of these proteins, and the six missing proteins were all in the lowest abundance classes. All of the proteins in the category “less than 50 copies/cell” were identified, but they may have been misclassified (18). Together, these results indicate that our data set covered a remarkably large dynamic range.
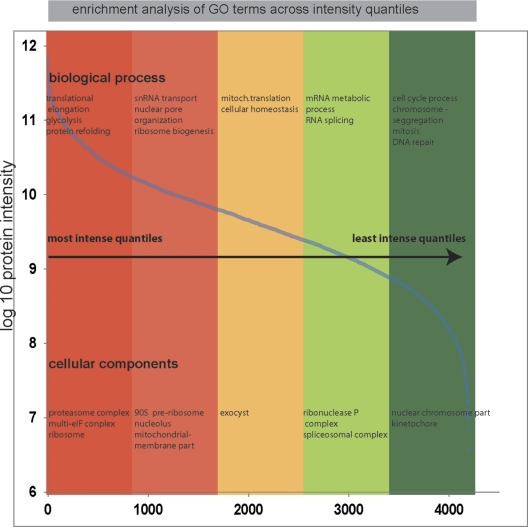
Fig. 3.
Dynamic range of the identified proteome. Expression levels of identified proteins were roughly estimated using their summed peptide intensities. The proteins were ranked into five quantiles based on their abundance. A Fisher exact test extracted enriched GO terms in each quantile (false discovery rate < 0.02 after Benjamini-Hochberg correction).
Bioinformatic enrichment analysis of GO terms in the most abundant quantiles of the distribution, as expected, placed the cell cytoskeleton and biogenesis-related functions among the functions carried out by the most abundant proteins. Cell cycle-related functions are diluted down in nonsynchronized cells and accordingly were enriched in the lowest quantile.
Performance of a Spike-in SILAC Standard
Although SILAC has become a standard and highly accurate quantification method in many systems, the requirement for metabolic labeling prevents some researchers from adopting this technology. Furthermore, in some systems the requirement for media free of external amino acids may impose restrictions on the intended experiments. These issues are addressed by a spike-in SILAC approach (22). In that strategy, a standard representing the proteome of interest is heavy lysine-labeled and serves as a reference across diverse experiments. Biological experiments can be performed as usual, and the spike-in standard is mixed in before sample preparation.
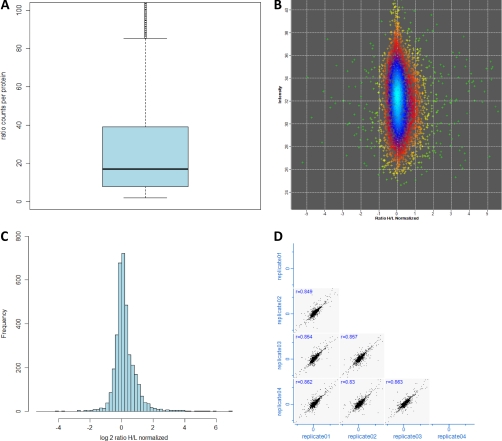
To enable a spike-in strategy for yeast, we SILAC-labeled the W303 MATα strain in which the Lys2 gene was knocked out by homologous recombination. A relatively small amount of standard is sufficient for a large number of experiments (“Experimental Procedures”). It is advantageous to choose the standard so that it represents diverse conditions. Therefore we also cultured yeast under a different growth condition (2% ethanol) and a temperature stress condition. The spike-in mix was prepared by combining all three conditions in equal amounts. To test quantification with the spike-in SILAC standard in single-run conditions, we mixed it into yeast growing under normal laboratory conditions in rich media. Quadruplicate single-run analyses together identified 3794 yeast proteins (supplemental Table III). This number is somewhat lower than in the above “label-free” experiments because SILAC doubles the complexity of the peptide mixtures and because the number of runs was lower. Of these proteins, 3656 and 3553 were quantified with two and three “ratio counts,” respectively, which designates valid SILAC quantification ratios in the MaxQuant analysis. The median number of ratio counts/protein was 16 (Fig. 4A). Despite using a spike-in SILAC standard including several conditions, the distribution of the ratios in these single-run experiments was very narrow, with 89% of the protein ratios within a 2-fold change (Fig. 4, B and C). Furthermore, correlation analysis between all of the individual replicates resulted in R values of at least 0.83 (Fig. 4D). Remarkably, inclusion of the ethanol growth condition in the mix now enabled complete identification of the glycolysis and gluconeogenesis pathways, TCA cycle, and glyoxylate cycle (45 of 45 proteins) as targeted in the recent multiple reaction monitoring study (15). These results demonstrate that the yeast spike-in SILAC adequately represents the yeast proteome and that it performs well in single-run quantification analysis.
Fig. 4.
Quantification of the yeast proteome using spike-in SILAC labeling. From four single-shot runs, more than 3200 proteins were quantified with respect to the spike-in SILAC mix. A, box plot of the number of ratio counts contributing to quantification of each protein. B and C, distribution of protein ratios to the spike-in SILAC standard. D, the reproducibility of spike-in SILAC quantification of the biological replicates as illustrated by the protein ratio correlations as shown here.
Systems-wide Response to Heat Shock
To test the single-run workflow in a systems biology context, we chose to investigate the heat shock response. This is a much studied stress response in yeast. Despite many microarray studies (31, 32), no in-depth proteomic study of this process has been reported. In addition, heat shock is an inevitable component of experiments involving temperature-sensitive mutants, and it would therefore be interesting to know how heat shock modulates the proteome.
The heat shock experiment was performed by shifting the yeast cultures from 24 to 37 °C, taking time points at 0 min and after 30 min at 37 °C (Fig. 5A). The samples were combined with the spike-in standard and analyzed by 4-h single runs in quadruplicates. After MaxQuant analysis with the “matching between run” feature, we identified 4072 proteins. The heat shock data set had an overlap of 3708 proteins with the core proteome depicted in Fig. 2D. We filtered for proteins that had at least been quantified twice at both time points and obtained 3152 yeast proteins (supplemental Table IV).
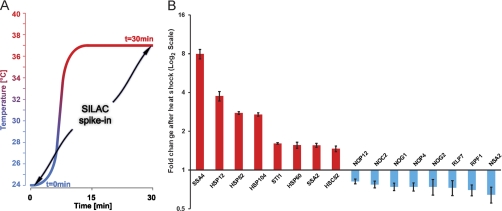
Fig. 5.
Quantitation of heat shock response by spike in SILAC strategy. A, schematic representation of the heat shock experiment. Samples at t = 0 and 30 min after incubation at 37 °C were mixed 1:1 with spike-in SILAC standard grown at 30 °C. B, fold change represented in log2 ratios is shown for selected proteins.
Fig. 5B shows the fold change of proteins with significant change upon heat shock on a log2 scale. For every protein, these fold changes were calculated as “ratios of ratios” by dividing the ratios of the unlabeled samples to spike-in SILAC standard (light to heavy ratio) for control (t = 0) and heat shock (t = 30 min). One of the proteins with the highest fold change (close to 4-fold induction) was HSP12 (heat shock protein 12), which is known to be highly induced by heat shock as well as other stress factors (33). Other heat shock proteins were also up-regulated, including SSA4, SSA2, HSP104, HSP82, and HSP60 (Fig. 5B), and this group displayed the highest fold changes overall. Among the down-regulated factors, we noticed a prominent group of proteins involved in ribosomal biogenesis. For example, NSA2, NOG1, RPF1, NOP4, and NOP12 were all down-regulated significantly. The fold changes of these proteins were between 0.6 and 8.0, which was still reliably quantified by MaxQuant (see error bars in Fig. 5B).
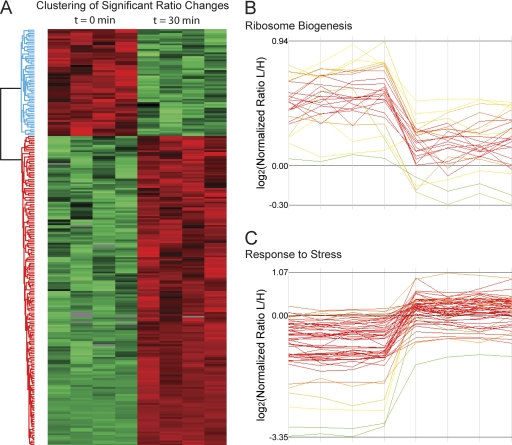
Next we explored the global proteomics response using the Perseus bioinformatics environment that is part of MaxQuant. We performed one-way analysis of variance between the quadruplicates at t = 0 and t = 30 min and Benjamini-Hochberg correction for multiple hypothesis testing with a cutoff false discovery rate value of 0.02. This yielded 234 proteins that were significantly changing in expression (supplemental Table V). More than half of these proteins were up-regulated (Fig. 6A). Enrichment analysis of either set revealed the GO terms “nucleolus” and “ribosome biogenesis” as highly significantly down-regulated (p < 10−16). Among the up-regulated proteins, the GO categories “response to stress” and “catabolic process” were most dominant. The profiles of the proteins responsible for these effects are plotted in Fig. 6 (B and C). As a control, we inspected the profiles in the category “transport,” which is not significantly changing upon heat shock. These profiles do not display a coherent trend upon heat shock.
Fig. 6.
Hierarchical clustering of significantly changing proteins. A, clustering of significantly up- and down-regulated proteins upon heat shock. Significance was determined by analysis of variance with correction for multiple hypothesis testing. B and C, expression patterns for clusters enriched for ribosome biogenesis (B) and response to stress (C) show the two major trends of protein regulation.
Closer inspection of the down-regulated processes highlighted additional categories related to the regulation of translation. For example, proteins belonging to “tRNA metabolic processes”, which are needed for translation initiation and elongation, are all significantly down-regulated during heat shock (p < 10−5). By the same token, rRNA transcription, maturation, and ribosome assembly would be expected to be down-regulated, and this is indeed what our bioinformatics analysis shows. The nucleolus itself is the site for many of these processes and is independently known to be a key sensor of cellular stress (34). Our analysis now pinpoints proteins responsible for this interesting connection.
Conclusion and Outlook
Here we have devised a minimalistic proteomic workflow consisting only of pipette-based preparation of digested yeast cell lysate, spike-in SILAC as the quantification technology, single UHPLC-runs on a bench top mass spectrometer and data analysis by the freely available MaxQuant framework. Despite its simplicity, this technology reaches very large coverage of the yeast proteome and readily allows system-wide analysis of a perturbation such as stress response.
Attractive features of our workflow include its sensitivity and rapid analysis times. Because there are no requirements for labeling, experiments can be performed according to standard protocols, and standard yeast strains can be employed. We believe that the single-shot system is indeed a valid third approach between in-depth shotgun proteomics employing fractionation and targeted approaches. That said, there are many applications of proteomics where the single shot technology as described here would not be the ideal approach. For example, very large sequence coverage of the proteome, as needed to distinguish all isoforms, cannot be expected of this strategy. Likewise, analysis of post-translational modifications usually requires enrichment and fractionation steps. However, almost all the improvements made to enable nearly complete coverage of the yeast proteome would carry over to the analysis of fractions in a standard shotgun proteomics approach.
Here we have applied the single-shot technology to the yeast model system. The human proteome is much more complex than the yeast proteome, but with further advances in technology, it is possible that much of that proteome will also be analyzable by single-shot approaches.
Footnotes
* This work was supported by European Commission's 7th Framework Programme Grant Agreement HEALTH-F4-2008-201648/PROSPECTS. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains Tables I–V and Figs. 1 and 2.
This article contains Tables I–V and Figs. 1 and 2.
1 The abbreviations used are:
- UHPLC
- ultra HPLC
- SILAC
- stable isotope labeling of amino acids in cell culture
- FASP
- filter-aided sample preparation
- GO
- gene ontology.
REFERENCES
- 1. Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., Oliver S. G. (1996) Life with 6000 genes. Science 274, 546, 563–567 [DOI] [PubMed] [Google Scholar]
- 2. Bader G. D., Heilbut A., Andrews B., Tyers M., Hughes T., Boone C. (2003) Functional genomics and proteomics: Charting a multidimensional map of the yeast cell. Trends Cell Biol. 13, 344–356 [DOI] [PubMed] [Google Scholar]
- 3. Jorgensen P., Breitkreutz B. J., Breitkreutz K., Stark C., Liu G., Cook M., Sharom J., Nishikawa J. L., Ketela T., Bellows D., Breitkreutz A., Rupes I., Boucher L., Dewar D., Vo M., Angeli M., Reguly T., Tong A., Andrews B., Boone C., Tyers M. (2003) Harvesting the genome's bounty: Integrative genomics. Cold Spring Harb. Symp. Quant. Biol. 68, 431–443 [DOI] [PubMed] [Google Scholar]
- 4. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 5. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 6. Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Ménard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A. M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., Boone C. (2004) Global mapping of the yeast genetic interaction network. Science 303, 808–813 [DOI] [PubMed] [Google Scholar]
- 7. Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Boucherie H., Mann M. (1996) Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U.S.A. 93, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Figeys D., Ducret A., Yates J. R., 3rd, Aebersold R. (1996) Protein identification by solid phase microextraction-capillary zone electrophoresis-microelectrospray-tandem mass spectrometry. Nat. Biotechnol. 14, 1579–1583 [DOI] [PubMed] [Google Scholar]
- 9. Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 10. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 11. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 12. de Godoy L. M., Olsen J. V., de Souza G. A., Li G., Mortensen P., Mann M. (2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 7, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swaney D. L., Wenger C. D., Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 15. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Köcher T., Swart R., Mechtler K. (2011) Ultra-high-pressure RPLC hyphenated to an LTQ-Orbitrap Velos reveals a linear relation between peak capacity and number of identified peptides. Anal. Chem. 83, 2699–2704 [DOI] [PubMed] [Google Scholar]
- 17. Michalski A., Cox J., Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 18. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Frohlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell Proteomics 10, 10.1074/mcp.M110.003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, 10.1074/mcp.M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 21. Walther T. C., Olsen J. V., Mann M. (2010) Yeast expression proteomics by high-resolution mass spectrometry. Methods Enzymol. 470, 259–280 [DOI] [PubMed] [Google Scholar]
- 22. Geiger T., Wisniewski J. R., Cox J., Zanivan S., Kruger M., Ishihama Y., Mann M. (2011) Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protocols 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 23. Futcher B. (1999) Cell cycle synchronization. Methods Cell Sci. 21, 79–86 [DOI] [PubMed] [Google Scholar]
- 24. Walker G. M. (1999) Synchronization of yeast cell populations. Methods Cell Sci. 21, 87–93 [DOI] [PubMed] [Google Scholar]
- 25. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 26. Wiśniewski J. R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 27. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 28. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 30. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 31. Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Praekelt U. M., Meacock P. A. (1990) HSP12, a new small heat shock gene of Saccharomyces cerevisiae: Analysis of structure, regulation and function. Mol. Gen. Genet. 223, 97–106 [DOI] [PubMed] [Google Scholar]
- 34. Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I. (2010) The nucleolus under stress. Mol. Cell 40, 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]