Abstract
Deep proteomic analysis of mammalian cell lines would yield an inventory of the building blocks of the most commonly used systems in biological research. Mass spectrometry-based proteomics can identify and quantify proteins in a global and unbiased manner and can highlight the cellular processes that are altered between such systems. We analyzed 11 human cell lines using an LTQ-Orbitrap family mass spectrometer with a “high field” Orbitrap mass analyzer with improved resolution and sequencing speed. We identified a total of 11,731 proteins, and on average 10,361 ± 120 proteins in each cell line. This very high proteome coverage enabled analysis of a broad range of processes and functions. Despite the distinct origins of the cell lines, our quantitative results showed surprisingly high similarity in terms of expressed proteins. Nevertheless, this global similarity of the proteomes did not imply equal expression levels of individual proteins across the 11 cell lines, as we found significant differences in expression levels for an estimated two-third of them. The variability in cellular expression levels was similar for low and high abundance proteins, and even many of the most highly expressed proteins with household roles showed significant differences between cells. Metabolic pathways, which have high redundancy, exhibited variable expression, whereas basic cellular functions such as the basal transcription machinery varied much less. We harness knowledge of these cell line proteomes for the construction of a broad coverage “super-SILAC” quantification standard. Together with the accompanying paper (Schaab, C. MCP 2012, PMID: 22301388) (17) these data can be used to obtain reference expression profiles for proteins of interest both within and across cell line proteomes.
Mammalian cell lines are the basis of much of the biological work that examines protein function and cell response to perturbations and they have been indispensable for many of the biological insights obtained in the last decades. In the majority of cases these cell lines were extracted from tumors of different origins, and were then adapted to growth in vitro. These cell lines serve as proxies not only of the original tumors or tissues but also for fundamental biological processes. A system-wide and comparative view of the proteomes of such cell lines can reveal commonalities and discrepancies between cell lines in general and highlight the biological processes and their variations across the cells.
So far only very few proteomic studies have attempted to determine shared and distinct features of different cell lines. Burkard et al. defined a “central proteome” in a comparison of seven cell lines (1). It consisted of the 1124 proteins that were identified in all these cell systems and that were preferentially involved in protein expression, metabolism and proliferation. This study identified 2000–4000 proteins per cell line, and was therefore limited to the more abundant proteins in the cell. It also did not attempt to quantify expression differences between the proteomes. With Uhlen and coworkers we recently analyzed gene expression in three distinct human cell lines by next generation sequencing, quantitative proteomics and the antibodies provided by the Human Protein Atlas. RNA-seq, stable isotope labeling with amino acid in cell culture (SILAC)-based1 proteomics and antibody-based confocal microscopy all found a high degree of similarity in expressed genes (2). In that study, the depth of our proteomic analysis was limited to about 5000 proteins raising the question whether this limitation contributed to the high resemblance of the cell lines at the protein level. This issue could be addressed by performing more comprehensive mass spectrometric analysis of cell lines, and by increasing the number of analyzed cell lines to examine the generality of the large overlap of proteomes.
Rapid developments in MS-based proteomics have enabled identification of increasing proportions of analyzed proteomes, aiding in the attempt to reach a comprehensive view of the system (3–6). In the yeast model, which has a genome of 6000 genes, such a comprehensive proteomic analysis identified 4400 proteins (7). The same degree of coverage has not yet been reached for human cells, whose genome consists of about 20,000 genes and whose proteomes are much more complex. Routine analyses of mammalian systems currently can lead to the identification of 4000–6000 proteins in a few days of analysis (8–10), which corresponds to about 50% of the expressed proteome based on the common estimate that a single cell type expresses 10,000 proteins. Significantly higher numbers of identified proteins were so far only achieved by combining multiple diverse cell lines or tissues in one analysis (11), or by investing weeks of measurement for single samples (12, 13).
Here we employ the latest proteomics technology in order to achieve a very extensive proteomic coverage of multiple human cell lines. The linear trap quadrupole (LTQ)-Orbitrap Velos mass spectrometer has improved higher-energy collisional dissociation (HCD) capabilities, and therefore enables acquisition of high resolution tandem MS (MS/MS) spectra without compromising the depth of analysis (14). Here, we additionally make use of a novel “high field” Orbitrap analyzer with higher resolution and higher sequencing speed (15). This Orbitrap mass spectrometer is described in detail in another manuscript in this issue (16). We performed deep analysis of 11 cell lines in relatively short analysis time and obtained very extensive characterization of their proteomes. The data is deposited in the MaxQB database, which is the subject of an accompanying manuscript and which allows sophisticated analysis and visualization of these reference proteomes [www.biochem.mpg.de/maxqb] (17).
EXPERIMENTAL PROCEDURES
Cell Culture Sample Preparation
Cell cultures of A549, GAMG, HEK293, HeLa, HepG2, K562, MCF7, RKO, and U2OS cells were grown with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. LnCap and Jurkat cells were cultured with RPMI supplemented with 10% fetal bovine serum and antibiotics. Three separate cell pellets from subconfluent cultures were flash frozen in liquid nitrogen and stored at −80 °C. Cells were lysed with a buffer consisting of 0.1 m Tris-HCl, pH 7.5, 0.1 m dithiothreitol and 4% SDS, and incubated at 95 °C for 5 min. Lysates were sonicated using a Branson type sonicator and were then clarified by centrifugation at 16,100 × g for 10 min. For SILAC experiments Jurkat, HEK293, LnCap, HeLa, and K562 cells were cultured in medium containing Lys8 and Arg10 instead of the natural amino acids, and supplemented with 10% dialyzed serum. Cells were cultured for about eight doublings in the SILAC medium to reach complete labeling. For the preparation of super-SILAC mix (18) equal amounts of heavy lysates were mixed and then combined with nonlabeled cells as described in the RESULTS section.
Protein Digestion and Fractionation
Cell lysates (100 μg) were diluted in 8 m urea in 0.1 m Tris-HCl followed by protein digestion with trypsin according to the FASP protocol (8). After an overnight digestion peptides were eluted from the filters with 25 mm ammonium bicarbonate buffer. The yield of the FASP digestion was more than 50%. From each sample 40 μg of peptides were separated into six fractions by strong anion exchange as described previously (19). Eluted peptides were concentrated and purified on C18 StageTips (20).
LC-MS/MS Analysis
Peptides were separated by reverse-phase chromatography on in-house made 20-cm columns (inner diameter 75 μm, 1.8 μm ReproSil-Pur C18-AQ media), using a nano-flow HPLC (Easy nanoLC, Thermo Fisher Scientific). The high performance liquid chromatography (HPLC) was coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) (14). Peptides were loaded onto the column with buffer A (0.5% acetic acid) and eluted with a 200 min linear gradient from 2 to 30% buffer B (80% acetonitrile, 0.5% acetic acid). After the gradient the column was washed with 90% buffer B and re-equilibrated with buffer A.
Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS scans using a top 10 method. MS spectra were acquired in the Orbitrap analyzer, with a mass range of 300–1650 Th and a target value of 106 ions. Peptide fragmentation was performed with the HCD method (21) and MS/MS spectra were acquired in the Orbitrap analyzer and a target value of 40,000 ions. Ion selection threshold was set to 5000 counts. Two of the data sets were acquired with a high field Orbitrap cell in which the resolution was 60,000 instead of 30,000 (at 400 m/z) for the MS scans. In the first of the two replicates with the high field Orbitrap MS/MS scans were acquired with 15,000 resolution, and in the second with 7500 resolution, which is the same as in the standard Orbitrap, but with shorter transients.
Data Analysis
Raw MS files were analyzed by MaxQuant (22) version 1.2.0.28. MS/MS spectra were searched by the Andromeda search engine (23) against the decoy IPI-human database version 3.68 containing forward and reverse sequences (total of 174,166 entries including forward and reverse sequences). Additionally the database included 248 common contaminants. MaxQuant analysis included an initial search with a precursor mass tolerance of 20 ppm the results of which were used for mass recalibration (24). In the main Andromeda search precursor mass and fragment mass had an initial mass tolerance of 6 ppm and 20 ppm, respectively. The search included variable modifications of methionine oxidation and N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Minimal peptide length was set to six amino acids and a maximum of two miscleavages was allowed. The false discovery rate (FDR) was set to 0.01 for peptide and protein identifications. In the case of identified peptides that are all shared between two proteins, these are combined and reported as one protein group. For comparison between samples we used label-free quantification with a minimum of two ratio counts to determine the normalized protein intensity (25). For ranking of the absolute abundance of different proteins within a single sample we used the iBAQ algorithm (9). Protein table were filtered to eliminate the identifications from the reverse database, and common contaminants. Peptide and protein tables are given as supplemental Tables S2 and S3. The MS raw files associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash: zb+OvxkUhczlFM0ja4v6fVi5BxsP9TolWFPI8qmtu4SUgyEzuQUaCHr0×89Q1A8fuLx9nGN/5ka7y+OgtOxWQg1MheoAAAAAAABhOg==.
Bioinformatic Analysis
Categorical annotation was supplied in the form of Gene Ontology (GO) biological process, molecular function, and cellular component, the TRANSFAC database (26) as well as participation in a KEGG pathway and membership in a protein complex as defined by CORUM (27). The annotation matrix algorithm tests the difference of any protein annotations from the overall intensity distribution. The specific test we used is a two-dimensional version of the nonparametric Mann-Whitney test. Multiple hypothesis testing was controlled by using a Benjamini-Hochberg FDR threshold of 0.05. Fisher exact test was performed with an FDR value of 0.02.
Hierarchical clustering of proteins was performed on logarithmized intensities after filtration of the data to have at least six valid values and z-score normalization of the data, using Euclidean distances between averages. For multiple t-tests (ANOVA) replicates were grouped and the statistical test was performed with an FDR value of 0.05 and S0 = 0.5. The S0 factor was described by Tusher et al. for t test (28) and was here generalized for ANOVA test.
RESULTS
Proteomic Analysis of Cell Lines
For the comparison of cell line proteomes we selected a panel of eleven commonly used cell lines from distinct origins and performed deep proteomic analyses (Fig. 1A). These included mainly cancer cells from various origins: A549 from lung carcinoma, GAMG from glioblastoma, HeLa from cervical carcinoma, HepG2 from hepatoma, Jurkat from acute T-cell leukemia, K562 from chronic myeloid leukemia, LnCap from prostate carcinoma, MCF7 from mammary carcinoma, RKO from colon carcinoma, U2OS from osteosarcoma and the noncancerous cell line HEK293 (Table I). We performed triplicate analysis of each cell line and in each replicate we separated the peptides into six fractions using strong anion exchange in a StageTip format (19). One replicate of the LC-MS/MS analyses was done on the LTQ-Orbitrap Velos mass spectrometer, which enabled acquisition of high resolution MS/MS spectra (14). In the other two replicates we used a novel LTQ-Orbitrap family mass spectrometer with a high-field Orbitrap mass analyzer that has doubled resolution (16). This instrument was run in one replicate with cycles consisting of a survey scan with a resolution of 60,000 at 400 Th (transient length 384 ms) followed by 10 MS/MS scans with 15,000 resolution. In the other replicate MS/MS scans were acquired with 7500 resolution to reduce the scan times. The project produced at total of 198 raw files (11 cell lines × 6 fractions × 3 replicates) and each cell line required 3 days of instrument time. These files were analyzed together by the MaxQuant software using the Andromeda search engine and relative abundance of proteins was determined by label-free quantification (EXPERIMENTAL PROCEDURES). Comparison of the three replicates of each cell line indicated that the use of the high-field Orbitrap increased the number of MS scans by 25% and MS/MS scans by 33%, multiplying the number of detected isotope clusters by two- to threefold, and improving the mass accuracy (supplemental Table S1).
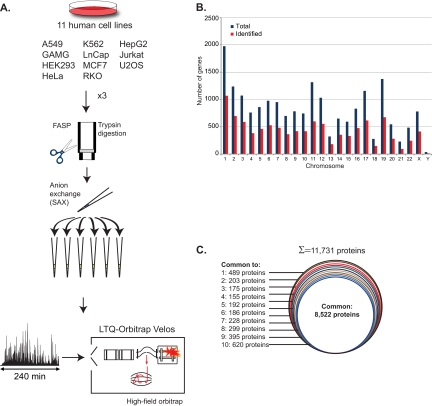
Fig. 1.
Deep proteome analysis of eleven cell lines. A, Eleven commonly used human cell lines were cultured, lysed in SDS based buffer, trypsin digested according to the FASP protocol, and separated into six fractions using SAX in a StageTip format. The analysis was performed in triplicate (1-day measurement per proteome each). LC MS/MS with 4-h runs and HCD fragmentation were performed in the LTQ-Orbitrap Velos equipped with a high field Orbitrap analyzer. B, Distribution of the identified genes (red) in relation to total genes (blue) for each chromosome. Slightly more than half the genome was covered in total and in each chromosome. C, Proportion of proteins identified in all or various subsets of cell lines.
Table I. Protein identifications in eleven cell lines. Summary of the number of identified proteins from triplicate analysis of each cell line. Using the “match between runs” option in MaxQuant enabled transfer of identification between LC MS/MS runs based on retention time and accurate mass and therefore increased the number of identifications per cell line.
| Cell line | Origin | Identified proteins |
|
|---|---|---|---|
| Triplicate analyses | ‘Match between runs’ | ||
| A549 | Lung carcinoma | 8008 | 10,432 |
| GAMG | Glioblastoma | 8292 | 10,503 |
| HEK293 | Embryonic kidney cells | 8543 | 10,504 |
| Hela | Cervical carcinoma | 7781 | 10,371 |
| HepG2 | Hepatoma | 7131 | 10,204 |
| Jurkat | Acute T-Cell Leukemia | 7804 | 10,443 |
| K562 | Chronic Myeloid Leukemia | 7395 | 10,156 |
| LnCap | Prostate carcinoma | 7630 | 10,369 |
| MCF7 | Mammary carcinoma | 7836 | 10,411 |
| RKO | Colon carcinoma | 7336 | 10,252 |
| U2OS | Osteosarcoma | 8025 | 10,402 |
Combined analysis of the triplicates of all cell lines together, using a peptide and protein FDR of 1%, identified 158,294 sequence unique and fully tryptic peptides (supplemental Table S2). These peptides assembled to 11,731 protein groups (proteins distinguishable by MS). Average Andromeda identification score was 113, average number of peptide per protein was 17 (median 11), leading to an average sequence coverage of 35% (supplemental Table S3). Only 2.5% of the total cell line proteome was identified by a single peptide. The 11,731 identified proteins correspond to 10,216 ENSEMBL genes, which represent 50.5% of the complete human genome. Protein identification spread evenly across the chromosomes, with an average of 52% of the genes identified in each chromosome (Fig. 1B). Interestingly, we found four proteins that are encoded exclusively by Y chromosome genes, and the unique peptides of these proteins were expressed only in the cell lines of male origin (LnCap, Jurkat, HepG2, and A549). When the cell lines were analyzed separately 7000–8500 proteins were identified in each of them (Table I). MaxQuant can transfer peptide identifications between matching runs based on the retention time and the very accurate masses determined in the Orbitrap analyzer. Using this “match between runs” option added 25–30% protein identifications, which resulted in the identification of more than 10,000 proteins in each cell line (Table I). A total of 8522 proteins were identified in all cell lines (73% of all identified proteins) and an average of 96% protein identifications were shared between at least two proteomes (Fig. 1C). This overlap is much higher than that typically reported in proteomic studies. In those studies, the proteome was not sampled as deeply, in which case stochasticity in the selection of peptides for sequencing can lead to substantial differences in the identified proteins even for identical samples. This illustrates that deep proteome coverage is necessary for accurate comparison between cells, at least when not employing quantification by stable isotope labeling.
To control for potential false matches between runs, we constructed a method for estimating the FDR of the transfer of peptide identifications from an LC-MS run in which the peptide has been identified by MS/MS to another LC-MS run where either no MS/MS spectrum has been acquired for that MS peptide feature or the peptide could not be identified based on the MS/MS spectrum. The FDR is calculated after the retention times of all LC-MS runs have been aligned to each other. To generate input data for the FDR calculation, each run is matched to a randomly chosen other run based on its pre-fractionation index. The likelihood of wrongly matching MS features based on accurate mass and retention time is lower for peptides with high signal intensity of the two involved MS isotope patterns. For all of these alignments we collect all number pairs Δt, log(I), where Δt is the difference in retention times between the two matched isotope patterns, and log(I) is the logarithm of a suitable combination of the intensities of the two isotope patterns. From these number pairs we estimate the density of matches in the Δt-log(I) plane by Voronoi tessellation. We decompose this density into a true positive and a false positive contribution based on the assumption that the background density of false matches does not depend on Δt whereas the density of true matches does not depend on log(I). By applying suitable scaling factors we can calculate from the intensity-dependent false-positive density the intensity-dependent probability that a matching try leads to a false positive match. To estimate the matching FDR in our data we applied the procedure described above to the 18 LC-MS runs corresponding to the MCF7, A549, and U2OS cell lines. The FDR estimate based on the fraction-restricted matching between these three cell lines is 0.12%. Because in reality we are matching between 11 cell lines we estimate the FDR to be lower than 11/3*0.12% = 0.44%.
The complete dataset of the eleven proteomes is deposited in the MaxQB database, which is the subject of another paper in this issue (17). These data can serve as a library of protein identifications and quantifications, as well as a resource for high-resolution MS/MS spectra that can be used for targeted proteomic analyses (5, 29, 30).
Label-free Quantification of the Cell Line Proteomes
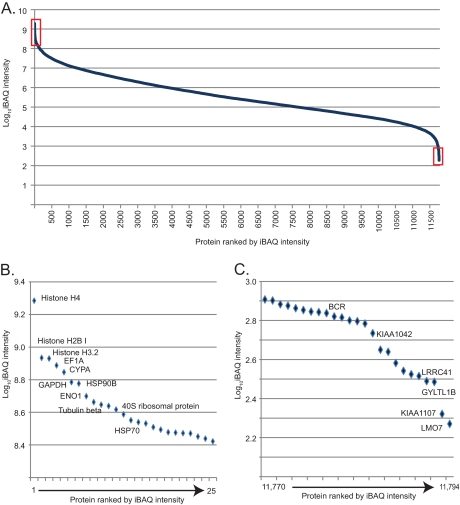
To quantify the proteins and compare their levels across the various cell lines we used label-free quantification. For this analysis MS signals of the same peptides detected in different cell lines are quantified relative to each other (EXPERIMENTAL PROCEDURES). In contrast, to roughly estimate the abundance of proteins within a proteome, the MS signals of the peptides identifying a protein can be summed and normalized to the size, length, or number of theoretical peptides (31–33). Here we use the iBAQ algorithm, which essentially normalizes the summed peptide intensities by the number of theoretically observable peptides of the protein. In each of the 11 cell line proteomes, the iBAQ values varied over about seven orders of magnitude. We calculated the median values across the cell lines and plotted the estimated absolute abundance of the roughly 12,000 proteins of the composite cell lines proteome. Like the individual proteomes, the apparent dynamic range of protein expression was seven orders of magnitude (Fig. 2A). The 25 most highly expressed proteins contain the core histones, enolase, GAPDH, tubulin and heat shock proteins as well as proteins of the ribosome (Fig. 2B). The 25 proteins with the lowest MS signals included several novel and uncharacterized proteins as well as isoforms of well-known proteins (Fig. 2C). In this expression range, the in silico abundance estimation is only a very rough indication of actual expression levels. In particular, the expression levels may be underestimated for the apparently least expressed proteins. Conversely, the proteomes measured here are not complete and the missing proteins presumably have very low expression values.
Fig. 2.
Dynamic range of the composite cell line proteome. A, The median absolute expression value of each protein in the eleven cell lines was estimated by iBAQ revealing the typical S-shaped distribution over the seven orders of dynamic range of MS signals. B, The 25 most abundant proteins (left red box in A) are structural constituents of chromatin or the cytoskeleton or they are abundant enzymes, chaperones, and constituents of the translational apparatus. C, The 25 proteins with the lowest abundance (right red box in A) include uncharacterized proteins and isoforms of proteins. Note that in-silico estimation of protein amount is not accurate in this abundance range.
Comparison of Cell Line Proteomes
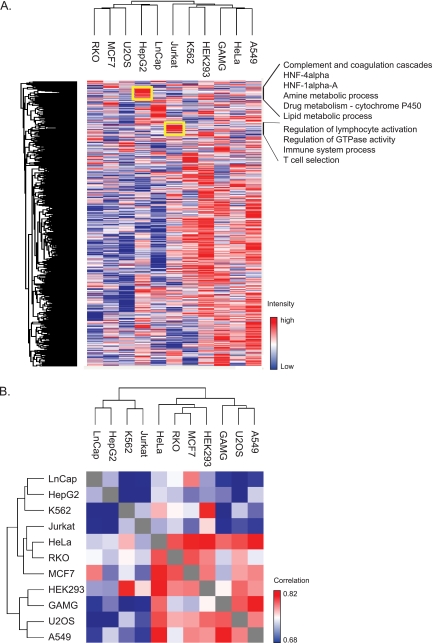
We examined the similarity of the cell lines using the median value of the normalized protein intensities from the triplicate analyses. Unsupervised hierarchical clustering created four clusters of cell lines encompassing all but the Jurkat cells. (Fig. 3A). The first included RKO, MCF7, and U2OS, the second LnCap and HepG2, the third K562, HEK293, and the fourth cluster consisted of A549, HeLa and GAMG cells. Surprisingly, there was little connection between the clustering of the cell lines and their tissue of origin. Cell lines from epithelial origin, for example, did not necessarily co-cluster, but rather grouped with cells originating from distinct tissue types. Nevertheless, most cell lines displayed protein clusters that were typical of the original function of the cell type. For example, the liver cell line HepG2 coordinately expressed proteins involved in the complement system (normally secreted from the liver), multiple metabolic processes, and targets of HNF1 and HNF4 as annotated by the TRANSFAC database (25) (Fig. 3A). Similarly, in Jurkat cells, which derive from T-cell leukemia, clusters of highly expressed proteins were enriched for the protein annotations “positive regulation of lymphocyte activation” and “immune system process” (Fig. 3A). Despite these differences the Pearson correlation between cell lines was relatively high—on average 0.74 (Fig. 3B). At 0.67 Jurkat and HepG2 cells had the lowest correlation whereas the proteomes of HeLa and A549 cells were the most similar ones (r = 0.83).
Fig. 3.
Hierarchical clustering based on label-free proteome quantification. A, Two-way unsupervised hierarchical clustering of the median protein expression values of all proteins in a cell line does not group the cell lines according to the tissue of origin. This indicates dedifferentiation compared with the in vivo cell type. However, clusters of coregulated proteins carrying out cell type specific functions are retained (see yellow boxes for examples). Listed functions have FDR values (greatly) below 0.02 by Fishers exact test. B, Matrix representation of Pearson correlation values of the label-free protein abundances of each cell line proteome against the others. Correlations are uniformly high, varying only between r = 0.68 and 0.83.
Two Thirds of the Proteome Varies Across Cell Lines
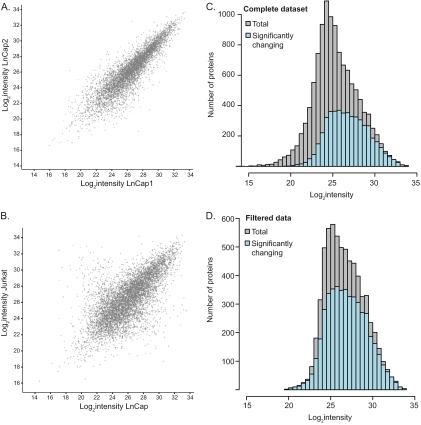
Next we wished to extract proteins that varied significantly between cell lines. We first examined the reproducibility of the label-free protein quantification between replicates and found an average Pearson correlation coefficient of 0.83. Scatter plots of the intensity distribution in one replicate relative to another show the typical non-uniform spread, which is wider at low intensities than at the high ones. As an example, Fig. 4A compares replicate 1 against replicate 2 of the LnCap cell line. This non-uniform distribution is a result of reduced accuracy and reproducibility of the measurements of peptides that are closer to the background level, and this effect needs to be taken into account properly as it is done in a t test. We therefore determined the proteins that are significantly changing between cell lines by performing a multiple t test (ANOVA). This test statistically determines the reproducibility among the triplicate measurements in relation to the difference between the cell lines. Fig. 4B shows that the difference of protein expression values between the cell lines is significantly larger than within the replicates. Note that in the comparison between cell lines, the median of the triplicate values is used, further increasing the accuracy of the comparison. A comparison of two triplicates of the same cell line typically results in a correlation greater than 0.9, whereas a comparison of single replicates has a correlation of 0.8–0.9.
Fig. 4.
Significance of proteomic changes across the abundance range. A, Scatter plot of label-free protein intensities between the first and second replicate of the prostate cancer cell line LnCap. B, The spread in the scatter plot of the median of triplicates of two different cell lines LnCap and the T-cell leukemia derived Jurkat cells is larger than in A. C, Significantly changing proteins in at least one of the eleven cell lines appear to be more abundant compared with the entire proteome. D, Filtering for robustly quantified proteins (a minimum of 16 valid quantification values) reveals that more than two thirds of the proteome are changing significantly and that this proportion does not depend on protein abundance.
Examination of the complete dataset in this way extracted 4881 proteins that had statistically significant changes in their expression values in at least one of the cell lines (FDR 0.05; EXPERIMENTAL PROCEDURES). Surprisingly, the majority of them were of high abundance (Fig. 4C). This result is explained by the fact that proteins of very low abundance are more challenging to quantify accurately by label-free MS analysis, and they therefore do not reach statistical significance. To eliminate this effect, we filtered the data for the proteins that were identified in at least half of the 33 proteome measurements. Examination of this more stably quantified proteome now showed that more than two-thirds of the proteins (4753 of 6630) varied significantly and that their intensity distribution did not differ from the overall distribution (Fig. 4D). Because there is no correlation between the intensity of the protein and its likelihood to be differentially expressed between cells, we conclude that two-thirds of the entire proteome are likely to be changing significantly. These results suggest that the differential expression of proteins between cell types relates to their function rather than to their abundance.
Functional Analysis of Cell Lines Across the Abundance Range
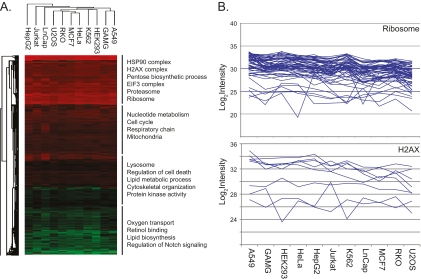
To examine the functional differences between cell lines we used multiple protein annotation databases: GO, KEGG pathways, the CORUM molecular complexes database (27), and the TRANSFAC transcription factor targets database (26). For this analysis we used a recently developed algorithm that determines the significance of the difference between the mean expression level of any annotation and the overall protein distribution (34). Here we generalized this algorithm to work with more than two dimensions and created an annotation matrix of annotations versus the eleven cell lines (Fig. 5A; supplemental Table S4; EXPERIMENTAL PROCEDURES). Distribution of annotations across the 11 cell lines was relatively uniform, suggesting generally similar functional profiles. We divided the annotation distribution into four groups according to the mean abundance of their constituent proteins. The major annotations in the most abundant quartile were HSP90 complex, H2AX complex, proteasome, and ribosome and in the second most abundant quartile they were cell cycle, nucleotide metabolism, and respiratory chain. The third group included lysosomal proteins, proteins involved in cell death, and protein kinases and in the least abundant quartile major annotations were oxygen transport, retinol binding, lipid biosynthesis, and Notch signaling.
Fig. 5.
Variability of cellular functions in dependence of protein abundance. A, Annotation matrix of protein attributes, such as pathways, complexes and gene ontologies versus the eleven cell lines. Color code indicates the normalized median abundance of the proteins belonging to the category (red most abundant; green least abundant). Protein annotations are separated into four blocks and labeled with representative individual annotations. B, Proteins comprising the “ribosomal” category are highly and uniformly expressed across the cell lines (upper panel). Proteins comprising the category “H2AX” are highly abundant but their expression levels vary dramatically across the cell lines.
The fact that the annotations belong to the same intensity block does not imply that the proteins involved are always expressed at the same levels. While this is the case for ribosomal proteins in our cell lines (Fig. 5B), it is not true of the equally abundant histone complex H2AX. As depicted in the figure, proteins involved in this function often vary more than fivefold in expression between cell lines.
To determine more generally which annotations differ between cells and which ones are constant, we performed a Fisher exact test for the enrichment of protein annotations in the set of significantly changing proteins. Among the highly variable annotations we found multiple metabolic pathways, including fatty acid metabolism, amino acid metabolism and glutathione metabolism (supplemental Table S5). This demonstrates the diversity of energy production and biosynthetic pathways that are employed by different cells. Large differences were also present in the actomyosin cytoskeleton, in accordance with their different morphology. The processes that were highly constant were related to the basal transcription machinery and to protein translation. Interestingly oxidative phosphorylation was also relatively constant between the cells, being the common end point of the energy production machinery of multiple catabolic pathways. These results suggest that fundamental processes and nonredundant pathways retain equal protein levels, whereas, for instance, cell metabolism and some structural proteins exhibit high diversity of protein expression values.
Use of Cell Lines as “Spike-In” SILAC Standards
Apart from shedding light on basic cell biological processes, the deep proteomic data of the diverse cell lines can also be put to practical purposes. Here we illustrate its use in the construction of a spike-in standard for relative quantification. We have previously shown that a super-SILAC mix of five breast cancer cell lines employed as a spike-in standard enables accurate and precise quantification of breast cancer tissue samples (18). The high overall similarity of cell line proteomes found in this investigation suggested that a set of relatively few heavy SILAC labeled cell lines could likewise serve as a standard for a broad range of common cell lines. We used the label-free data and the correlation matrix previously determined by label-free quantification (Fig. 3B), to select five cell lines, chosen to be as dissimilar to each other as possible (Jurkat, HEK293, LnCap, HeLa, and K562).
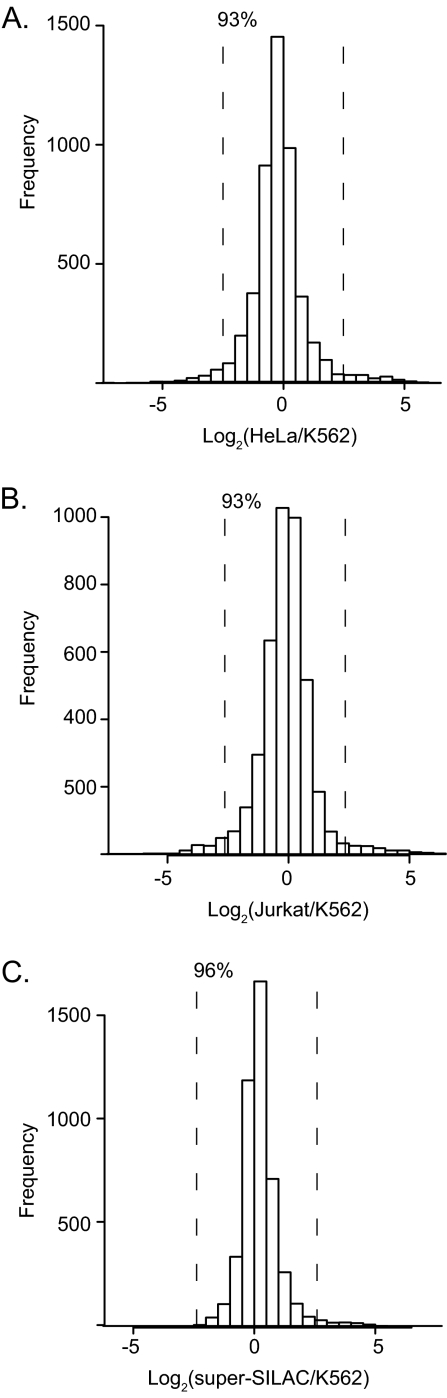
As expected from the high correlations, even the SILAC quantification experiments between binary combinations of heavy and light labeled cell lines resulted in narrow ratio distributions (Fig. 6A, 6B). This was further improved in the quantification of the heavy super-SILAC mix against a single cell line (Fig. 6C). These experiments also suggest a general strategy to construct super-SILAC mixes and to evaluate their suitability for quantifying any cell line (or tissue) of interest. Importantly, this only involves label free quantification of the various cell lines that are suggested for the super-SILAC mix (heavy or light) against the label-free proteome of interest.
Fig. 6.
Use of cell lines as a spike-in SILAC standard. Knowledge of the overall similarities of the proteomes was used to construct a five cell line, heavy labeled “super-SILAC” reference standard. A, HeLa and K562 as well as Jurkat and K562 (B) proteins are sufficiently similar that most of the SILAC ratios are within a fivefold ratio above and below the one to one ratio. C, Quantification of the heavy super-SILAC mix consisting of five cell lines quantifies 96% of the K562 proteome within a fivefold ratio.
DISCUSSION AND OUTLOOK
Here we employed state of the art mass spectrometric technology and characterized the proteome of eleven common cell lines to a depth of over 10,000 proteins in each case. Remarkably, this depth of coverage was achievable without extensive fractionation and with a relatively straightforward proteomic workflow. Total measuring time at one proteome per day added up to only 33 days for triplicates of all eleven cell lines, comparing favorably even to transcriptome measurements using next generation sequencing technology. Although the depth of analysis shown here is currently limited to specialized groups, there are no principal obstacles to its application to a broad range of scientists. An interesting corollary of our study is that MS-based proteomics is not limited by the rate of protein identification per se. If diverse samples could be found in which all of the proteins were present in the 10,000 most abundant proteins, then proteins for all genes in the human genome could be sequenced rather quickly. Therefore, the major difficulty in obtaining an identified protein for every gene, as called for by the Human Proteome Organization (35), is in a suitable supply of proteomes. Already the single project described here can serve as a resource to determine expression of proteins of interest in specific cell lines for proteins from half of the human genome. It also provides reference peptides and their high resolution HCD fragmentation spectra for these proteins (see accompanying paper Schaab et al.). Furthermore, we expect our data to have many practical applications, such as in creating proteome standards as already demonstrated here.
In biological research specific cell lines are chosen to investigate cellular processes that occur in their tissue of origin. In view of that fact, an unexpected finding of this study was the high degree of overall similarity of the proteomes of the diverse cell lines. For instance, the depth of the proteome detectable by our technology was very similar in all cases and label-free proteome correlations ranged from 0.68 to 0.83. Our findings do agree, however, with recent studies at the transcriptome and proteome levels that also found a large overlap of expressed genes in different cell types (2, 35, 36). This high commonality of the proteome presumably results in part from the adaptation of cell lines to the in vitro growth. In this situation, cellular clones that proliferate rapidly are selected for whereas many cell type and tissue specific functions that are not crucial to their growth and survival may be lost. We have previously addressed this question directly by quantifying the proteome of a liver cell line against primary hepatocytes and our results support the above conclusions (38).
Despite the overall proteomics similarities, cell type specific clusters of protein expression are clearly present in the cell lines (Fig. 3A). Furthermore, statistical analysis of the expression profiles showed that a large proportion of the proteins changes significantly in at least one of the cell lines. Interestingly, when filtering for a robust number of quantification events per protein, we found that more than two thirds of the proteome is likely to change significantly and that this is not affected by protein abundance. Bioinformatic analysis of protein function revealed higher variability in redundant pathways whereas basal functions such as gene and protein expression tended to be more uniformly represented across the cell lines. These analyses can guide researchers in the choice of the optimal cell line for the biological interests. Our data shows that even functions carried out by abundant and ubiquitous proteins do not necessarily imply that these proteins need to be expressed at the same levels in all cell lines. Instead they often vary several fold.
What do these results mean for the common notion of a “core” or “household” proteome composed of proteins that are needed by every cell type and that are highly abundant? At a minimum, deep proteomics reveals that a household proteome, is not as straightforward a concept as frequently believed. For instance, at least in cell lines, proteins tend to be present in very diverse cell types, not only for very common but also for more specialized functions. Furthermore, the household proteins themselves are not necessarily uniformly expressed. For a biologically desirable definition of the household proteome it may be necessary to study the proteomes of cell types in vivo, an undertaking that we expect to become technological possible within the next few years.
Acknowledgments
We thank the members of the Department of Proteomics and Signal Transduction for fruitful discussions. Especially, we thank Annette Michalski for MS expertise and Bianca Splettstöβer for help with the cell culture and Nadin Neuhauser for computational support.
Footnotes
* This project was supported by the European Commission's 7th Framework Program PROteomics SPECificat ion in Time and Space (PROSPECTS, HEALTH-F4-2008-021,648).
 This article contains supplemental Tables S1 to S5.
This article contains supplemental Tables S1 to S5.
1 The abbreviations used are:
- SILAC
- stable isotope labeling with amino acid in cell culture
- FDR
- false discovery rate
- MS/MS
- tandem mass spectrometry
- HCD
- Higher energy Collisional Dissociation
- LTQ
- Linear trap quadrupole.
REFERENCES
- 1. Burkard T. R., Planyavsky M., Kaupe I., Breitwieser F. P., Bürckstummer T., Bennett K. L., Superti-Furga G., Colinge J. (2011) Initial characterization of the human central proteome. BMC Syst. Biol. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lundberg E., Fagerberg L., Klevebring D., Matic I., Geiger T., Cox J., Algenäs C., Lundeberg J., Mann M., Uhlen M. (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 4. Swaney D. L., Wenger C. D., Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mallick P., Kuster B. (2010) Proteomics: a pragmatic perspective. Nat. Biotechnol. 28, 695–709 [DOI] [PubMed] [Google Scholar]
- 6. Beck M., Claassen M., Aebersold R. (2011) Comprehensive proteomics. Current Opin. Biotechnol. 22, 3–8 [DOI] [PubMed] [Google Scholar]
- 7. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 8. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 9. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 10. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal 4, rs3. [DOI] [PubMed] [Google Scholar]
- 11. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makarov A., Denisov E., Lange O. (2009) Performance evaluation of a high-field Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 20, 1391–1396 [DOI] [PubMed] [Google Scholar]
- 16. Michalski A., Damoc E., Lange O., Denisov E., Nolting D., Mueller M., Viner R., Schwartz J., Remes P., Belford M., Dunyach J. J., Cox J., Horning S., Mann M., Makarov A. (2011) Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics DOI 10.1074/mcp.O111.013698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaab C., Geiger T., Stoehr G., Cox J., Mann M. (2012) Analysis of high-accuracy, quantitative proteomics data in the MaxQB database. Mol. Cell. Proteomics DOI 10.1074/mcp.M111.014068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 19. Wiśniewski J. R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 20. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 21. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 22. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 23. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Michalski A., Mann M. (2011) Software Lock Mass by Two-Dimensional Minimization of Peptide Mass Errors. J. Am. Soc. Mass Spectrom. 22, 1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 [DOI] [PubMed] [Google Scholar]
- 26. Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., Wingender E. (2006) TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruepp A., Waegele B., Lechner M., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Mewes H. W. (2010) CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 38, D497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt A., Claassen M., Aebersold R. (2009) Directed mass spectrometry: towards hypothesis-driven proteomics. Curr. Opin. Chem. Biol. 13, 510–517 [DOI] [PubMed] [Google Scholar]
- 30. Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 32. Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shinoda K., Tomita M., Ishihama Y. (2010) emPAI Calc–for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics 26, 576–577 [DOI] [PubMed] [Google Scholar]
- 34. Geiger T., Cox J., Mann M. (2010) Proteomic Changes Resulting from Gene Copy Number Variations in Cancer Cells. PLoS Genet 10.1371/journal.pgen.1001090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Legrain P., Aebersold R., Archakov A., Bairoch A., Bala K., Beretta L., Bergeron J., Borchers C. H., Corthals G. L., Costello C. E., Deutsch E. W., Domon B., Hancock W., He F., Hochstrasser D., Marko-Varga G., Salekdeh G. H., Sechi S., Snyder M., Srivastava S., Uhlen M., Wu C. H., Yamamoto T., Paik Y. K., Omenn G. S. (2011) The human proteome project: current state and future direction. Mol. Cell. Proteomics 10, M111 009993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pontén F., Gry M., Fagerberg L., Lundberg E., Asplund A., Berglund L., Oksvold P., Björling E., Hober S., Kampf C., Navani S., Nilsson P., Ottosson J., Persson A., Wernérus H., Wester K., Uhlén M. (2009) A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 5, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramskold D., Wang E. T., Burge C. B., Sandberg R. (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Computat. Biol. 5, e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]