Abstract
Influenza virus encodes only 11 viral proteins but replicates in a broad range of avian and mammalian species by exploiting host cell functions. Genome-wide RNA interference (RNAi) has proven to be a powerful tool for identifying the host molecules that participate in each step of virus replication. Meta-analysis of findings from genome-wide RNAi screens has shown influenza virus to be dependent on functional nodes in host cell pathways, requiring a wide variety of molecules and cellular proteins for replication. Because rapid evolution of the influenza A viruses persistently complicates the effectiveness of vaccines and therapeutics, a further understanding of the complex host cell pathways coopted by influenza virus for replication may provide new targets and strategies for antiviral therapy. RNAi genome screening technologies together with bioinformatics can provide the ability to rapidly identify specific host factors involved in resistance and susceptibility to influenza virus, allowing for novel disease intervention strategies.—Meliopoulos, V. A., Andersen, L. E., Birrer, K. F., Simpson, K. J., Lowenthal, J. W., Bean, A. G. D., Stambas, J., Stewart, C. R., Tompkins, S. M., van Beusechem, V. W., Fraser, I., Mhlanga, M., Barichievy, S., Smith, Q., Leake, D., Karpilow, J., Buck, A., Jona, G., Tripp, R. A. Host gene targets for novel influenza therapies elucidated by high-throughput RNA interference screens.
Keywords: antivirals, HTS, genome, pathway, meta-analysis
Many emerging infectious diseases (EIDs) in humans are zoonotic: deriving from animals or animal products. EIDs are broadly defined to include new agents, existing yet previously undetected agents, the reemergence of known agents, and/or expansion of a known agent into a new geographic range (1). Influenza viruses are well-documented examples of EIDs. In 2009, the H1N1 pandemic virus erupted from the swine population and quickly spread to over 200 countries, overtaking preexisting H1N1 and H3N2 viruses in the human population and becoming the dominant circulating strain (2). Similarly, outbreaks of highly pathogenic H5N1 influenza viruses continue to emerge and exhibit up to 60% mortality rates in humans (3). These outbreaks highlight the persistent and devastating nature of influenza infections, and increase the risk of new pandemics.
Despite scientific advancements over the past 3 decades, EIDs continue to inflict substantial social and economic costs. For example, influenza is a leading cause of morbidity and mortality in the world (4) with seasonal viruses affecting up to 15% of the human population, causing severe illness in 3–5 million people and fatality of ∼500,000 individuals/yr (5). Coupled with this is the economic burden that is associated with widespread influenza infection. In the United States alone, financial losses, resulting from seasonal influenza infection are estimated to exceed $87 billion annually (6).
The ability of influenza viruses to continuously mutate has made control strategies based on vaccination difficult. Simultaneously, resistance to current antivirals is increasing. For these reasons, new strategies to combat EIDs such as influenza are required, and an expanded knowledge of host-virus interactions is a crucial first step. Despite the complexity of influenza biology, viruses of this class contain only 8 gene segments (7) and therefore lack the full complement of proteins required to produce infectious virus. As such, influenza, like all viruses, must coopt a wide array of host proteins, noncoding RNAs, and cellular processes (for example, transport vesicles) to generate infectious viral particles. The replicative cycle of orthomyxoviruses can be divided into early, middle, and late stages (8). Early stages involve events linked to virus binding via hemagglutinin (HA) to sialic acid moieties found on the host cell membrane and internalization of the viral particles (9). Midstage events include the expression and translation of viral genes (10), while late events comprise virus protein trafficking to the cell membrane, virion assembly, and budding from the cell (11, 12). Understanding the host contribution to viral replication and immune evasion is essential for discovering new therapeutic strategies. Genome screening technologies that use RNA interference (RNAi), together with bioinformatics, provide the ability to rapidly identify the complement of essential host functions and pathways that are essential to the virus.
THE RNAi PATHWAY
RNAi is a natural, sequence-specific post-transcriptional gene silencing pathway (Fig. 1) present in most eukaryotes (13, 14). Components of the RNAi pathway serve multiple roles that can vary in different organisms. On the one hand, elements of the pathway participate in the innate immune response by recognizing and degrading long double-stranded RNAs (dsRNAs) typically associated with viral infection (15–17). Simultaneously, the RNAi pathway has a recognized role in regulating host gene expression related to a myriad of cell functions by guiding the maturation and transcript targeting capabilities of a unique class of noncoding RNAs called microRNAs (miRNAs; refs. 18, 19). This aspect of RNAi is particularly intriguing given that some viruses, in particular herpesviruses, encode miRNAs that regulate host gene expression and, therefore, have a critical role in the dynamics of host-pathogen interactions (20–22).
Figure 1.
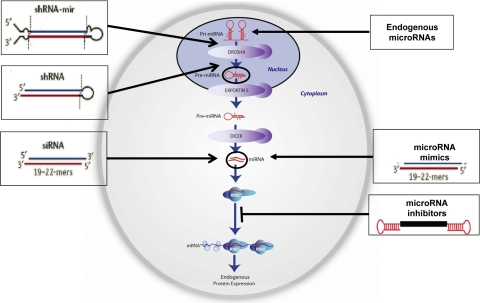
RNAi pathway. Endogenous miRNAs are transcribed as Pri-miRNAs that are subsequently processed by Drosha and Dicer to generate mature miRNAs. The guide strand is then incorporated into the RNA-induced silencing complex (RISC) to facilitate gene silencing. Reagents that have been developed for screening include expressed constructs (shRNA and shRNA-miR), as well as synthetic constructs: siRNA, miRNA mimics, and miRNA inhibitors.
Outside the important role in gene modulation, the RNAi pathway is now recognized as a preeminent means for studying gene function (23, 24). In this regard, researchers have created entire libraries of gene-specific targeting reagents that silence individual gene function by entering the pathway at one of two distinct positions. In one instance, synthetic dsRNA reagents referred to as small interfering RNAs (siRNAs) have been generated. When introduced into cells, siRNAs associate with the RNA-induced silencing complex (RISC), where the siRNA guide strand anneals to its complementary target mRNA (25, 26) and facilitates cleavage by Argonaute 2, a core endonuclease of RISC (Fig. 1). In an alternative approach, researchers have developed collections of gene-targeting short hairpin RNAs (shRNAs) that can be generated from plasmid or viral expression constructs (27–29). Like their endogenous miRNA counterparts, shRNAs are expressed in the nucleus and are predicted to associate with RISC only after being processed by two separate endonucleases, Drosha and Dicer (Fig. 1 and refs. 18, 19).
GENERAL WORKFLOW OF HOST-PATHOGEN SCREENING USING RNAi TECHNOLOGY
The complexity of RNAi screens for host-pathogen interactions requires thoughtful consideration of the silencing reagent, the cell line, and virus to be used. RNAi screening typically begins with the plating of the target cells and introduction of the silencing reagent. For siRNA-based studies, screens are generally performed in an arrayed format where individual genes are targeted with anywhere from 1 to 4 siRNAs/well. In most cases, pooled reagents are preferred, as they provide a cumulative silencing effect that greatly increases the efficacy of gene knockdown. Synthetic silencing reagents can be introduced via forward or reverse transfection protocols (30–33), which, in many cell types, lead to efficient intracellular delivery of the siRNA. For cases in which shRNAs are used and expressed from a viral construct, one of two approaches can be employed. In the first, shRNA workflows can parallel siRNA screening processes by adopting a 1-gene-1-well arrayed platform. Alternatively, shRNAs screens often make use of pools of viral particles that contain expression constructs targeting hundreds to thousands of genes. In this scenario, cell plating is followed by virus transduction at low multiplicities of infection, thus ensuring that each infected cell receives only one viral particle. Drug selection follows transduction to eliminate cells that do not carry a gene-targeting construct (27–29). Pooled shRNA screens are best suited to output assays that permit selection or outgrowth of cells that exhibit the phenotype of interest, where specific shRNAs, which influence the phenotype (and are consequently enriched or depleted from the population) can be identified by either microarray or next-generation sequencing (NGS). For an overview of the comparative workflows for siRNA vs. shRNA, see Table 1.
Table 1.
Comparison of screen stages for shRNA and siRNA screens
| Screen stage | shRNA | siRNA |
|---|---|---|
| 1. Cell-based optimization | Define MOI for cell line | Establish transfection conditions |
| Identify +ve, −ve controls | Identify +ve, −ve controls | |
| Make virus for individual shRNAs | siGLO and nontargeting control | |
| 2. Assay development | Establish screen phenotype | Define assay parameters |
| Verify phenotype of controls in dilution of pools | Robustness, Z′ factor | |
| Develop automation | ||
| Identify analysis rules | ||
| 3. Screen | Transduce cells with library pool | Primary SMARTpool screen |
| Select with GFP/puromycin | Duplicate or triplicate technical replicates | |
| Assay: select reference population | Analysis ongoing | |
| Freeze cells, extract genomic DNA | ||
| Amplify gDNA and NGS analysis | ||
| 4. Bioinformatics | Process NGS data | Statistical analysis |
| Statistically rank shRNAs | Define hit list | |
| 5. Validation | Identify individual shRNA hits | Secondary validation screen |
| Make virus for all constructs per target | Deconvolute SMARTpools | |
| Rescreen using same assay | Same assay or different assay | |
| Verify knockdown | Additional cell lines | |
| 6. Bioinformatics | Pathway analysis | Pathway analysis |
| Data mining | Data mining | |
| Tertiary analysis, more cell lines, different assays |
shRNA and siRNA screen stages broken into chronological order. Stages can take different times depending on the assay and quantitation method. Bioinformatics analysis is an ongoing effort that intervenes in the screen process at several points. Central to each screening platform is identification of robust positive (+ve) and negative controls (−ve). For the siRNA platform, siGLO, a fluorescent reporter, is used to indicate transfection efficiency.
Regardless of the type of silencing reagent employed, the cells are subsequently cultured for a period of time that allows for gene knockdown and accounts for any unique attributes of the screen. For instance, if the intent of the screen is to study viral uptake, infection may be delayed to avoid lipid-mediated siRNA transfection affecting the ability of the virus to bind the cell. Cells are then infected with the virus of choice, cultured for an additional period of time, and assayed by one or more techniques (e.g., transcriptional reporters, viral titers) to determine the effect of gene knockdown on various aspects of the viral life cycle. It is worth noting that different silencing reagents permit different types of assays to be incorporated in the workflow. Although the period of gene silencing provided by siRNA can vary with the gene and the cell type being studied, synthetic siRNAs generally provide 3–7 d of gene knockdown (34, 35). As such, these reagents are compatible with assays that are performed within this brief window. In contrast, shRNAs expression constructs, delivered by a lentivirus that stably integrates into the host genome permit assays that extend beyond the transient (siRNA) window to be performed.
RNAi screens that utilize siRNA technology are greatly aided by automation, in which extreme accuracy, reproducibility, and, to some extent, speed are necessary for managing the siRNA library resource, dispensing and aspirating cells and medium, and performing lipid-mediated transfection. For screens that involve infectious reagents, liquid handling platforms that are modular are attractive, as they facilitate the separation of steps that require high-level biocontainment from those that are common to all RNAi screens.
Both screening processes are iterative, and full genome-wide screens can take anywhere from weeks to months, depending on the assay complexity, the level of automation, the particulars of the pathogen being investigated, and the logistics used to divide and screen the gene-silencing collection. With regard to hit identification, a term used to identify genes that when silenced have a significant effect on the function being assayed, a number of bioinformatic approaches have been described. In one instance, a Z′ factor is calculated to determine how distinct the positive and negative controls are from each other; subsequently, a Z score, which is a method of normalization to the mean value of the samples on each plate, is commonly used to rank genes (36). Candidate genes are then categorized for defining features before being validated. A statistical enrichment of targets relative to a reference population is then ranked for validation. Many parameters may be captured in a screen, and these are weighted on the importance to the principle screen phenotype and classified into distinct bins to refine the gene list prior to validation. For example, for an shRNA screen, following NGS analysis, data are subjected to a statistical enrichment of targets relative to a reference population, which results in a ranked list to identify targets for validation.
siRNAS AND shRNAS
siRNA libraries have been constructed with a variety of designs, and while these reagents have been used in a range of studies and cell types, it is important to consider which of these tools is best suited for host-pathogen screens. One potentially relevant consideration centers on activation of cellular immunity pathways. It is well documented that viral infection is monitored by pathogen recognition receptors (PRRs) that trigger an innate immune response upon identifying an evolutionarily conserved pathogen-associated molecular pattern (PAMP) (37). For example, recognition of influenza single-stranded RNA genome by the endosomal PRR, Toll-like receptor 7 (TLR7), leads to a strong antiviral cytokine response (38–40). Previous studies have also shown that transfection of short dsRNA (such as siRNA) can also perturb cellular immunity. For example, it has been demonstrated that siRNAs longer than ∼23–25 bp strongly up-regulate interferon (IFN)-stimulated genes in HeLa cells (41). Other groups have shown that siRNAs can activate a type I interferon response, particularly through TLR7 and TLR8 (42, 43). Thus, if both siRNA and virus can independently stimulate TLR pathways, the question arises of how one can effectively screen for host genes that play a role in virus replication. One solution may lie in reagent selection. For example, while most siRNAs are designed to have complete complementarity to the target transcript, one design variable is duplex length. Previous studies have shown that in some cell types, short (19 bp) siRNAs effectively silence genes but are weaker activators of TLR signaling pathways (44). As such, under certain circumstances, these reagents can minimize the immune stimulatory effects that would otherwise complicate RNAi screens involving pathogens. In addition, several groups have demonstrated that modification of particular nucleotides with defined chemical groups (e.g., 2′-O-methyl,2′OMe) can greatly reduce the immunostimulatory effects of siRNA (45–47). Such modifications, which can act in either cis or trans, limit TLR7/8 activation and thereby prevent the triggering of downstream interferon and cytokine pathways.
Separately, another noteworthy caveat of all RNAi screening is siRNA specificity. Extensive studies have shown that in addition to targeting the mRNA of interest, siRNAs can act in a similar fashion to miRNAs and attenuate the translation of dozens of genes (48, 49). False positives (referred to as “off-targets”) induced by this mechanism are frequently mediated by the seed region of the siRNA (nt 2–8), and may result in a 2- to 4-fold down-regulation of unintended targets (49). Notably, while off-targets are generally thought to be genes encoded by the cell, in the case of RNAi screens designed to identify host-pathogen interactions, off-targets can also include genes encoded by the pathogen.
Multiple approaches have been developed to minimize the effects of off-targets, including chemical modification of the siRNA duplex to reduce seed-mediated gene silencing. For example, 2′-O-methyl modification of the second nucleotide of the siRNA antisense strand can greatly reduce seed-mediated off-target effects (50). Use of these reagents during primary screening can minimize the number of false positives and thereby reduce the time and costs associated with downstream validation. Alternatively, in cases where pools of siRNA were used in the primary screen, deconvolution of the pools and demonstration that multiple individual siRNAs (having different seed sequences) give the same phenotype increases the level of confidence that the phenotype is related to the knockdown of the targeted gene. Lastly, bioinformatics can be effectively used to identify off-target effects. Cross-checking the seed sequence of siRNAs that induce a phenotype with the seeds of known miRNAs (host and pathogen) can often identify instances in which one or more siRNAs are mimicking a miRNA. Although these hits should not be excluded from further study (due to potentially interesting host miRNA-pathogen interactions), they may indicate that the observed phenotypes are the result of events unrelated to the knockdown of the intended target gene.
Similar considerations may be relevant when performing screens with shRNA expression cassettes. In most cases, shRNAs are generated from DNA-based expression constructs, and the predominant collections of shRNA libraries use either lentiviral or retroviral delivery systems to facilitate the entry and integration of the cassette into the host genome (27, 28). Previous genome profiling studies of lentiviral (HIV)-infected CD4+ T cells have shown that infection leads to differential expression of multiple gene categories, including those related to complement activation, actin filaments, and proteasome cores (51). Thus, although the current shRNA platforms are essential for performing host-pathogen studies in cases where the cell type of choice is refractory to lipid-mediated transfection or an extended period of gene knockdown is required, one must consider the potential contributions that the delivery platform has on the outcome of host-pathogen screens.
miRNAS
miRNAs have emerged as essential regulators of eukaryotic gene expression (52–55). Mature miRNA sequences have been found in >150 species, including viruses, with >1000 identified in humans (52, 54, 56, 57). Recent studies have identified both host and viral encoded miRNAs as critical elements regulating virus replication. For example, human miR-122 has been identified as an essential component affecting the biology of hepatitis C virus replication (58). Interestingly, both procytomegalovirus (pro-CMV) and anti-CMV miRNAs have been identified as encoded by the mouse genome (59). These and other studies emphasize the need for miRNA screening to accompany siRNA/shRNA screens that target protein-coding genes.
Identifying host-encoded miRNAs that are relevant to viral infection generally involves a two-pronged approach. Synthetic miRNAs mimics (or equivalent sets of reagents expressed from e.g., a plasmid or viral-based vector) can be used to increase the cellular concentration of any given miRNA. In contrast, miRNA inhibitors are designed against mature miRNAs and can act as artificial targets to prevent the endogenous miRNA from interacting with its natural targets (60). A library of mimics and inhibitors was recently used to identify miRNAs that play a role in diverse herpes virus infections [murine CMV (MCMV), murine γ herpes virus-68 (MHV-68), and herpes simplex virus-1 (HSV-1); ref. 59]. By evaluating the phenotypic effect induced by mimics and corresponding inhibitors, the study was able to identify 4 antiviral and 3 proviral miRNAs that acted across diverse β-herpes viruses. Further analysis implicated the miRNAs in a variety of host signaling networks, including extracellular regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/AKT signaling, among others (59). Because these pathways are also implicated in influenza infection (61, 62), these findings suggest that miRNAs might have broad therapeutic potential in prospective disease intervention strategies.
CELL TYPES
As the scope of many screens is translational, the cell type chosen often has qualities consistent with evaluating druggable, pharmacokinetic, and cytotoxicity properties. As far as is practical, researchers try to use cell types that are most representative of their biology of interest (for example a lung cell line to investigate influenza infection), but full genome-wide RNAi screens with primary cells is often impractical. For this reason, most screens utilize immortalized cell lines. In general, these cells are amenable to the standard transfection/transduction procedures used in RNAi screening, but it is recognized that cell lines maintained in culture for long periods of time have significantly altered genomes, epigenomes, and transcriptomes. In truth, these differences may be one of the largest contributors to screen-to-screen variation observed across host-pathogen studies. For this reason, putative hits identified in the primary screen are often validated using counterscreens that employ alternative cell types, related viruses, or siRNAs (or shRNAs) targeting different seed sites on the same gene, or small-molecule inhibitors known to target the gene of interest. This iterative process is designed to identify candidate genes for in vivo testing and (ultimately) clinical studies.
Finally, it is worth noting that the quality assurance or quality control of cell lines is particularly important in host-pathogen screening. It is recognized that cell lines derived from a range of sources can be contaminated with pathogens, such as mycoplasma. Given the focus of host-pathogen screens and the possibility that contaminant pathogens can augment the cellular physiology, frequent testing of cell cultures over the course of screening and validation is necessary to minimize the possibility that underlying contamination alters screen outputs.
VIRUSES
Viruses grow rapidly and are capable of accumulating mutations (e.g., point mutations, deletions) in very short period of time. While DNA viruses are generally more genetically stable than RNA viruses (due to the cellular error correction mechanism for DNA), mutations in either viral class can result in defective progeny [referred to as defective interfering (DI) particles] that lack the necessary complement of genes for infection and replication. As these features can dramatically alter the output and interpretation of an RNAi screen, studies designed to identify host-pathogen interactions should use low-passage viral particles from recently isolated stocks.
In one scenario, the virus under investigation may be of serious concern to human and/or animal health, and is therefore subject to high-level biosafety containment. In this case, the virus may require enhanced containment, such as biosafety level (BSL)-3 or BSL-4 that uses biosafety cabinets, isolators, and personal protection equipment, including HEPA-filtered exhaust air. To minimize the impact of these restrictions on assay workflow, RNAi screens that utilize high-level pathogens limit the amount of work performed at the highest level of containment by performing siRNA transfections and cell handling in regular BSL-2 laboratories, and moving cells to high biocontainment for the infection phases only. Once the infection period is complete, protocols allow for virus-infected plates to be decontaminated for removal, allowing functional readouts to be performed outside containment. Such an approach has been taken to screen chemical compound libraries for inhibitors of Hendra virus (63).
In the absence of biocontainment facilities, one way to study high-risk viruses at lower biosafety levels, e.g., BSL-2 is to use pseudotype viruses. These recombinant model viruses are constructed by replacing the native envelope glycoprotein of a BSL-2 level virus (e.g., vesicular stomatitis virus) with the envelope glycoprotein of the high-risk virus of interest (e.g., Ebola). Pseudotype viruses mimic the viral entry process of the original virus and are competent for a single cycle of infection. The shortcomings associated with using pseudoviral particles are that not all envelope proteins can be incorporated in the carrier virus and their value in investigating postentry processes is limited. As such, researchers can gain valuable information regarding host-encoded viral receptors and endocytic pathways that facilitate viral entry, but subsequent steps in replication may be masked. Such viruses have been used in vaccine development for highly pathogenic H5N1 influenza virus, Ebola, and Lassa hemorrhagic fever virus as means to work around biosafety issues in working with wild-type viruses (64–66).
META-ANALYSIS OF RNAi SCREENS FOR INFLUENZA VIRUS
To date, 6 RNAi screens incorporating variable methodologies and endpoints have been performed to detect host contributions to influenza replication. Some screens focused on early replication events only (31, 33), while others included both early and late events (30, 32, 67). Furthermore, each screen examined different subsets of host genes and utilized unrelated cell models ranging from permissive cells (32, 33) to cells that influenza does not naturally infect (30, 31, 67). The 6 screens generated a list of potentially relevant host genes that represented ∼2% of the screened genes. An exception was Shapira et al. (67), which focused on genes previously implicated in a yeast 2-hybrid (Y2H) study, and therefore observed a significantly higher (35%) hit rate. As has been observed in several host-pathogen RNAi screens, the candidates identified across all of the screens had little overlap. Overall, only 3 genes were independently validated in 4 of the 6 screens, 9 genes were validated in 3 of 6 screens, and 86 genes were validated across 2 screens (Table 2). The absence of overlap in influenza screen hit lists suggests that the identity of specific cellular factors involved in the response to an influenza virus infection is context dependent and influenced by experimental factors that vary from screen to screen. This is supported by the observation that hit lists identified by Karlas et al. (32) and Konig et al. (33), both of which used A549 cells and WSN/33 virus, show the highest degree of overlap. Given that other factors, including differences in sources of media and serum, variability in the chromosomal insertion position of reporter constructs, and inconsistency in the silencing efficiency of different siRNAs collections, may all contribute to the observed disparities, a more global approach to screen analysis is required. In the context of host-pathogen RNAi screens, this has generally come in the form of meta-analysis that includes in-depth literature review and programs such as ingenuity pathway analysis (68) and others that can identify significant networks, top functions and canonical pathways associated with the different gene hits uncovered by the screen and provide biological insight into the interactions between genes, proteins, chemicals, pathways, cellular phenotypes, and disease processes (69–76). The observed pathway fidelity from such analysis suggests a conserved set of core processes that are robust to experimental variability and may be coopted during influenza virus infection and replication in a mammalian cell. Examples of a subset of processes identified by these means include the following host cell signaling pathways:
Table 2.
Results from host-pathogen RNAi screens
| Screen | Cell line | Influenza virus | Readout | Genes screened | Validated hits | Validation | Reference |
|---|---|---|---|---|---|---|---|
| siRNA screen (Ambion, Austin, TX, USA) | Drosophilad-Mel2 | Recombinant A/WSN/33 | Luciferase activity | 13,071 | 121 (110↓; 11↑) | Decreased luciferase expression in 2 replicates, inhibiting ≥mean ± 2.5 sd in ≥1 replicate, and phenotype consistent when targeted with an alternate dsRNA amplicon | 31 |
| siRNA screen (Dharmacon, Lafayette, CO, USA) | Human U2OS | A/PR/8/34 H1N1 | HA immunostain | 17,877 | 260 (250↓; 10↑) | Rescreen with individual siRNAs from pool | 30 |
| Virus-host direct interactions (Y2H), transcriptional responses (microarray), and pathway association (IPA) | Human HBEC | A/PR/8/34, ΔNS1,a or vRNA | Infectious virus or IFNβ production | 1,745 | 616 | siRNA to candidate gene affected the phenotype in ≥1 of 3 functional assays | 67 |
| siRNA screen (Qiagen, Valencia, CA, USA; Invitrogen, Carlsbad, USA; IDT, Coralville, IA, USA) | Human A549 | Recombinant A/WSN/33, SOIV A/NL/602/09 | Luciferase activity | 19,628 | 295 (295 WSN, 12 SOIV) | ≥2 unique siRNAs to candidate gene reduced viral infection ≥35% | 33 |
| siRNA screen (Qiagen) | Human A549 | A/WSN/33, SOIV A/Hamburg/04, HPAI A/VN/1203/04 | Infectious virus quantified using a 293T cell reporter system and NP immunostain; viral replication measured by titrating A549 supernatant on MDCK cells | 22,843 | 168 (119 WSN, 121 SOIV, 6 HPAI) | ≥2 unique siRNAs to candidate gene decreased virus replication >5-fold | 32 |
| siRNA screen (Dharmacon) | Human A549 | A/WSN/33 | Amount of infectious virus, NP expression, M gene levels | 1,201 | 28 (25↓; 3↑) | Phenotype is emulated using a novel siRNA targeting a different seed region of the hit gene | Unpublished results |
↓, hits that decreased virus replication; ↑, hits that increased virus replication; HA, hemagglutinin; NA, neuraminidase; VSV-G, vesicular stomatitis virus glycoprotein G; eGFP, enhanced green fluorescence protein; HBEC, human bronchial epithelial cells; NP, influenza virus nucleoprotein; M, influenza virus matrix protein; Y2H, yeast 2-hybrid; IPA, ingenuity pathway analysis; vRNA, viral RNA; IFN, interferon; SOIV, swine-origin influenza virus; HPAI, highly pathogenic avian-origin influenza virus; MDCK, Madin-Darby canine kidney.
ΔNS1, PR8 virus lacking the nonstructural gene.
RECEPTOR TYROSINE KINASE (RTK) SIGNALING
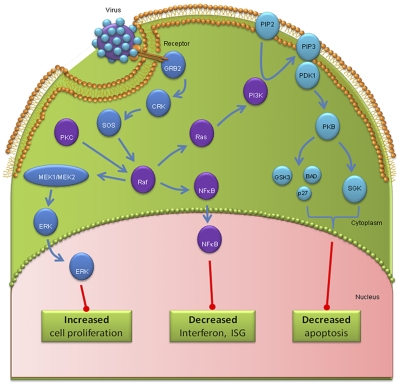
Within the host cell, a major mechanism that transmits extracellular signals to intracellular signaling is the engagement of RTKs (Fig. 2). Among the family of RTKs are the group of epidermal growth factor receptors (EGFRs), consisting of four members (EGF, ErbB2, ErbB3, and ErbB4 receptors; ref. 77). ErbB expression has been associated with a multitude of cellular functions and responses, including proliferation, cell migration, differentiation, and apoptosis (78–81). Cellular endocytosis of influenza coopts pathways used by EGFRs, resulting in protein ubiquitinylation and sorting into the vacuolar pathway (82). In addition, influenza virus particles are sorted into the same population of late endosomes as EGFRs (83). Specific inhibition of tyrosine kinases by small-molecule inhibitors, as well as specific EGFR inhibition via RNAi, reduces virus uptake and subsequent virus titers (84). Furthermore, attachment of influenza virus to the host cell causes clustering of plasma membrane lipids, similar to that seen following EGF stimulation. Therefore, on influenza virus binding to host cell sialic acids, it is able to cluster and activate EGFRs and other RTKs to form a lipid raft-based signaling platform (84). This leads to receptor-mediated signaling events, which enhance influenza virus uptake and subsequent viral replication. It is thought that this activation is not mediated by viral engagement of a particular receptor kinase but is a more general phenomenon that affects several RTKs (84). This is additionally supported by results of a recent siRNA screening study, which identified the involvement of fibroblast growth factor receptors FGFR 1, 2, and 4 as RTKs in the very early steps of viral infection (33). Therefore, influenza virus entry accompanied by down-regulation of signaling receptors promotes coendocytosis of the virus into the host cell.
Figure 2.
Host signaling factors. On binding to host cell sialic acids, influenza virus activates RTKs to form lipid raft-based signaling platforms that enhance influenza uptake and cell proliferation. HA-mediated activation of protein kinase C (PKC) also aids viral entry and cellular proliferation. RTKs activate phosphatidylinositol 3-kinase (PI3K) signaling, inhibiting cellular apoptosis. The Raf/MEK/ERK cascade downstream of RTKs is required for efficient nuclear export of viral ribonucleoproteins. Viral manipulation of NF-κB inhibits antiviral type I interferon production and resultant interferon-stimulated gene (ISG) production.
PROTEIN KINASE C (PKC) SIGNALING
PKC belongs to large family of serine/threonine kinases involved in a multitude of physiological processes (85). PKC plays an integral role in sodium ion transport, important for maintaining the low pH in the endosome (86–89). PKC has also been shown to be critical for the entry of enveloped viruses via receptor-mediated endocytosis (90). Upon influenza virus infection, the HA rapidly activates PKC (87, 90), and it has been shown that a specific inhibitor of PKC prevents influenza virus replication by inhibiting the entry of the virus. Similarly, influenza virus replication has also been reported in cells expressing a phosphorylation-deficient form of PKC (91).
PI3K SIGNALING
The family of PI3Ks regulates various cellular events, such as cell metabolism, proliferation, and survival (92, 93). The major function of the PI3K is to phosphorylate membrane phospholipids. On PI3K activation, phosphatidylinositol-3,4,5-triphosphate (PIP3) is generated by phosphorylation of phosphatidylinositol-4,5-bisphosphate, which functions as a second messenger through interaction with pleckstrin homology domain-containing proteins such as Akt/PKB and phosphoinositide-dependent kinase (PDK)-1 (92). Cells treated with inhibitors of PI3K or PIP3 show significantly decreased influenza virus titers (94), suggesting that PI3K performs influenza-supportive functions.
Raf/MEK/ERK SIGNALING
Infection with influenza virus leads to activation of a variety of different MAPK cascades (95–98). They are activated by a variety of extracellular stimuli, such as growths factors, cytokines, and environmental stress factors like osmotic stress or ultraviolet light. Downstream substrates for MAPKs are transcription factors (e.g., ATF2, ELK-1, or c-Jun), and other protein kinases, such as the MAP kinase-activated protein kinases MK2 and MK3. Thereby, MAPK pathways regulate a variety of cellular responses such as gene expression, proliferation, differentiation, apoptosis, and immune responses (62, 99, 100).
Influenza ribonucleoprotein (RNP) formation and nuclear export are important steps in the life cycle of influenza virus, and data indicate that Raf/MEK/ERK cascade is required for an efficient nuclear RNP export, as indicated by several studies (98, 101). Inhibition of Raf signaling results in nuclear retention of viral RNP and the concomitant inhibition of virus production (98). Influenza virus HA membrane accumulation and its tight association with lipid raft domain trigger the activation of MAPK cascades via PKC-α activation and RNP export (102). HA membrane accumulation is enhanced by the higher polymerase activity of influenza virus, resulting in up-regulation of the MAPK cascade and more efficient nuclear RNP-export, along with virus production (103). In addition, p38 MAPK and JNK have been shown to regulate the expression of proinflammatory cytokines in influenza virus-infected cells (95, 104–106).
NUCLEAR FACTOR κB (NF-κB) SIGNALING
An important influenza virus-induced signaling mediator is the transcription factor NF-κB. This factor regulates expression of a variety of antiviral cytokines, including IFN-β, which is the initiator of a strong type I IFN defense program (107). Although NF-κB is generally regarded as a central factor in the innate immune defense (108), two independent studies demonstrated that replication of influenza viruses were impaired rather than enhanced in cells where this pathway was blocked (109, 110). NF-κB acts via induction of proapoptotic factors, such as TNF-related apoptosis-inducing ligand (TRAIL) or FasL (109), and subsequent activation of caspases (111). This results in an enhanced export of viral RNPs from the nucleus, presumably by specific cleavage of nuclear pore proteins, resulting in an enhanced diffusion of the RNP through the pores (112, 113). Lastly, NF-κB differentially regulates viral RNA synthesis (10). Each of these mechanisms may contribute to a different extent to the enhancing effect of NF-κB on virus propagation, thereby identifying the factor as a potential target for antiviral intervention. Besides the direct antiviral action, NF-κB inhibition may also indirectly influence and, in fact, exacerbate the pathogenesis of influenza virus, since the majority of cytokines/chemokines induced during infection with highly pathogenic viruses are regulated by NF-κB (107).
To summarize, the host cell pathway overlap identified among RNAi screens for influenza virus (Table 2) indicates that core cell signaling processes are coopted by influenza virus. Variability in specific gene hit lists from different screens is likely explained by variations in the efficacy of knockdown of specific gene targets by different RNAi libraries, variation in the timing of individual screen assays, and other factors related to the assays.
CHALLENGES OF RNAi SCREENING
While RNAi screening has demonstrated enormous potential in improving our understanding of host-pathogen interactions, several challenges remain. As just described, one issue focuses on the lack of overlap associated with hits identified across different screens. Differences in viral subtypes, host cells, assay types, and reagents may be responsible for some of the differences, but, to date, there has been no definitive study to address these discrepancies. Several practices could offset the challenges associated with disparate hit lists. One would be to follow the direction of researchers in the microarray field who have developed a consistent set of minimum information standards complemented with a centralized, publicly accessible portal for data submission and review (114, 115). Minimum information about an RNAi experiment (MIARE; http://miare.sourceforge.net/) is a standards concept for RNAi screening that advocates all published screening data include extensive experimental details regarding target cell sources, delivery methods, assay design, plate layout, reagent composition, controls, and metrics of data analysis (116). In 2008, a database was established for submission of MIARE-compliant RNAi screens. While the ongoing development of the MIARE standards and the codevelopment of the PubMed database for RNAi screens represents important progress, community-wide adoption of MIARE standards and submission policies has not yet been achieved. A revitalized collaborative effort by academic, government, and industrial partners would greatly accelerate the acceptance and implementation of MIARE and thereby facilitate the community's ability to address questions regarding screen reproducibility.
A second practice that could address the issue of disparate hit lists pertains to efforts in hit validation. In cases in which siRNA pools are used in the primary screen, deconvolution of the pool and identification of instances in which ≥2 individual siRNAs from a single pool induce the same phenotype have been used in validation (30). In other cases, validation employs the use of completely different sets of siRNAs targeting a different seed site on the same gene (32, 33), the use shRNAs (28), multiple screening assays (67), multiple viral strains (32, 33, 67), bioinformatics (29), and completely different gene-silencing technologies, for example small molecules (117). Whether hits validated by each of these procedures should be considered equal is open to debate; however, adopting a standard validation method may bring greater parity between screens.
Related to techniques employed for hit validation are the criteria used to identify a primary hit. Most screens to date have defined a hit based on a plate mean, or standard deviation from the mean, or relative to a negative control (118). However, in a recent study where 15 separate parameters were used to compile a hit signature, there was no improvement in the concordance with previous screens or increase in validation of the hits during secondary experiments (119, 120).
COMPARING RNAi SCREENING TO ANALOGOUS TECHNOLOGIES
Because a major goal of RNAi host-pathogen screens is to identify host genes and pathways that facilitate viral replication, it is important to note other analogous technologies that have been used to address this question, such as compound screens, microarrays, and NGS and Y2H screens.
Compound library screens
Small-molecule inhibitor and drug compound library screens involve high-throughput analysis of potential pharmacological agents that modulate biological pathways and sometimes specific host genes. A major advantage of compound library screens is the large number of candidate compounds, sometimes in the millions, which come from bioactive, commercial, academic and natural extract libraries, which can be screened for antiviral potential, often without the need for delivery reagents. Although a major limitation is often the lack of information that compound screens provide on the cellular target and mechanisms linked to antiviral activity, advances in screening technology platforms can compensate. For example, high-content analysis methods that use images of living cells as the basic unit for molecule discovery can track, quantitate, and provide qualitative information of the proteins of interest present in the cells using fluorescent tags, such as the green fluorescent protein, or by fluorescent antibodies. Image analysis is then used to measure changes in properties of the cells caused by treatment with candidate compounds, such as chemical inhibitors or RNA interference. However, it is important to note that compound library screens are often complicated by the need to perform analyses at multiple compound concentrations to address toxicity concerns, as cell death could be mistakenly identified as efficacy. Other problems associated with compound and small-molecule library screens include poor solubility of drugs, poor and/or variable cellular uptake, and drug specificity issues (121). Despite these concerns, a number of compound library screens have been conducted on Madin-Darby canine kidney (MDCK) cells to identify influenza inhibitors, and these results have led to follow-up validation studies in human cells (121, 122).
Microarrays and NGS
Microarrays have been used extensively to study host pathways implicated in virus life cycles and to inform our understanding of viral pathogenesis (123). In the first reported microarray study of host-influenza virus interaction, the expression levels of over 4600 genes were measured in HeLa cells in response to live or heat-inactivated influenza A/WSN/33 (H1N1) exposure (124). Of the 329 differentially expressed genes identified within 8 h of influenza infection, the majority were down-regulated and could be classified into 5 main groups—protein synthesis, cytokine signaling, ubiquitin pathway factors, mRNA processing, export proteins, and transcription factors. Broad comparisons of the gene families and pathways show a reasonable level of complementarity with those identified more recently by RNAi screening. Experiments performed using microarray technology have identified a number of host pathways that are impacted by a range of viruses, including HIV (125), human papillomavirus (126), and herpes simplex virus (127), and by treatments with IFNs -α, -β, and -γ (128). These studies have provided valuable information regarding host pathways and have provided new candidates for antiviral therapies.
Despite the large amount of microarray data and infrastructure available to researchers, it appears that NGS may supersede this technology due mainly to improvements in transcriptome coverage, sensitivity, and resolution. Since microarrays are limited by the genome coverage of their probe sets, NGS has the advantage of identifying expression changes of poorly characterized transcripts, particularly those from the small noncoding RNA families. NGS-generated genome-wide expression profiles of host cells are emerging for a number of viruses, including Mimivirus (129), vaccinia virus (130), and HIV (131). These studies provide a global view of how the host genome is impacted by viral infection, in addition to changes in virus gene expression levels during infection. NGS analyses of host cells in response to influenza infection are eagerly anticipated. An important distinction is the information based on loss-of-function resulting from RNAi-mediated gene silencing. Microarray and sequencing approaches, while providing detailed readouts on fold changes in gene expression, may not determine the importance of specific genes in biological processes, and may ignore important genes and pathways if low level changes in expression are observed following infection.
Y2H screens
Y2H technology allows screening of protein-protein and protein-DNA interactions (132). Y2H screens have identified a protein-protein “interactome” network for Epstein-Barr virus (133), SARS-like coronavirus (134), and hepatitis C virus (135) infections. A comprehensive study has recently been conducted in which intraviral protein-protein interactions were analyzed by Y2H technology for 10 influenza virus (A/PR/8/34) proteins, in addition to interactions between these viral proteins with 12,000 human proteins from the Human ORFeome (67). While the influenza NS1 gene segment is known to interact with host pathways to disrupt antiviral actions (reviewed in ref. 17), numerous novel contacts of host proteins with PB1, PB2, and NP influenza gene segments were identified. This study elegantly combined Y2H and RNAi technology to identify host proteins and pathways involved in physical recognition of influenza proteins and the resultant pathways mediating antiviral responses.
CONCLUSIONS
The importance of influenza virus to global human and animal health, coupled with the current limitations associated with vaccine production and antiviral therapeutic pipelines, underscores the need for novel solutions to accelerate the development of virus intervention strategies. Genome-wide RNAi screening is an emerging technology with the power to detail host-virus interactions, furthering our understanding of virus pathogenesis and thereby driving the development of next-generation antivirals. On the basis of the findings from the meta-analyses of influenza screens (31, 67, 136–138), it is likely that influenza virus principally coopts host genes late during virus replication. Succinctly, influenza virus carries all necessary proteins to infect a cell and deliver its vRNP into the cell nucleus. However, viral proteins are synthesized in the cytoplasm; thus, the virus must use host proteins and pathways to import NP and RNA polymerases back into the nucleus to form new vRNP. Likewise, the assembly of influenza components occurs at the plasma membrane, requiring host mechanisms to move new vRNP from the nucleus into the cytoplasm, and then traffic the viral protein components toward the membrane for assembly and packaging of new virions. This hypothesis is consistent with the subtle differences that have been observed in viral genomic expression relative to the huge differences observed in high-content analysis and in virus replication (33, 136). In addition, in the meta-analyses studies (31, 67, 136–138), host factors linked with regulating cell death are affected by influenza virus, a feature likely required to facilitate replication.
Given the scale of information generated by RNAi screens, it is critically important and beneficial to collaborate in this research arena. There are admirable examples of how scientific consortia and centers composed of dispersed networks of laboratories have successfully worked together to achieve significant goals. The U.S. National Institute of Allergy and Infectious Diseases Centers for Excellence in Influenza Research and Surveillance and the Human Genome Project provide two examples where intervention strategies are tackled by academic, government, and industrial entities worldwide and represent good examples as to how disparate entities can work in partnership toward a common goal. Similarly, the efforts by organizations such as the RNAi Global Initiative are designed to tackle the challenges represented by EIDs, such as influenza, to benefit human health. It will be vital to the success of this emerging field that such collaborative initiatives promote the development of data standards and best practices that facilitate the comparison of data from complementary screens. This should lead to a better understanding of the factors that most strongly influence screen reproducibility and data quality, and ultimately improve the potential of RNAi technology to identify optimal antiviral-drug candidates.
Footnotes
- BSL
- biosafety level
- CMV
- cytomegalovirus
- dsRNA
- double-stranded RNA
- EGFR
- epidermal growth factor receptor
- EID
- emerging infectious disease
- ERK
- extracellular regulated kinase
- HA
- hemagglutinin
- MAPK
- mitogen-activated protein kinase
- MIARE
- minimum information about an RNAi experiment
- miRNA
- microRNA
- NF-κB
- nuclear factor-κB
- NGS
- next-generation sequencing
- PI3K
- phosphatidylinositol 3-kinase
- PKC
- protein kinase C
- RISC
- RNA-induced silencing complex
- RNAi
- RNA interference
- RNP
- ribonucleoprotein
- RTK
- receptor tyrosine kinase
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- TLR
- Toll-like receptor
- Y2H
- yeast 2-hybrid.
REFERENCES
- 1. Daszak P., Cunningham A. A., Hyatt A. D. (2000) Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science 287, 443–449 [DOI] [PubMed] [Google Scholar]
- 2. Taubenberger J. K., Morens D. M. (2010) Influenza: the once and future pandemic. Public Health Rep. 125(Suppl. 3), 16–26 [PMC free article] [PubMed] [Google Scholar]
- 3. Komar N., Olsen B. (2008) Avian influenza virus (H5N1) mortality surveillance. Emerg. Infect. Dis. 14, 1176–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization (2009) Overview of pandemic (H1N1) 2009 in the WHO European Region. Retrieved March 11, 2010, from http://www.euro.who.int/influenza/AH1N1/20091026_1
- 5. World Health Organization (2009) Seasonal influenza. Retrieved March 10, 2010, from http://www.euro.who.int/influenza/20080618_1
- 6. Molinari N.-A. M., Ortega-Sanchez I. R., Messonnier M. L., Thompson W. W., Wortley P. M., Weintraub E., Bridges C. B. (2007) The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 [DOI] [PubMed] [Google Scholar]
- 7. Nagata K., Kawaguchi A., Naito T. (2008) Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18, 247–260 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Z., Xue Q., Wan Y., Yang Y., Wang J., Hung T. (2011) Lysosome-associated membrane glycoprotein 3 is involved in influenza A virus replication in human lung epithelial (A549) cells. Virol. J. 8, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su B., Wurtzer S., Rameix-Welti M. A., Dwyer D., van der Werf S., Naffakh N., Clavel F., Labrosse B. (2009) Enhancement of the influenza A hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA). PloS One 4, e8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar N., Xin Z. T., Liang Y., Ly H. (2008) NF-κB signaling differentially regulates influenza virus RNA synthesis. J. Virol. 82, 9880–9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruce E. A., Digard P., Stuart A. D. (2010) The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 84, 5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rossman J. S., Lamb R. A. (2011) Influenza virus assembly and budding. Virology 411, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leung R. K., Whittaker P. A. (2005) RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol. Ther. 107, 222–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tripp R. A., Tompkins S. M. (2009) Therapeutic applications of RNAi for silencing virus replication. Methods Mol. Biol. 555, 43–61 [DOI] [PubMed] [Google Scholar]
- 15. Bagasra O., Prilliman K. R. (2004) RNA interference: the molecular immune system. J. Mol. Histol. 35, 545–553 [DOI] [PubMed] [Google Scholar]
- 16. Blair C. D. (2011) Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 6, 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hale B. G., Randall R. E., Ortin J., Jackson D. (2008) The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89, 2359–2376 [DOI] [PubMed] [Google Scholar]
- 18. Kim D. H., Rossi J. J. (2007) Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 8, 173–184 [DOI] [PubMed] [Google Scholar]
- 19. Kim V. N., Han J., Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 20. Held K., Junker A., Dornmair K., Meinl E., Sinicina I., Brandt T., Theil D., Derfuss T. (2011) Expression of herpes simplex virus 1-encoded microRNAs in human trigeminal ganglia and their relation to local T-cell infiltrates. J. Virol. 85, 9680–9685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tirabassi R., Hook L., Landais I., Grey F., Meyers H., Hewitt H., Nelson J. (2011) Human cytomegalovirus US7 is regulated synergistically by two virally-encoded miRNAs and by two distinct mechanisms. J. Virol. 85, 11938–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfeffer S., Zavolan M., Grasser F. A., Chien M., Russo J. J., Ju J., John B., Enright A. J., Marks D., Sander C., Tuschl T. (2004) Identification of virus-encoded microRNAs. Science 304, 734–736 [DOI] [PubMed] [Google Scholar]
- 23. Hughes R. A., Miklos A. E., Ellington A. D. (2011) Gene synthesis methods and applications. Methods Enzymol. 498, 277–309 [DOI] [PubMed] [Google Scholar]
- 24. Zhang W., Tripp R. A. (2008) RNA interference inhibits respiratory syncytial virus replication and disease pathogenesis without inhibiting priming of the memory immune response. J. Virol. 82, 12221–12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakurai K., Chomchan P., Rossi J. J. (2010) Silencing of gene expression in cultured cells using small interfering RNAs. Curr. Protoc. Cell Biol. 47, 27.1.1–27.1.28 [DOI] [PubMed] [Google Scholar]
- 26. Snead N. M., Rossi J. J. (2010) Biogenesis and function of endogenous and exogenous siRNAs. Wiley Interdiscip. Rev. RNA 1, 117–131 [DOI] [PubMed] [Google Scholar]
- 27. Blakely K., Ketela T., Moffat J. (2011) Pooled lentiviral shRNA screening for functional genomics in mammalian cells. Methods Mol. Biol. 781, 161–182 [DOI] [PubMed] [Google Scholar]
- 28. Gobeil S., Zhu X., Doillon C. J., Green M. R. (2008) A genome-wide shRNA screen identifies GAS1 as a novel melanoma metastasis suppressor gene. Genes Dev. 22, 2932–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh M. H., Tsai T. C., Kuo H. P., Chang N. W., Lee M. R., Chung J. G., Tsai M. H., Liu J. Y., Kao M. C. (2011) Lentiviral short hairpin RNA screen of human kinases and phosphatases to identify potential biomarkers in oral squamous cancer cells. Intl. J. Oncol. 39, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 30. Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C. A., Newton M. A., Ahlquist P., Kawaoka Y. (2008) Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., Maurer A. P., Muller E., Wolff T., Rudel T., Meyer T. F. (2010) Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 [DOI] [PubMed] [Google Scholar]
- 33. Konig R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Gao Q., Andrews S. E., Bandyopadhyay S., De Jesus P., Tu B. P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., Garcia-Sastre A., Young J. A., Palese P., Shaw M. L., Chanda S. K. (2009) Human host factors required for influenza virus replication. Nature 463, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kisielow M., Kleiner S., Nagasawa M., Faisal A., Nagamine Y. (2002) Isoform-specific knockdown and expression of adaptor protein ShcA using small interfering RNA. Biochem. J. 363, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holen T., Amarzguioui M., Wiiger M. T., Babaie E., Prydz H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 30, 1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Birmingham A., Selfors L. M., Forster T., Wrobel D., Kennedy C. J., Shanks E., Santoyo-Lopez J., Dunican D. J., Long A., Kelleher D., Smith Q., Beijersbergen R. L., Ghazal P., Shamu C. E. (2009) Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6, 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu S., Metcalf J. P., Wu W. (2011) Innate immune response to influenza virus. Curr. Opin. Infect. Dis. 24, 235–240 [DOI] [PubMed] [Google Scholar]
- 38. Barchet W., Krug A., Cella M., Newby C., Fischer J. A., Dzionek A., Pekosz A., Colonna M. (2005) Dendritic cells respond to influenza virus through TLR7- and PKR-independent pathways. Eur. J. Immunol. 35, 236–242 [DOI] [PubMed] [Google Scholar]
- 39. Lund J. M., Alexopoulou L., Sato A., Karow M., Adams N. C., Gale N. W., Iwasaki A., Flavell R. A. (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 101, 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hayden F. G., Fritz R., Lobo M. C., Alvord W., Strober W., Straus S. E. (1998) Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J. Clin. Invest. 101, 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim D. H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J. J. (2004) Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 22, 321–325 [DOI] [PubMed] [Google Scholar]
- 42. Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., Endres S., Hartmann G. (2005) Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 11, 263–270 [DOI] [PubMed] [Google Scholar]
- 43. Sioud M. (2005) Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 348, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 44. Reynolds A., Anderson E. M., Vermeulen A., Fedorov Y., Robinson K., Leake D., Karpilow J., Marshall W. S., Khvorova A. (2006) Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA 12, 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robbins M., Judge A., Liang L., McClintock K., Yaworski E., MacLachlan I. (2007) 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 15, 1663–1669 [DOI] [PubMed] [Google Scholar]
- 46. Sioud M., Furset G., Cekaite L. (2007) Suppression of immunostimulatory siRNA-driven innate immune activation by 2′-modified RNAs. Biochem. Biophys. Res. Commun. 361, 122–126 [DOI] [PubMed] [Google Scholar]
- 47. Eberle F., Giessler K., Deck C., Heeg K., Peter M., Richert C., Dalpke A. H. (2008) Modifications in small interfering RNA that separate immunostimulation from RNA interference. J. Immunol. 180, 3229–3237 [DOI] [PubMed] [Google Scholar]
- 48. Hirsch A. J. (2010) The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol. 5, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sigoillot F. D., King R. W. (2011) Vigilance and validation: Keys to success in RNAi screening. ACS Chem. Biol. 6, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jackson A. L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J. M., Lim L., Karpilow J., Nichols K., Marshall W., Khvorova A., Linsley P. S. (2006) Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12, 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu J. Q., Dwyer D. E., Dyer W. B., Yang Y. H., Wang B., Saksena N. K. (2011) Genome-wide analysis of primary CD4+ and CD8+ T-cell transcriptomes shows evidence for a network of enriched pathways associated with HIV disease. Retrovirology 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee R. C., Feinbaum R. L., Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 53. Lee R. C., Ambros V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 [DOI] [PubMed] [Google Scholar]
- 54. Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 [DOI] [PubMed] [Google Scholar]
- 55. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001) Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 [DOI] [PubMed] [Google Scholar]
- 56. Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Griffiths-Jones S. (2004) The microRNA registry. Nucleic Acids Res. 32, D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581 [DOI] [PubMed] [Google Scholar]
- 59. Santhakumar D., Forster T., Laqtom N. N., Fragkoudis R., Dickinson P., Abreu-Goodger C., Manakov S. A., Choudhury N. R., Griffiths S. J., Vermeulen A., Enright A. J., Dutia B., Kohl A., Ghazal P., Buck A. H. (2010) Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc. Natl. Acad. Sci. U. S. A. 107, 13830–13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vermeulen A., Robertson B., Dalby A. B., Marshall W. S., Karpilow J., Leake D., Khvorova A., Baskerville S. (2007) Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA 13, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ehrhardt C., Wolff T., Pleschka S., Planz O., Beermann W., Bode J. G., Schmolke M., Ludwig S. (2007) Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81, 3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pleschka S. (2008) RNA viruses and the mitogenic Raf/MEK/ERK signal transduction cascade. Biol. Chem. 389, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 63. Porotto M., Orefice G., Yokoyama C. C., Mungall B. A., Realubit R., Sganga M. L., Aljofan M., Whitt M., Glickman F., Moscona A. (2009) Simulating henipavirus multicycle replication in a screening assay leads to identification of a promising candidate for therapy. J. Virol. 83, 5148–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bredenbeek P. J., Molenkamp R., Spaan W. J., Deubel V., Marianneau P., Salvato M. S., Moshkoff D., Zapata J., Tikhonov I., Patterson J., Carrion R., Ticer A., Brasky K., Lukashevich I. S. (2006) A recombinant Yellow Fever 17D vaccine expressing Lassa virus glycoproteins. Virology 345, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Geisbert T. W., Bailey M., Hensley L., Asiedu C., Geisbert J., Stanley D., Honko A., Johnson J., Mulangu S., Pau M. G., Custers J., Vellinga J., Hendriks J., Jahrling P., Roederer M., Goudsmit J., Koup R., Sullivan N. J. (2011) Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge. J. Virol. 85, 4222–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang W., Xue T., Wu X., Zhang P., Zhao G., Peng D., Hu S., Wang X., Liu X., Liu W. (2011) Increase in viral yield in eggs and MDCK cells of reassortant H5N1 vaccine candidate viruses caused by insertion of 38 amino acids into the NA stalk. Vaccine 29, 8032–8041 [DOI] [PubMed] [Google Scholar]
- 67. Shapira S. D., Gat-Viks I., Shum B. O., Dricot A., de Grace M. M., Wu L., Gupta P. B., Hao T., Silver S. J., Root D. E., Hill D. E., Regev A., Hacohen N. (2009) A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ganter B., Giroux C. N. (2008) Emerging applications of network and pathway analysis in drug discovery and development. Curr. Opin. Drug Disc. Dev. 11, 86–94 [PubMed] [Google Scholar]
- 69. Friedman A. A., Tucker G., Singh R., Yan D., Vinayagam A., Hu Y., Binari R., Hong P., Sun X., Porto M., Pacifico S., Murali T., Finley R. L., Jr., Asara J. M., Berger B., Perrimon N. (2011) Proteomic and functional genomic landscape of receptor tyrosine kinase and ras to extracellular signal-regulated kinase signaling. Sci. Signal. 4, rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yore M. M., Kettenbach A. N., Sporn M. B., Gerber S. A., Liby K. T. (2011) Proteomic analysis shows synthetic oleanane triterpenoid binds to mTOR. PloS One 6, e22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ruan L., Li X. H., Wan X. X., Yi H., Li C., Li M. Y., Zhang P. F., Zeng G. Q., Qu J. Q., He Q. Y., Li J. H., Chen Y., Chen Z. C., Xiao Z. Q. (2011) Analysis of EGFR signaling pathway in nasopharyngeal carcinoma cells by quantitative phosphoproteomics. Proteome Sci. 9, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Satoh J., Tabunoki H. (2011) Comprehensive analysis of human microRNA target networks. BioData Mining 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Glaab E., Baudot A., Krasnogor N., Valencia A. (2010) Extending pathways and processes using molecular interaction networks to analyse cancer genome data. BMC Bioinformatics 11, 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jianu R., Yu K., Cao L., Nguyen V., Salomon A. R., Laidlaw D. H. (2010) Visual integration of quantitative proteomic data, pathways, and protein interactions. IEEE Trans. Vis. Comput. Graphics 16, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bankhead A., 3rd, Sach I., Ni C., LeMeur N., Kruger M., Ferrer M., Gentleman R., Rohl C. (2009) Knowledge-based identification of essential signaling from genome-scale siRNA experiments. BMC Systems Biol. 3, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Fraser I. D., Germain R. N. (2009) Navigating the network: signaling cross-talk in hematopoietic cells. Nat. Immunol. 10, 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Prenzel N., Fischer O. M., Streit S., Hart S., Ullrich A. (2001) The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr. Relat. Cancer 8, 11–31 [DOI] [PubMed] [Google Scholar]
- 78. Weiss F. U., Wallasch C., Campiglio M., Issing W., Ullrich A. (1997) Distinct characteristics of heregulin signals mediated by HER3 or HER4. J. Cell. Physiol. 173, 187–195 [DOI] [PubMed] [Google Scholar]
- 79. Jones F. E., Welte T., Fu X. Y., Stern D. F. (1999) ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 147, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kainulainen V., Sundvall M., Maatta J. A., Santiestevan E., Klagsbrun M., Elenius K. (2000) A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J. Biol. Chem. 275, 8641–8649 [DOI] [PubMed] [Google Scholar]
- 81. Long W., Wagner K. U., Lloyd K. C., Binart N., Shillingford J. M., Hennighausen L., Jones F. E. (2003) Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130, 5257–5268 [DOI] [PubMed] [Google Scholar]
- 82. Khor R., McElroy L. J., Whittaker G. R. (2003) The ubiquitin-vacuolar protein sorting system is selectively required during entry of influenza virus into host cells. Traffic 4, 857–868 [DOI] [PubMed] [Google Scholar]
- 83. Lakadamyali M., Rust M. J., Zhuang X. (2006) Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Eierhoff T., Hrincius E. R., Rescher U., Ludwig S., Ehrhardt C. (2010) The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 6, e1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Toker A. (1998) Signaling through protein kinase C. Front. Biosci. 3, D1134–D1147 [DOI] [PubMed] [Google Scholar]
- 86. Hoffmann H. H., Palese P., Shaw M. L. (2008) Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antiviral Res. 80, 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kunzelmann K., Beesley A. H., King N. J., Karupiah G., Young J. A., Cook D. I. (2000) Influenza virus inhibits amiloride-sensitive Na+ channels in respiratory epithelia. Proc. Natl. Acad. Sci. U. S. A. 97, 10282–10287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lazrak A., Iles K. E., Liu G., Noah D. L., Noah J. W., Matalon S. (2009) Influenza virus M2 protein inhibits epithelial sodium channels by increasing reactive oxygen species. FASEB J. 23, 3829–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sieczkarski S. B., Whittaker G. R. (2005) Characterization of the host cell entry of filamentous influenza virus. Arch. Virol. 150, 1783–1796 [DOI] [PubMed] [Google Scholar]
- 90. Constantinescu S. N., Cernescu C. D., Popescu L. M. (1991) Effects of protein kinase C inhibitors on viral entry and infectivity. FEBS Lett. 292, 31–33 [DOI] [PubMed] [Google Scholar]
- 91. Root C. N., Wills E. G., McNair L. L., Whittaker G. R. (2000) Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J. Gen. Virol. 81, 2697–2705 [DOI] [PubMed] [Google Scholar]
- 92. Neri L. M., Borgatti P., Capitani S., Martelli A. M. (2002) The nuclear phosphoinositide 3-kinase/AKT pathway: a new second messenger system. Biochim. Biophys. Acta 1584, 73–80 [DOI] [PubMed] [Google Scholar]
- 93. Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L. C. (2005) Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem. Sci. 30, 194–204 [DOI] [PubMed] [Google Scholar]
- 94. Ehrhardt C., Marjuki H., Wolff T., Nurnberg B., Planz O., Pleschka S., Ludwig S. (2006) Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 8, 1336–1348 [DOI] [PubMed] [Google Scholar]
- 95. Kujime K., Hashimoto S., Gon Y., Shimizu K., Horie T. (2000) p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 164, 3222–3228 [DOI] [PubMed] [Google Scholar]
- 96. Ludwig S., Pleschka S., Planz O., Wolff T. (2006) Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell. Microbiol. 8, 375–386 [DOI] [PubMed] [Google Scholar]
- 97. Ludwig S., Planz O., Pleschka S., Wolff T. (2003) Influenza-virus-induced signaling cascades: targets for antiviral therapy? Trends Mol. Med. 9, 46–52 [DOI] [PubMed] [Google Scholar]
- 98. Pleschka S., Wolff T., Ehrhardt C., Hobom G., Planz O., Rapp U. R., Ludwig S. (2001) Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3, 301–305 [DOI] [PubMed] [Google Scholar]
- 99. Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 100. Dong C., Davis R. J., Flavell R. A. (2002) MAP kinases in the immune response. Annu. Rev. Immunol. 20, 55–72 [DOI] [PubMed] [Google Scholar]
- 101. Ludwig S., Wolff T., Ehrhardt C., Wurzer W. J., Reinhardt J., Planz O., Pleschka S. (2004) MEK inhibition impairs influenza B virus propagation without emergence of resistant variants. FEBS Lett. 561, 37–43 [DOI] [PubMed] [Google Scholar]
- 102. Marjuki H., Alam M. I., Ehrhardt C., Wagner R., Planz O., Klenk H. D., Ludwig S., Pleschka S. (2006) Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J. Biol. Chem. 281, 16707–16715 [DOI] [PubMed] [Google Scholar]
- 103. Marjuki H., Yen H. L., Franks J., Webster R. G., Pleschka S., Hoffmann E. (2007) Higher polymerase activity of a human influenza virus enhances activation of the hemagglutinin-induced Raf/MEK/ERK signal cascade. Virol. J. 4, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Guillot L., Le Goffic R., Bloch S., Escriou N., Akira S., Chignard M., Si-Tahar M. (2005) Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J. Biol. Chem. 280, 5571–5580 [DOI] [PubMed] [Google Scholar]
- 105. Karin M., Liu Z., Zandi E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 106. Ludwig S., Wang X., Ehrhardt C., Zheng H., Donelan N., Planz O., Pleschka S., Garcia-Sastre A., Heins G., Wolff T. (2002) The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76, 11166–11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pahl H. L. (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 108. Chu W. M., Ostertag D., Li Z. W., Chang L., Chen Y., Hu Y., Williams B., Perrault J., Karin M. (1999) JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity 11, 721–731 [DOI] [PubMed] [Google Scholar]
- 109. Wurzer W. J., Ehrhardt C., Pleschka S., Berberich-Siebelt F., Wolff T., Walczak H., Planz O., Ludwig S. (2004) NF-κB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J. Biol. Chem. 279, 30931–30937 [DOI] [PubMed] [Google Scholar]
- 110. Nimmerjahn F., Dudziak D., Dirmeier U., Hobom G., Riedel A., Schlee M., Staudt L. M., Rosenwald A., Behrends U., Bornkamm G. W., Mautner J. (2004) Active NF-κB signalling is a prerequisite for influenza virus infection. J. Gen. Virol. 85, 2347–2356 [DOI] [PubMed] [Google Scholar]
- 111. Wurzer W. J., Planz O., Ehrhardt C., Giner M., Silberzahn T., Pleschka S., Ludwig S. (2003) Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 22, 2717–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kramer A., Liashkovich I., Oberleithner H., Ludwig S., Mazur I., Shahin V. (2008) Apoptosis leads to a degradation of vital components of active nuclear transport and a dissociation of the nuclear lamina. Proc. Natl. Acad. Sci. U. S. A. 105, 11236–11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Faleiro L., Lazebnik Y. (2000) Caspases disrupt the nuclear-cytoplasmic barrier. J. Cell Biol. 151, 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C. A., Causton H. C., Gaasterland T., Glenisson P., Holstege F. C., Kim I. F., Markowitz V., Matese J. C., Parkinson H., Robinson A., Sarkans U., Schulze-Kremer S., Stewart J., Taylor R., Vilo J., Vingron M. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29, 365–371 [DOI] [PubMed] [Google Scholar]
- 115. Brazma A. (2009) Minimum information about a microarray experiment (MIAME)–successes, failures, challenges. Sci. World J. 9, 420–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Haney S. A. (2007) Increasing the robustness and validity of RNAi screens. Pharmacogenomics 8, 1037–1049 [DOI] [PubMed] [Google Scholar]
- 117. Schulte J., Sepp K. J., Wu C., Hong P., Littleton J. T. (2011) High-content chemical and RNAi screens for suppressors of neurotoxicity in a Huntington's disease model. PloS One 6, e23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pache L., Konig R., Chanda S. K. (2011) Identifying HIV-1 host cell factors by genome-scale RNAi screening. Methods 53, 3–12 [DOI] [PubMed] [Google Scholar]
- 119. Genovesio A., Giardini M. A., Kwon Y. J., de Macedo Dossin F., Choi S. Y., Kim N. Y., Kim H. C., Jung S. Y., Schenkman S., Almeida I. C., Emans N., Freitas-Junior L. H. (2011) Visual genome-wide RNAi screening to identify human host factors required for Trypanosoma cruzi infection. PLoS One 6, e19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Genovesio A., Kwon Y. J., Windisch M. P., Kim N. Y., Choi S. Y., Kim H. C., Jung S., Mammano F., Perrin V., Boese A. S., Casartelli N., Schwartz O., Nehrbass U., Emans N. (2011) Automated genome-wide visual profiling of cellular proteins involved in HIV infection. J. Biomol. Screen. 16, 945–958 [DOI] [PubMed] [Google Scholar]
- 121. Severson W. E., McDowell M., Ananthan S., Chung D. H., Rasmussen L., Sosa M. I., White E. L., Noah J., Jonsson C. B. (2008) High-throughput screening of a 100,000-compound library for inhibitors of influenza A virus (H3N2). J. Biomol. Screen. 13, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Shih S. R., Chu T. Y., Reddy G. R., Tseng S. N., Chen H. L., Tang W. F., Wu M. S., Yeh J. Y., Chao Y. S., Hsu J. T., Hsieh H. P., Horng J. T. (2010) Pyrazole compound BPR1P0034 with potent and selective anti-influenza virus activity. J. Biomed. Sci. 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Borjabad A., Morgello S., Chao W., Kim S. Y., Brooks A. I., Murray J., Potash M. J., Volsky D. J. (2011) Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders. PLoS Pathog. 7, e1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Geiss G. K., An M. C., Bumgarner R. E., Hammersmark E., Cunningham D., Katze M. G. (2001) Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75, 4321–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Geiss G. K., Bumgarner R. E., An M. C., Agy M. B., van 't Wout A. B., Hammersmark E., Carter V. S., Upchurch D., Mullins J. I., Katze M. G. (2000) Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266, 8–16 [DOI] [PubMed] [Google Scholar]
- 126. Chang Y. E., Laimins L. A. (2000) Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74, 4174–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Khodarev N. N., Advani S. J., Gupta N., Roizman B., Weichselbaum R. R. (1999) Accumulation of specific RNAs encoding transcriptional factors and stress response proteins against a background of severe depletion of cellular RNAs in cells infected with herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 96, 12062–12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Der S. D., Zhou A., Williams B. R., Silverman R. H. (1998) Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95, 15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Legendre M., Audic S., Poirot O., Hingamp P., Seltzer V., Byrne D., Lartigue A., Lescot M., Bernadac A., Poulain J., Abergel C., Claverie J. M. (2010) mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 20, 664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yang Z., Bruno D. P., Martens C. A., Porcella S. F., Moss B. (2010) Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 107, 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lefebvre G., Desfarges S., Uyttebroeck F., Munoz M., Beerenwinkel N., Rougemont J., Telenti A., Ciuffi A. (2010) Analysis of HIV-1 expression level and sense of transcription by high-throughput sequencing of the infected cell. J. Virol. 85, 6205–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Fields S., Song O. (1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 133. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U. S. A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. von Brunn A., Teepe C., Simpson J. C., Pepperkok R., Friedel C. C., Zimmer R., Roberts R., Baric R., Haas J. (2007) Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PloS One 2, e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugue S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., Penin F., Jacob Y., Vidalain P. O., Vidal M., Andre P., Rabourdin-Combe C., Lotteau V. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., Maurer A. P., Muller E., Wolff T., Rudel T., Meyer T. F. (2010) Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 [DOI] [PubMed] [Google Scholar]
- 137. Konig R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Gao Q., Andrews S. E., Bandyopadhyay S., De Jesus P., Tu B. P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., Garcia-Sastre A., Young J. A., Palese P., Shaw M. L., Chanda S. K. (2010) Human host factors required for influenza virus replication. Nature 463, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 [DOI] [PubMed] [Google Scholar]