Abstract
In laboratory rodents, caloric restriction (CR) retards several age-dependent physiological and biochemical changes in skeletal muscle, including increased steady-state levels of oxidative damage to lipids, DNA, and proteins. We have previously used high-density oligonucleotide arrays to show that CR can prevent or delay most of the major age-related transcriptional alterations in the gastrocnemius muscle of C57BL/6 mice. Here we report the effects of aging and adult-onset CR on the gene expression profile of 7,070 genes in the vastus lateralis muscle from rhesus monkeys. Gene expression analysis of aged rhesus monkeys (mean age of 26 years) was compared with that of young animals (mean age of 8 years). Aging resulted in a selective up-regulation of transcripts involved in inflammation and oxidative stress, and a down-regulation of genes involved in mitochondrial electron transport and oxidative phosphorylation. Middle-aged monkeys (mean age of 20 years) subjected to CR since early adulthood (mean age of 11 years) were studied to determine the gene expression profile induced by CR. CR resulted in an up-regulation of cytoskeletal protein-encoding genes, and also a decrease in the expression of genes involved in mitochondrial bioenergetics. Surprisingly, we did not observe any evidence for an inhibitory effect of adult-onset CR on age-related changes in gene expression. These results indicate that the induction of an oxidative stress-induced transcriptional response may be a common feature of aging in skeletal muscle of rodents and primates, but the extent to which CR modifies these responses may be species-specific.
Caloric restriction (CR) extends maximum lifespan and retards the development of a broad spectrum of pathophysiological changes in laboratory rodents (1). We have previously used high-density oligonucleotide arrays to demonstrate that aging is associated with specific transcriptional alterations in the gastrocnemius muscle, cerebral cortex, and cerebellum of C57BL/6 mice and that CR can prevent or delay most of the largest age-related transcriptional alterations (2, 3). These studies suggest that monitoring the effects of CR on multiple tissues is feasible through the use of DNA microarrays. Regarding human health, investigation of the ability of CR to retard aging and diseases in a long-lived nonhuman primate species may be more germane than rodent studies. A long-term CR study in adult (8- to 14-year-old) male rhesus monkeys, the maximum lifespan of which is ≈40 years, was initiated in 1989 at the Wisconsin Regional Primate Research Center (4). This ongoing study, along with a similar study at the National Institute on Aging (5), provides opportunities to investigate the ability of adult-onset CR to influence longevity, disease patterns, and other consequences of biological aging in primates.
To examine the molecular events associated with aging in this nonhuman primate species and the influence of CR on these events, we used oligonucleotide-based arrays to define the transcriptional response to the aging process in vastus lateralis muscle. Our choice of tissue was guided by skeletal muscle being primarily composed of long-lived, high oxygen-consuming postmitotic cells, a feature shared with other critical aging targets such as heart and brain. Also, we have previously used oligonucleotide microarrays to investigate the influences of aging and CR in mouse skeletal muscle (gastrocnemius) (2). The loss of skeletal muscle mass during aging, often referred to as sarcopenia, leads to frailty and is of great public health significance but of unknown etiology (6).
Methods
Animals and Experimental Design.
This study involved 12 male rhesus monkeys (Macaca mulatta), born and housed indoors with complete clinical and experimental histories available. The influence of aging was studied in two groups (n = 3) of conventionally maintained monkeys given free access to Purina (St. Louis, MO) Monkey Chow (no. 5038). These monkeys were either young (7–11 years old) or old (25–27 years old). For the CR study, normally fed (n = 3) and calorie-restricted (n = 3) middle-aged (19–21 years old) monkeys from our long-term CR study were examined. These animals had been subjected to CR for 9 years when biopsies of vastus lateralis were obtained for this study. No animal had any clinical or experimental history expected to differentially affect skeletal muscle. This protocol was carried out with the approval of the Institutional Animal Care and Use Committee of the University of Wisconsin.
The methods used to house and feed the animals are described in detail elsewhere (4, 7). To summarize briefly for the long-term CR study, both control and calorie-restricted animals were fed a purified diet (no. 85387, Teklad, Madison, WI) and housed individually for accurate measurement of daily food intake, but they had extensive auditory and visual contact with other monkeys housed in the same room. For all monkeys, temperature was maintained at ≈21°C with average relative humidity of 50–65%. Room lighting was automatically controlled to provide alternating 12-hr periods of light and darkness. The CR animals had ≈30% lower food intakes than the controls. This restriction was achieved by randomly assigning animals to a treatment group after a 3- to 5-month period of baseline assessment, during which food intake of the experimental diet was determined for individual animals. Food intake of the animals assigned to CR was reduced from their baseline period averages by 10% per month for 3 months and then maintained at this 30% restriction level. The controls continued to have free access to food. In 1994, the CR monkeys were switched to the caloric equivalent of a modified semipurified diet (Teklad no. 93131), which is enriched by 30% in vitamins and minerals.
Tissue Preparation for DNA Microarray Analysis.
Details for tissue preparation and procedures for the use of Affymetrix microarrays have been described (2). Total RNA was isolated from each biopsy and individual samples were used for gene expression profiles. Target RNA was prepared by converting 1 μg of mRNA into double-stranded cDNA (Superscript Choice System, GIBCO/BRL) with a T7-(dT)24 primer incorporating a T7 RNA polymerase promoter. Biotin-labeled cRNA was synthesized from cDNA by using an RNA transcript labeling kit (Enzo Biochem). After complementary RNA had been fragmented to sizes ranging from 35 to 200 bases by heating (35 min at 95°C), 10 μg of RNA fragments were hybridized (16 h at 45°C) to a HuGeneFL Array (Affymetrix, Santa Clara, CA). After hybridization, the gene chips were automatically washed and stained with streptavidin-phycoerythrin by using a fluidics system. The chips were scanned with a Hewlett Packard GeneArray Scanner.
Data Analysis.
The Affymetrix HuGeneFL Array contained about 7,070 human genes and expressed sequence tags (ESTs) from UniGene (Build 18), GenBank, and The Institute for Genomic Research (TIGR) databases. Each gene was represented in the array by 20 perfectly matched (PM) oligonucleotides and 20 mismatched (MM) control probes that contain a single central-base mismatch. Fluorescence intensity was read for each oligonucleotide to calculate the average signal intensity (SI) for each gene by subtracting the intensities of ≈20 PM oligonucleotides from the intensity of the MM control probes, after discarding the maximum, the minimum, and any outliers beyond three standard deviations. All calculations were performed by an Affymetrix algorithm. To determine the effects of aging, nine pairwise comparisons between young (n = 3) and old (n = 3) individuals were performed. Likewise, the effects of CR were determined by comparing each calorie-restricted (n = 3) to each control (n = 3) monkey, generating nine pairwise comparisons. The data reported in Tables 1–5 and in Tables 6–9, which are published as supplemental data on the PNAS web site, www.pnas.org, represent the average fold changes obtained through the nine pairwise combinations. To be acceptable for reporting, average fold changes had to exceed 1 SE from a 1.3-fold change.
Table 1.
Genes up-regulated with aging in vastus lateralis muscle
Table 5.
Effects of CR on age-associated changes in gene expression in middle-aged rhesus monkeys
Statistical Analysis of the Effect of CR on Changes in Gene Expression.
A statistical analysis was conducted for a subset of 34 genes that were either up-regulated or down-regulated in both aged and middle-aged monkeys. The analysis asked whether there was evidence of any effect of CR on expression level by regressing expression level on dummy codes for both CR (0 = no CR; 1 = CR) and Age (0 = young; 1 = middle-aged) by ordinary least-squares linear regression.
Results
Human High-Density Oligonucleotide Arrays Can Be Used to Monitor Gene Expression Patterns in Rhesus Monkeys.
To determine whether Affymetrix HuGeneFL arrays could be used to detect transcriptional changes in rhesus monkeys, we compared hybridization patterns of human and monkey vastus lateralis samples. From a total of 7,070 represented cDNAs, the number of transcripts resulting in positive signal intensities was 4,895 for humans (n = 5) and 4,677 for rhesus monkeys (n = 5). Average intragroup correlation coefficients were lower in the rhesus monkeys (0.83, n = 3) as compared with humans (0.92, n = 5). These observations indicate that the degree of sequence identity between humans and rhesus monkeys allows for oligonucleotide-based microarray analysis of rhesus monkeys by using human arrays, a finding that should be applicable to other nonhuman primate species.
Age-Related Changes in Gene Expression.
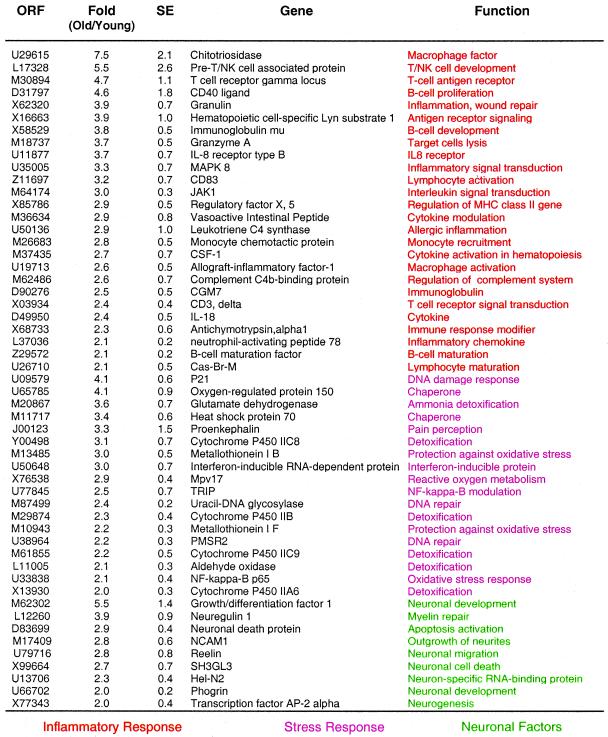
Comparison of hybridization patterns from the vastus lateralis of young adult (7–11 years old) and old (25–27 years old) monkeys revealed that aging is associated with a 2-fold or higher increase in expression of 300 genes, 4.2% of the total number of genes represented in the oligonucleotide array (for a full list of the 300 genes, see Table 6, which is published as supplemental data on the PNAS web site, www.pnas.org). Of these genes, 24 (7.9%) encode proteins that could be linked to an inflammatory/immune function as determined by database searches, representing the largest class of transcripts that display a large (2-fold or more) change in expression pattern with aging (Table 1). The transcript displaying the largest change in expression (7.5-fold) was chitotriosidase (Table 1). Chitotriosidase is a member of the chitinase family of proteins which are secreted by activated human macrophages (8) and is markedly elevated in plasma of Gaucher disease, a disorder characterized by the presence of large amounts of activated, lipid-laden macrophages in spleen, liver, and other tissues (9). Genes involved in B cell function included CD40 ligand, immunoglobulin μ chain, and B cell maturation factor. Genes involved in inflammation include granulin, allograft-inflammatory factor-1, neutrophil-activating peptide 78, and leukotriene C4 synthase. We also observed the concerted induction of genes involved in an oxidative stress response, including HSP-70, the 150-kDa oxygen-regulated protein ORP150, and the p65 subunit of NF-κB. Additionally, genes involved in neuronal death, remodeling, and repair were consistently activated, including reelin, glial growth factor-2 (neuregulin 1), and phogrin. This latter observation is consistent with the loss of motor neuron units and subsequent re-innervation of muscle fibers during aging in mammals (10, 11).
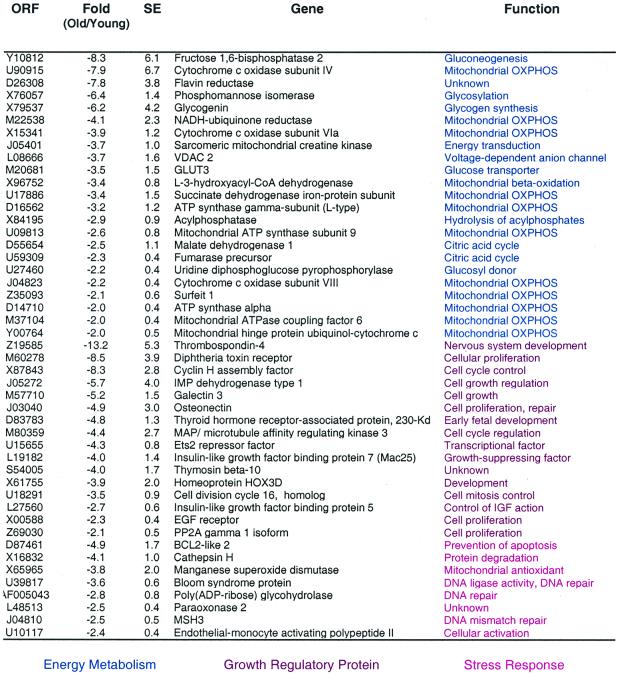
One hundred and forty-nine (2.1%) genes were down-regulated with aging in the vastus lateralis muscle of rhesus monkeys (see Table 7, which is published as supplemental data on the PNAS web site, www.pnas.org). These included genes involved in energy metabolism, cell growth, structural components, and stress response (Table 2). Striking among these was the abundance of genes related to energy metabolism, accounting for 15.8% of all down-regulated genes. These included several nuclear encoded proteins that function in mitochondrial bioenergetics, such as the sarcomeric mitochondrial creatine kinase, NADH:ubiquinone reductase, ATP synthase α and γ subunits, and cytochrome c oxidase subunit VIa. Recent studies strongly suggest that cytochrome c oxidase controls mitochondrial energy metabolism (12).
Table 2.
Genes down-regulated with aging in vastus lateralis muscle
Effect of Adult-Onset CR on Gene Expression Profiles.
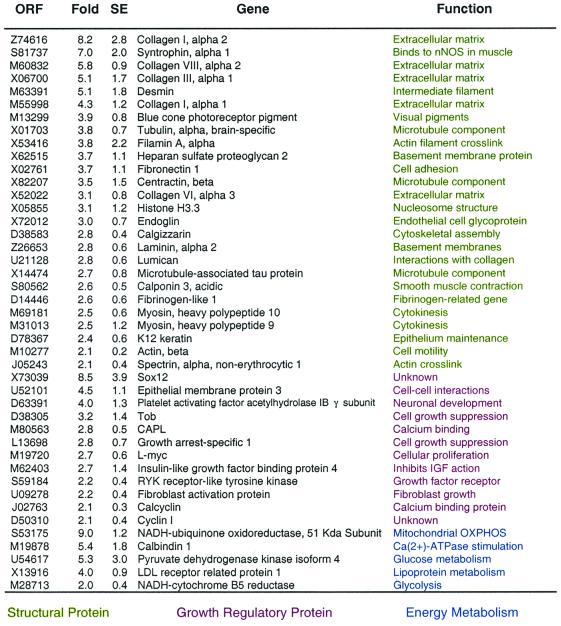
To determine the effect of CR on gene expression profile, we compared the transcriptional patterns of middle-aged monkeys (mean age of 20 years) receiving the control diet to aged-matched middle-aged monkeys that been subjected to CR since early adulthood. This comparison allows for the identification of transcriptional shifts that are diet-related, as opposed to age-related, and therefore represent a transcriptional reprogramming induced by CR. The expression of 107 genes (1.5%) was up-regulated by CR by 2-fold or more (for a full listing of these genes, see Table 8, which is published as supplemental data on the PNAS web site, www.pnas.org). The major transcriptional class induced by CR was composed by genes encoding structural proteins (Table 3). These included several collagens (collagen I α1 and α2 subunits, collagen VII α2 subunit, and collagen III α1 subunit). The extracellular matrix of muscle is composed mostly of collagens, and collagen synthesis in skeletal muscle is up-regulated by growth factors such as growth hormone (13, 14). In contrast, collagen genes are down-regulated in streptozotocin-induced diabetes in rats (15). Other cytoskeletal or extracellular matrix-encoding genes induced by CR include desmin, laminin, β-actin, and myosin heavy chain. CR also induced the expression of several genes that appear to have roles in cellular growth, such as L-myc, insulin-like growth factor binding protein 4, cyclin 1, and calcyclin (Table 3).
Table 3.
Genes up-regulated by CR in vastus lateraris muscle
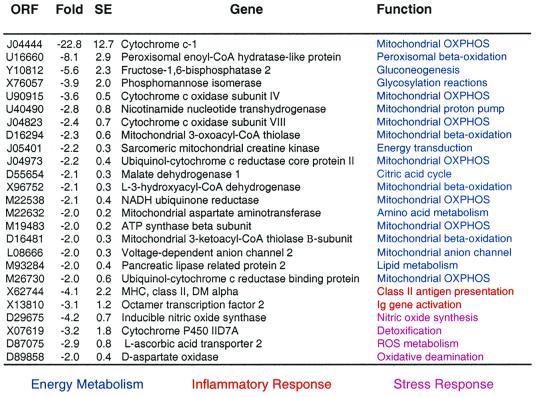
CR down-regulated 93 genes (1.3%) by 2-fold or more (see Table 9, which is published as supplemental data on the PNAS web site, www.pnas.org). The major transcriptional class in this group of genes encodes proteins involved in energy metabolism (Table 4). The largest decrease in expression (23-fold) was observed for cytochrome c1, a component of the electron-transport chain complex III. Interestingly, cytochrome c1 expression is extremely sensitive to thyroid status at both mRNA (16) and protein levels (17). Other alterations in gene expression linked to oxidative phosphorylation (OXPHOS) include the genes that encode the mitochondrially located cytochrome c oxidase subunit VII, cytochrome c subunit IV, sarcomeric mitochondrial creatine kinase, ATP synthase α and β subunits, and ubiquinol:cytochrome c reductase core protein II. Other energy-related transcripts reduced in expression by CR include fructose-1,6-bisphosphatase, malate dehydrogenase, NADH:ubiquinone oxidoreductase, and NADH reductase. These transcriptional alterations provide support for the concept that CR monkeys may be in a hypometabolic state associated with reduced activity of the mitochondrial electron transport system.
Table 4.
Genes down-regulated by CR in vastus lateraris muscle
Effect of Adult-Onset CR on Age-Associated Alterations.
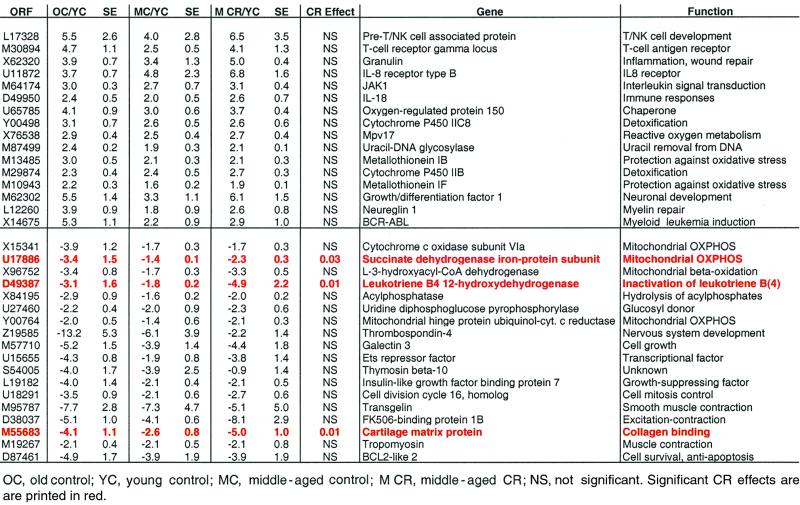
To investigate the effect of adult-onset CR on age-associated alterations in gene expression, we obtained muscle biopsies from rhesus monkeys that had been subjected to CR since early adulthood. The control group consisted of animals that had the same mean age and consumed a similar, but higher-calorie, semipurified diet since early adulthood. We first determined whether changes in gene expression observed in older monkeys (Tables 1 and 2) were either completely or partially established in middle-aged monkeys. We identified 34 transcripts that were elevated or decreased in expression with aging, and also by middle age (Table 5). The vast majority (32/34) displayed an intermediate level of expression between young and old animals.
Next, we determined how many of such transcripts were altered in expression as a result of caloric restriction. A statistical analysis was conducted for this subset of genes. The analysis asked whether there was any evidence of any effect of CR on expression level by regressing expression level on dummy codes for both CR (0 = no CR; 1 = CR) and Age (0 = young; 1 = middle aged) by ordinary least-squares linear regression. Surprisingly, only three genes appeared to show an effect, and it represented aggravation of down-regulations observed with aging (i.e., CR further reduced the level of expression). The two-tailed P values were U17886, P = 0.029; D49387, P = 0.027; and M55683, P = 0.009. These genes encoded, respectively, succinate dehydrogenase iron-protein subunit, a component of the complex II of the mitochondrial electron transport chain (18), and leukotriene B4 12-hydroxydehydrogenase, which converts leukotriene B4, a potent chemotactic and proinflammatory factor, into its biologically less active metabolite, 12-oxoleukotriene B4 (19), and a cartilage matrix protein (matrilin-1) (20).
Discussion
This large-scale gene expression analysis in a nonhuman primate underscores the potential of human high-density oligonucleotide arrays to monitor gene expression profiles of nonhuman primates. Because the DNA sequence homology between humans and rhesus monkeys is likely to be in the range of 95–98%, these results are not unexpected. In fact, a previous study used human high-density oligonucleotide array-based analysis to determine the distant history of single-nucleotide polymorphisms in chimpanzee, pygmy chimpanzee, and gorilla genomic DNA samples (21).
The analysis of age-associated alterations in gene expression in the vastus lateralis muscle of rhesus monkeys is consistent with our previous study in the gastrocnemius muscle of mice that demonstrated an age-associated induction of genes involved in stress responses and lowered expression of metabolic and biosynthetic genes (2). In particular, we have determined that transcripts induced by reactive oxygen species, such as oxygen-regulated protein 150, HSP70, metallothioneins IB and IIF, and NF-κB are up-regulated in aging skeletal muscle of rhesus monkeys (Table 1). This finding agrees with a recent study from our group that used electron microscopic (EM) techniques with antibodies raised against 4-hydroxy-2-nonenal (HNE)-modified proteins, dinitrophenol, and nitrotyrosine to quantify and localize the age-dependent accrual of oxidative damage in rhesus monkey vastus lateralis skeletal muscle. Using ImmunoGold EM analysis of muscle from rhesus monkeys ranging in age from 2 to 34 years old, we observed a 4-fold maximal increase in levels of HNE-modified proteins. Likewise, carbonyl levels increased approximately 2-fold with aging (22). The decrease in expression of transcripts involved in OXPHOS in aged rhesus monkeys is also in agreement with our previous observations in mice. Possibly, the reduction in gene expression of metabolic genes is due to an accrual of oxidative damage over the lifespan, leading to reduced mitochondrial function or biogenesis.
The effects of CR on gene expression profiles are striking, with a strong increase in the expression of structural and cytoskeletal genes and a reduction in the expression of mitochondria-related genes. Previous studies have determined that the expression of extracellular matrix genes in muscle is strongly modulated by growth factors such as growth hormone (13, 14). Therefore, it is plausible that alterations in hormonal levels may underlie the observed changes in gene expression. A recent study that has used high-density oligonucleotide arrays to investigate the gene expression profile of diabetes in mice has revealed that high glucose levels and poor insulin sensitivity result in lowered expression of cytoskeletal genes in adipocytes (Alan D. Attie, personal communication). In contrast, animals in our study display lower serum glucose levels and increased insulin sensitivity (23). An interesting observation of our study is the decrease in expression of genes involved in OXPHOS function in both aged animals receiving the control diets and middle-aged CR animals. We postulate that such alterations have a distinct molecular basis and are induced by mitochondrial dysfunction in aged animals as opposed to lower metabolic rate in CR animals. In particular, the 23-fold lower expression of cytochrome c1, which is particularly sensitive to thyroid status (16, 17), was observed in CR monkeys (Table 4) but not in aged animals (Table 2). Consistent with this hypothesis, long-term (2-year) CR leads to reduced energy expenditure and sleeping metabolic rate in humans (24). The concept that reduced OXPHOS gene expression activity with aging is caused by mitochondrial and metabolic dysfunction is supported by the induction of reactive oxygen species and inflammatory related transcripts in aged skeletal muscle of rhesus monkeys (Table 1), and by the accrual of oxidative damage (22) and high levels of mitochondrial DNA deletions in skeletal muscle of these animals (25).
Surprisingly, we did not observe beneficial effects of adult-onset CR on the progression of age-related transcriptional markers (Table 5). This finding is particularly striking, because this animal cohort is clearly showing the physiological beneficial effects of CR, such as increased insulin sensitivity, reduced blood glucose (23), and reduced oxidative stress-induced cytokine expression by peripheral blood mononuclear cells (26). This finding stands in contrast to our previous report that more than 80% of age-related changes in gene expression found in skeletal muscle are either partially or completely suppressed by early-onset CR in mice (2). Possibly, aging retardation at the transcriptional level can be observed only if CR is initiated early in life, or if tissues are profiled late in life. Alternatively, the ability of CR to suppress changes in gene expression associated with aging may be species-specific. The latter hypothesis suggests that despite beneficial effects on hormonal parameters and possible increases in survival rates, aging rates are not retarded at the molecular level by CR in primates. Testing these possibilities will require studies employing early-onset CR in primates and the examination of multiple tissues for age-related changes in gene expression in CR and control animals.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grants P01 AG 11915 (R.W.) and R01 CA 78732 (T.A.P.) T.A.P. is a recipient of the Shaw Scientist (Milwaukee Foundation), Burroughs Wellcome Young Investigator, and Basil O'Connor (March of Dimes) awards.
Abbreviations
- OXPHOS
oxidative phosphorylation
- CR
caloric restriction
References
- 1.Weindruch R, Walford R L. The Retardation of Aging and Disease by Dietary Restriction. Springfield, IL: Thomas; 1988. [Google Scholar]
- 2.Lee C-K, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 3.Lee C-K, Weindruch R, Prolla T A. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 4.Kemnitz J W, Weindruch R, Roecker E B, Crawford K, Kaufman P L, Ershler W B. J Gerontol A Biol Sci Med Sci. 1993;48:B17–B26. doi: 10.1093/geronj/48.1.b17. [DOI] [PubMed] [Google Scholar]
- 5.Ingram D K, Cutler R G, Weindruch R, Renquist D M, Knapka J J, April M, Belcher C T, Clark M A, Hatcherson C D, Marriott B M, Roth G S. J Gerontol A Biol Sci Med Sci. 1990;45:B148–B163. doi: 10.1093/geronj/45.5.b148. [DOI] [PubMed] [Google Scholar]
- 6.Kohrt W M, Holloszy J O. J Gerontol A Biol Sci Med Sci. 1995;50:68–72. doi: 10.1093/gerona/50a.special_issue.68. [DOI] [PubMed] [Google Scholar]
- 7.Ramsey J J, Roecker E B, Weindruch R, Kemnitz J W. Am J Physiol. 1997;272:E901–E907. doi: 10.1152/ajpendo.1997.272.5.E901. [DOI] [PubMed] [Google Scholar]
- 8.Boot R G, Renkema G H, Strijland A, van Zonneveld A J, Aerts J M. J Biol Chem. 1995;270:26252–26256. doi: 10.1074/jbc.270.44.26252. [DOI] [PubMed] [Google Scholar]
- 9.Hollak C E, van Weely S, van Oers M H, Aerts J M. J Clin Invest. 1994;93:1288–1292. doi: 10.1172/JCI117084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villani G, Attardi G. Free Radic Biol Med. 2000;29:202–210. doi: 10.1016/s0891-5849(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 11.Lexell J. J Nutr. 1997;127:1011S–1013S. doi: 10.1093/jn/127.5.1011S. [DOI] [PubMed] [Google Scholar]
- 12.Kadenbach B, Huttemann M, Arnold S, Lee I, Bender E. Free Radic Biol Med. 2000;29:211–221. doi: 10.1016/s0891-5849(00)00305-1. [DOI] [PubMed] [Google Scholar]
- 13.Wilson V J, Rattray M, Thomas C R, Moreland B H, Schulster D. Mol Cell Endocrinol. 1995;115:187–197. doi: 10.1016/0303-7207(95)03690-3. [DOI] [PubMed] [Google Scholar]
- 14.Wilson V J, Rattray M, Thomas C R, Moreland B H, Schulster D. Growth Horm IGF Res. 1998;8:431–438. doi: 10.1016/s1096-6374(98)80295-5. [DOI] [PubMed] [Google Scholar]
- 15.Han X, Karpakka J, Kainulainen H, Takala T E. Acta Physiol Scand. 1995;155:9–16. doi: 10.1111/j.1748-1716.1995.tb09941.x. [DOI] [PubMed] [Google Scholar]
- 16.Luciakova K, Nelson B D. Eur J Biochem. 1992;207:247–251. doi: 10.1111/j.1432-1033.1992.tb17044.x. [DOI] [PubMed] [Google Scholar]
- 17.Joste V, Goitom Z, Nelson B D. Eur J Biochem. 1989;184:255–260. doi: 10.1111/j.1432-1033.1989.tb15015.x. [DOI] [PubMed] [Google Scholar]
- 18.Au H C, Scheffler I E. Gene. 1994;149:261–265. doi: 10.1016/0378-1119(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 19.Yokomizo T, Ogawa Y, Uozumi N, Kume K, Izumi T, Shimizu T. J Biol Chem. 1996;271:2844–2850. doi: 10.1074/jbc.271.5.2844. [DOI] [PubMed] [Google Scholar]
- 20.Argraves W S, Deak F, Sparks K J, Kiss I, Goetinck P F. Proc Natl Acad Sci USA. 1987;84:464–468. doi: 10.1073/pnas.84.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacia J G, Fan J B, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer R A, Sun B, Hsie L, Robbins C M, et al. Nat Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- 22.Zainal T A, Oberley T D, Allison D B, Szweda L I, Weindruch R. FASEB J. 2000;14:1825–1836. doi: 10.1096/fj.99-0881com. [DOI] [PubMed] [Google Scholar]
- 23.Kemnitz J W, Roecker E B, Weindruch R, Elson D F, Baum S T, Bergman R N. Am J Physiol. 1994;266:E540–E547. doi: 10.1152/ajpendo.1994.266.4.E540. [DOI] [PubMed] [Google Scholar]
- 24.Weyer C, Walford R L, Harper I T, Milner M, MacCallum T, Tataranni P A, Ravussin E. Am J Clin Nutr. 2000;72:946–953. doi: 10.1093/ajcn/72.4.946. [DOI] [PubMed] [Google Scholar]
- 25.Schwarze S R, Lee C M, Chung S S, Roecker E B, Weindruch R, Aiken J M. Mech Ageing Dev. 1995;83:91–101. doi: 10.1016/0047-6374(95)01611-3. [DOI] [PubMed] [Google Scholar]
- 26.Kim M J, Aiken J M, Havighurst T, Hollander J, Ripple M O, Weindruch R. J Nutr. 1997;127:2293–2301. doi: 10.1093/jn/127.12.2293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.