Abstract
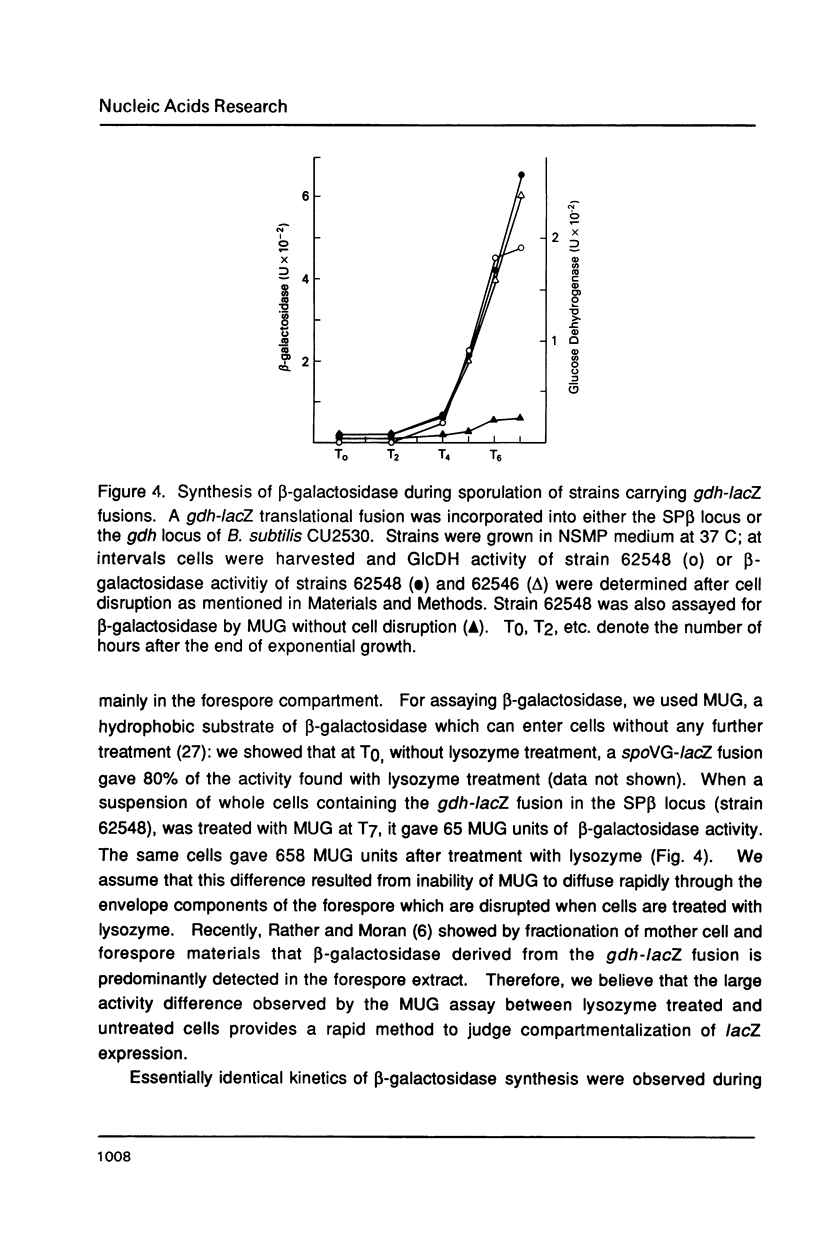
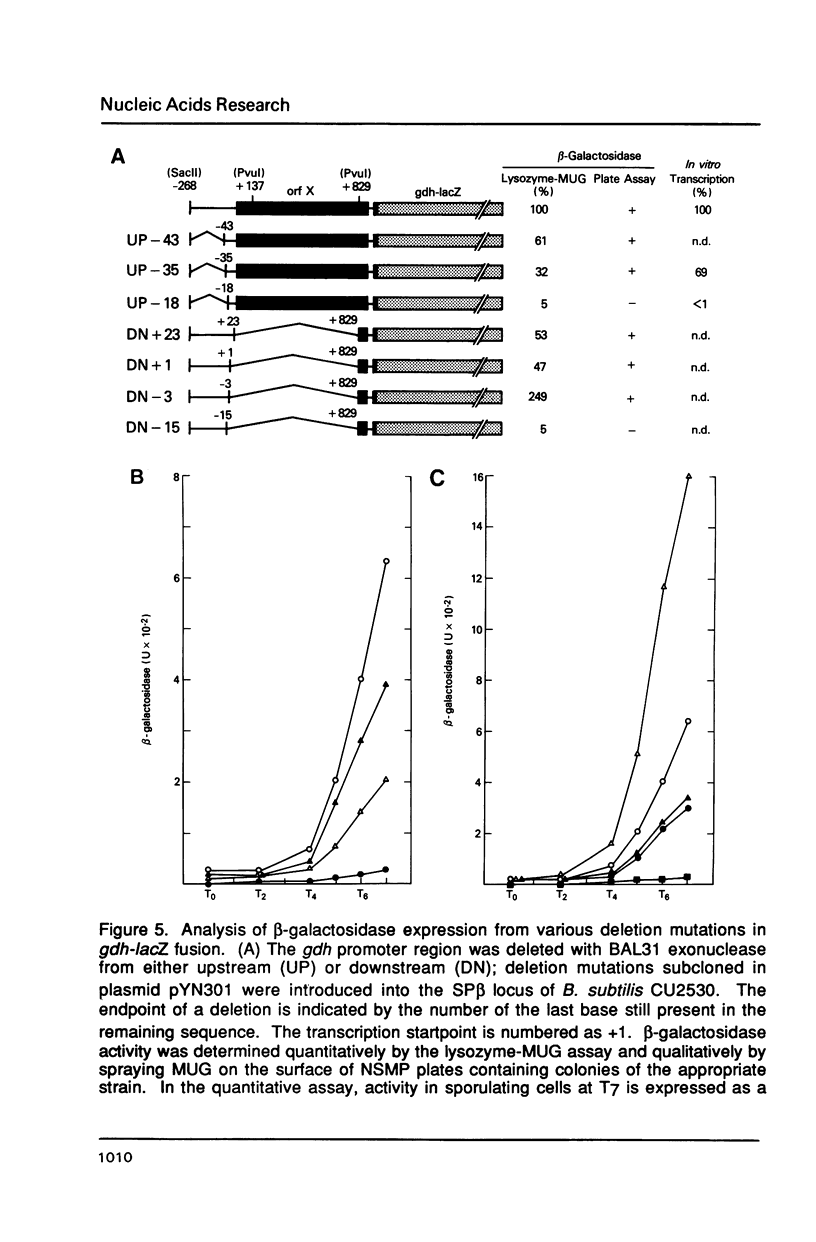
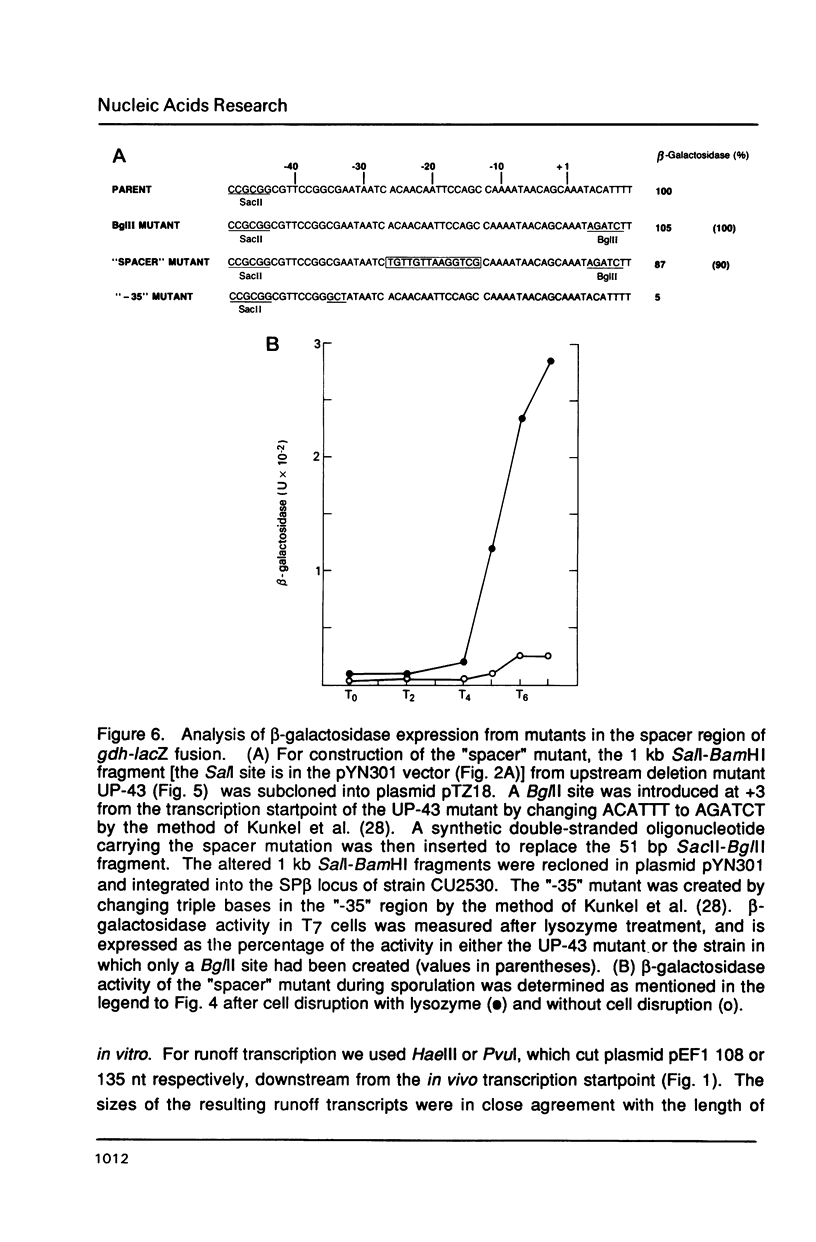
The gene encoding glucose dehydrogenase (gdh) is part of an operon whose expression is transcriptionally activated specifically in the developing forespore of Bacillus subtilis at stage III of sporulation. The in vivo startpoint of gdh transcription was determined using primer extension analysis. Deletion mapping and site-specific mutagenesis experiments indicated that the region responsible for regulated expression of gdh in vivo was limited to the "-35" and "-10" regions preceding the transcriptional start site. RNA polymerase containing omega G (E omega G) transcribed gdh in vitro with a start site identical to that found in vivo, and transcription of gdh by E omega G in vitro also did not require any specific sequences upstream from "-35" region. These results suggest that the appearance of E omega G in the forespore at stage III of sporulation is sufficient to cause temporal and compartment-specific expression of the gdh operon.
Full text
PDF



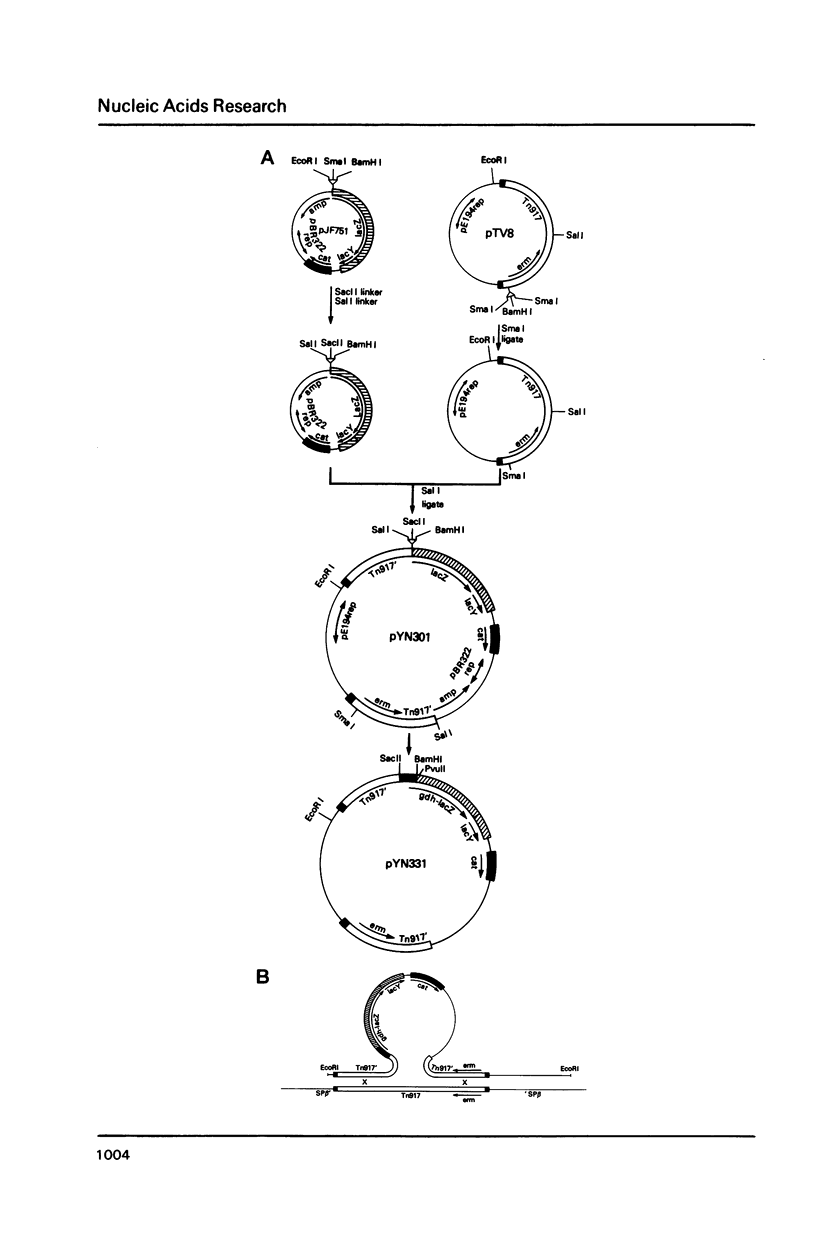


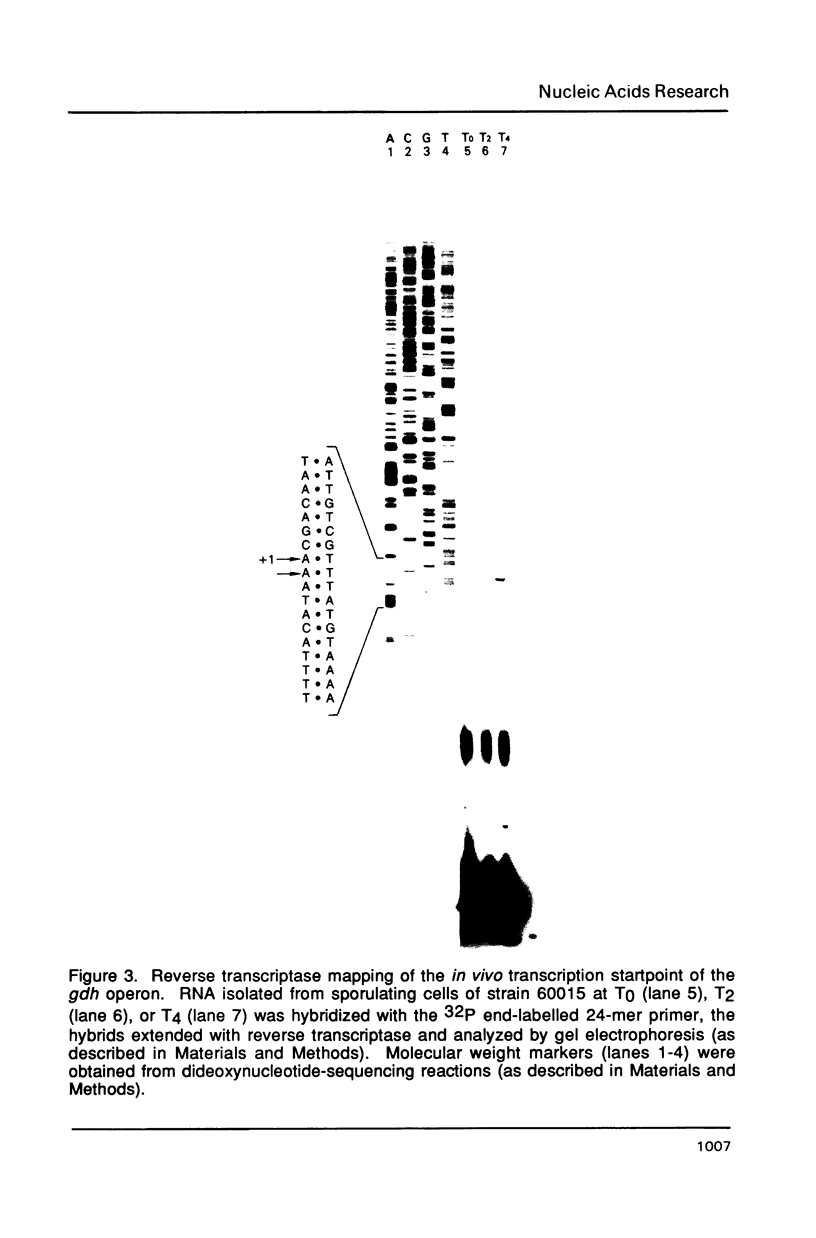







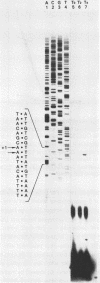
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., Hoch J. A. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J Bacteriol. 1985 Feb;161(2):556–562. doi: 10.1128/jb.161.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Ramaley R., Freese E. Location and properties of glucose dehydrogenase in sporulating cells and spores of Bacillus subtilis. J Bacteriol. 1977 Oct;132(1):282–293. doi: 10.1128/jb.132.1.282-293.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel K. A., Uratani B., Chaudhry G. R., Ramaley R. F., Rudikoff S. Characterization of the developmentally regulated Bacillus subtilis glucose dehydrogenase gene. J Bacteriol. 1986 Apr;166(1):238–243. doi: 10.1128/jb.166.1.238-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grice S. F., Shih C. C., Whipple F., Sonenshein A. L. Separation and analysis of the RNA polymerase binding sites of a complex Bacillus subtilis promoter. Mol Gen Genet. 1986 Aug;204(2):229–236. doi: 10.1007/BF00425503. [DOI] [PubMed] [Google Scholar]
- Leighton T. J., Doi R. H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971 May 25;246(10):3189–3195. [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Mason J. M., Hackett R. H., Setlow P. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J Bacteriol. 1988 Jan;170(1):239–244. doi: 10.1128/jb.170.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews C. R. Effect of point mutations on the folding of globular proteins. Methods Enzymol. 1987;154:498–511. doi: 10.1016/0076-6879(87)54092-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Minkley E. G., Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973 Jun 25;77(2):255–277. doi: 10.1016/0022-2836(73)90335-5. [DOI] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather P. N., Moran C. P., Jr Compartment-specific transcription in Bacillus subtilis: identification of the promoter for gdh. J Bacteriol. 1988 Nov;170(11):5086–5092. doi: 10.1128/jb.170.11.5086-5092.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980 Dec;144(3):1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Uratani B., Ramaley R. F., Freese E. Isolation of a developmental gene of Bacillus subtilis and its expression in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):785–789. doi: 10.1073/pnas.80.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura D. G., Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P. J., Perkins J. B., Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1983 Nov;35(1):275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]