Abstract
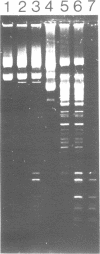
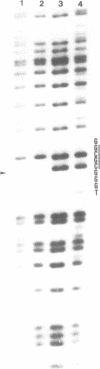
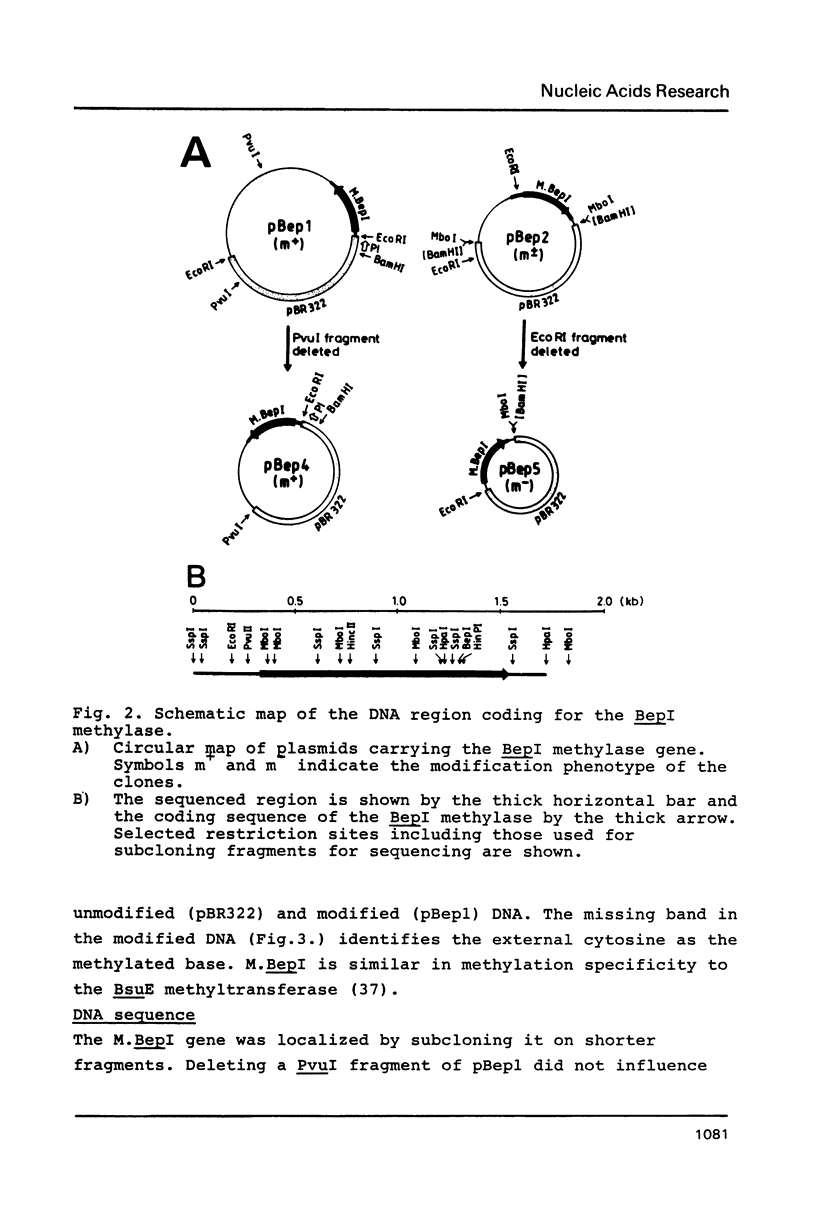
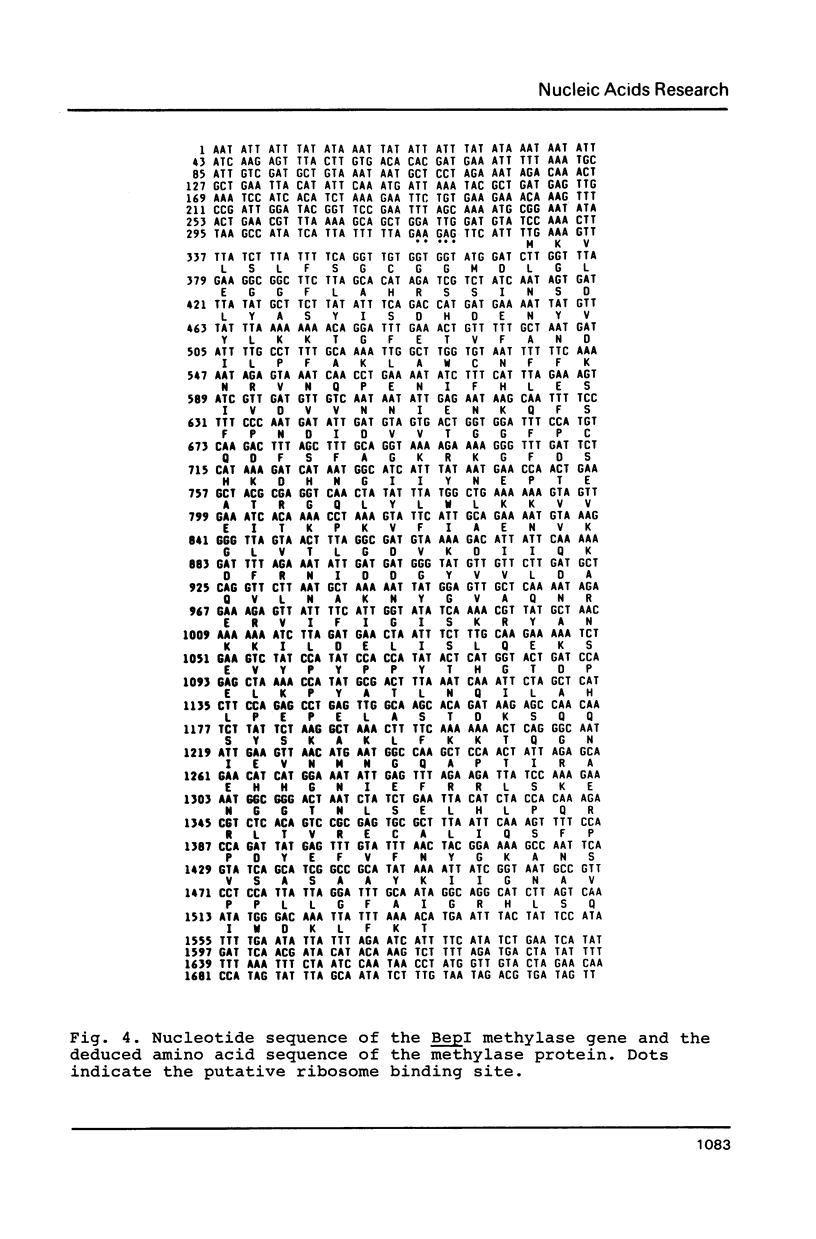
The gene coding for a CGCG specific DNA methylase has been cloned in E. coli from Brevibacterium epidermidis. The enzyme, named BepI methylase, is probably the cognate methylase of the FnuDII isoschizomer BepI endonuclease isolated from this strain. The expression of BepI methylase in E. coli is dependent on the orientation of the cloned fragment suggesting that the gene is transcribed from a promoter on the plasmid vector. No BepI endonuclease could be detected in the clones producing BepI methylase. The nucleotide sequence of the BepI methylase gene has been determined, it predicts a protein of 403 amino acids (MR: 45,447). Analysis of the amino acid sequence deduced from the nucleotide sequence revealed similarities between the BepI methylase and other cytosine methylases. M. BepI methylates the external cytosine in its recognition sequence.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balganesh T. S., Reiners L., Lauster R., Noyer-Weidner M., Wilke K., Trautner T. A. Construction and use of chimeric SPR/phi 3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J. 1987 Nov;6(11):3543–3549. doi: 10.1002/j.1460-2075.1987.tb02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R. M., Gregory S. A., Cooperider J. S. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J Bacteriol. 1985 Nov;164(2):501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bougueleret L., Schwarzstein M., Tsugita A., Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhk H. J., Behrens B., Tailor R., Wilke K., Prada J. J., Günthert U., Noyer-Weidner M., Jentsch S., Trautner T. A. Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene. 1984 Jul-Aug;29(1-2):51–61. doi: 10.1016/0378-1119(84)90165-3. [DOI] [PubMed] [Google Scholar]
- Caserta M., Zacharias W., Nwankwo D., Wilson G. G., Wells R. D. Cloning, sequencing, in vivo promoter mapping, and expression in Escherichia coli of the gene for the HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4770–4777. [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaido M. L., Prostko C. R., Strobl J. S. Isolation and characterization of BsuE methyltransferase, a CGCG specific DNA methyltransferase from Bacillus subtilis. J Biol Chem. 1988 Apr 5;263(10):4832–4836. [PubMed] [Google Scholar]
- Greene P. J., Gupta M., Boyer H. W., Brown W. E., Rosenberg J. M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981 Mar 10;256(5):2143–2153. [PubMed] [Google Scholar]
- Hattman S., Wilkinson J., Swinton D., Schlagman S., Macdonald P. M., Mosig G. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J Bacteriol. 1985 Nov;164(2):932–937. doi: 10.1128/jb.164.2.932-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman C., de Waard A. Cloning and complete nucleotide sequences of the type II restriction-modification genes of Salmonella infantis. J Bacteriol. 1988 Jun;170(6):2527–2532. doi: 10.1128/jb.170.6.2527-2532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A., Posfai G., Keller C. C., Venetianer P., Roberts R. J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985 Sep 25;13(18):6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A., Mannarelli B. M., Springhorn S. S., Greenberg B. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell. 1986 Sep 26;46(7):993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- Lauster R., Kriebardis A., Guschlbauer W. The GATATC-modification enzyme EcoRV is closely related to the GATC-recognizing methyltransferases DpnII and dam from E. coli and phage T4. FEBS Lett. 1987 Aug 10;220(1):167–176. doi: 10.1016/0014-5793(87)80897-9. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Mosig G. Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J. 1984 Dec 1;3(12):2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannarelli B. M., Balganesh T. S., Greenberg B., Springhorn S. S., Lacks S. A. Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4468–4472. doi: 10.1073/pnas.82.13.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Narva K. E., Wendell D. L., Skrdla M. P., Van Etten J. L. Molecular cloning and characterization of the gene encoding the DNA methyltransferase, M.CviBIII, from Chlorella virus NC-1A. Nucleic Acids Res. 1987 Dec 10;15(23):9807–9823. doi: 10.1093/nar/15.23.9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Baldauf F., Erdei S., Pósfai J., Venetianer P., Kiss A. Structure of the gene coding for the sequence-specific DNA-methyltransferase of the B. subtilis phage SPR. Nucleic Acids Res. 1984 Dec 11;12(23):9039–9049. doi: 10.1093/nar/12.23.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Kiss A., Erdei S., Pósfai J., Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J Mol Biol. 1983 Nov 5;170(3):597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner B., Kelly S., Smith H. O. The nucleotide sequence of the HhaII restriction and modification genes from Haemophilus haemolyticus. Gene. 1983 Oct;24(2-3):227–236. doi: 10.1016/0378-1119(83)90083-5. [DOI] [PubMed] [Google Scholar]
- Slatko B. E., Benner J. S., Jager-Quinton T., Moran L. S., Simcox T. G., Van Cott E. M., Wilson G. G. Cloning, sequencing and expression of the Taq I restriction-modification system. Nucleic Acids Res. 1987 Dec 10;15(23):9781–9796. doi: 10.1093/nar/15.23.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. M., Saunders J. R. Sequence analysis of the NgoPII methyltransferase gene from Neisseria gonorrhoeae P9: homologies with other enzymes recognizing the sequence 5'-GGCC-3'. Nucleic Acids Res. 1988 May 25;16(10):4369–4387. doi: 10.1093/nar/16.10.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznyter L. A., Slatko B., Moran L., O'Donnell K. H., Brooks J. E. Nucleotide sequence of the DdeI restriction-modification system and characterization of the methylase protein. Nucleic Acids Res. 1987 Oct 26;15(20):8249–8266. doi: 10.1093/nar/15.20.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault G., Roy P. H., Howard K. A., Benner J. S., Brooks J. E., Waters A. F., Gingeras T. R. Nucleotide sequence of the PaeR7 restriction/modification system and partial characterization of its protein products. Nucleic Acids Res. 1985 Dec 9;13(23):8441–8461. doi: 10.1093/nar/13.23.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Betcke A., Behrens B., Noyer-Weidner M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: comparison of their nucleotide sequences. Gene. 1986;42(1):89–96. doi: 10.1016/0378-1119(86)90153-8. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Balganesh T. S., Pawlek B. Chimeric multispecific DNA methyltransferases with novel combinations of target recognition. Nucleic Acids Res. 1988 Jul 25;16(14A):6649–6658. doi: 10.1093/nar/16.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetianer P., Orosz A. BepI restriction endonuclease, a new isoschisomer of FnuDII. Nucleic Acids Res. 1988 Jan 11;16(1):350–350. doi: 10.1093/nar/16.1.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Walder J. A., Donelson J. E. The organization and complete nucleotide sequence of the PstI restriction-modification system. J Biol Chem. 1984 Jun 25;259(12):8015–8026. [PubMed] [Google Scholar]